Abstract
Differential gene expression in biofilm cells suggests that adding the derepressed conjugative plasmid R1drd19 increases biofilm formation by affecting genes related to envelope stress (rseA and cpxAR), biofilm formation (bssR and cstA), energy production (glpDFK), acid resistance (gadABCEX and hdeABD), and cell motility (csgBEFG, yehCD, yadC, and yfcV); genes encoding outer membrane proteins (ompACF), phage shock proteins (pspABCDE), and cold shock proteins (cspACDEG); and phage-related genes. To investigate the link between the identified genes and biofilm formation upon the addition of R1drd19, 40 isogenic mutants were classified according to their different biofilm formation phenotypes. Cells with class I mutations (those in rseA, bssR, cpxA, and ompA) exhibited no difference from the wild-type strain in biofilm formation and no increase in biofilm formation upon the addition of R1drd19. Cells with class II mutations (those in gatC, yagI, ompC, cspA, pspD, pspB, ymgB, gadC, pspC, ymgA, slp, cpxP, cpxR, cstA, rseC, ompF, and yqjD) displayed increased biofilm formation compared to the wild-type strain but decreased biofilm formation upon the addition of R1drd19. Class III mutants showed increased biofilm formation compared to the wild-type strain and increased biofilm formation upon the addition of R1drd19. Cells with class IV mutations displayed increased biofilm formation compared to the wild-type strain but little difference upon the addition of R1drd19, and class V mutants exhibited no difference from the wild-type strain but increased biofilm formation upon the addition of R1drd19. Therefore, proteins encoded by the genes corresponding to the class I mutant phenotype are involved in R1drd19-promoted biofilm formation, primarily through their impact on cell motility. We hypothesize that the pili formed upon the addition of the conjugative plasmid disrupt the membrane (induce ompA) and activate the two-component system CpxAR as well as the other envelope stress response system, RseA-σE, both of which, along with BssR, play a key role in bacterial biofilm formation.
Conjugation transfers genetic material between bacteria through cell-to-cell contact (59); hence, it spreads virulence factors (20) and influences bacterial resistance to antibiotics (38). Conjugation is affected by growth conditions and biofilm structure (26), and biofilms promote conjugation (41). The reverse is also true, as conjugative plasmids promote biofilm formation (20, 47), and it has been proposed previously that conjugative pili act as adhesion factors (20). In addition, mature biofilms harboring conjugative plasmids were observed previously to be thicker than those not harboring such plasmids (46).
Plasmid R1, originally from the host Salmonella enterica serovar Paratyphi (20), is an F-like conjugative plasmid of the IncFII incompatibility group (13). The transfer region of R1 consists of four DNA transfer genes (traYALE); a large tra operon comprising at least 34 genes that have a high degree of homology to the F plasmid (35); finP and finO, which encode the fertility inhibition complex FinPO (66); and traM and traJ, which lie outside of the tra operon (4), as well as the conjugative transfer origin locus oriT (4). TraM is a positive regulator of the tra genes (43), and TraJ disrupts the host nucleoid-associated protein, a repressor of the tra operon (62). The tra promoter is repressed by FinO and FinP (36); therefore, the disruption of finO in plasmid R1drd19 promotes constitutive conjugation (46). Plasmid R1 represses conjugal pilus synthesis, but R1drd19 synthesizes these pili constitutively (20). Since the genetic mechanism by which conjugative plasmids control biofilm formation has not been elucidated (we found previously that the addition of R1drd19 increases biofilm formation by increasing aggregation and decreasing cell motility [21]), our goal here was to use DNA microarrays and isogenic mutants to investigate this mechanism.
Single-time-point DNA microarrays have been used previously to explore the genetic basis of Escherichia coli biofilm formation (5, 24, 32, 49, 54), and one temporal study has been completed (14); one common trend is that stress genes are induced. With DNA microarrays, we previously identified five stress response genes (hslST, hha, soxS, and ycfR) induced in 7-h E. coli biofilm cells harboring a conjugative plasmid compared to those in suspension cells harboring a conjugative plasmid (49), and recently, we showed how YcfR mediates this stress response in E. coli and how stress increases E. coli biofilm formation (67). In addition, the envelope stress response genes, such as pspABCDE, cpxAR, rpoE, and rseA, were previously found to be induced in E. coli 8-day-old biofilm cells compared to those in exponentially growing planktonic cells, regardless of the presence of a conjugative plasmid (5). rpoS plays a key role during biofilm formation because it encodes the sigma S factor, which regulates a number of stress-related genes (54). yeaGH have also been identified previously as putative stress response genes (54) since they are regulated by RpoS in S. enterica. In addition, cold shock protein regulators cspABFGI and the heat shock protein regulator htgA are induced in a temporal fashion during biofilm formation (14). In human urine, stress genes (e.g., cspAGH, ibpAB, pphA, soxS, and yfiD) in asymptomatic bacteriuria E. coli are also induced during biofilm formation (24).
CpxAR is a two-component system for response to cell envelope stress (10). CpxAR also controls the synthesis of adhesive organelles (45) and appears to help cells respond to adverse conditions (16). CpxA is a histidine kinase that functions in the inner membrane (12) as the sensor of envelope stress (e.g., cell invasion and high pH) (40) and unassembled P-pilus subunits (11). CpxR is the response regulator which resides in the cytoplasm (12); it is phosphorylated by CpxA, which autophosphorylates and then transfers the phosphate to CpxR (yielding CpxR-P). The accumulation of surface adherence factors such as pilus subunits in the cytoplasm or in the outer membrane leads to the activation of the Cpx system (16). The outer membrane protein NlpE activates the Cpx system when NlpE is overproduced, but the Cpx pathway is activated in an NlpE-independent manner in the presence of envelope stress (12). CpxR-P positively regulates virulence (39) and the porin OmpC (3) but negatively regulates both genes that encode adherence factors (e.g., csgBD) (31) and motility genes (e.g., motAB and cheAW) (10). At the posttranscription level, CpxR-P controls pilus monomer secretion (16).
σE responds specifically to cell envelope stress (1), and it is required for the expression of periplasmic folding catalysts, proteases, and other outer membrane components of the envelope (19). RseA is a 216-amino-acid, transmembrane, anti-sigma factor that can form an inhibitory complex that blocks σE from binding to RNA polymerase (7); hence, this anti-sigma factor can control envelope stress (9). The stability of RseA (and therefore its effect on σE) is based on outer membrane protein OmpC, which activates the protease DegS, which cleaves RseA (1). In the presence of envelope stress, the two-component system CpxAR is the dominant regulator over RseA-σE (16). Another outer membrane protein, OmpA, is linked to σE through the small RNA (sRNA) MicA (60); MicA is a negative antisense regulator of OmpA synthesis, and this sRNA is induced by the overexpression of σE (60).
Recently, we found that BssR (YliH) is a biofilm repressor because it represses the motility of E. coli in Luria-Bertani (LB) medium, induces indole, which is an inhibitor of biofilm formation, and represses autoinducer-2 (AI-2)-induced genes (15). DNA microarray analysis revealed that 13 stress response genes (e.g., sdiA, ydaD, and ydaK) are induced and that 51 stress response genes (e.g., yodC, yjbJ, and rpoS) are repressed by the deletion of bssR in the E. coli K-12 wild type (15).
Here, five pairs of DNA microarrays (corresponding to E. coli BW25113 with and without R1drd19 in complex medium at 7, 15, and 24 h, E. coli ATCC 25404 with and without R1drd19 in complex medium at 24 h, and E. coli MG1655 with and without R1drd19 in minimal medium at 24 h) were used to identify genes related to enhanced biofilm formation upon the addition of a conjugative plasmid. Based on the identified genes, 40 isogenic knockout mutations were investigated and classified. It was determined that R1drd19 mediates an increase in biofilm formation through its interaction with CpxAR, RseA, BssR, and OmpA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used are listed in Table 1. The following three E. coli strains were chosen since their biofilm formation has been studied previously in our lab: BW25113 (14), MG1655 (22), and ATCC 25404 (63). pCM18 (25) constitutively expresses the green fluorescent protein, so it was used to visualize the biofilms; this plasmid was maintained by the addition of 300 μg of erythromycin/ml. LB medium (52) was used for overnight cultures. LB medium and M9 minimal medium with 0.4% Casamino Acids and 0.4% glucose (M9C glu) (50) were used to form biofilms. To maintain R1drd19 (20), 30 μg of chloramphenicol/ml was added to the overnight cultures, and 50 μg of kanamycin/ml was added to the overnight cultures of the isogenic knockout strains (2). After the overnight culture, antibiotics were omitted for the crystal violet biofilm assay, the aggregation assay, and the motility assay. Vibrio harveyi was cultured in autoinducer bioassay (AB) medium for the AI-2 assay (58).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain(s) or plasmid | Descriptiona | Reference |
|---|---|---|
| E. coli strains | ||
| K-12 BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 8 |
| K-12 BW25113 mutants (all) | BW25113 Δ(gene) Ω Kmr | 2 |
| K-12 ATCC 25404 | Wild type | ATCCb |
| K-12 MG1655 | F− lambda−rfb-50 rph-1; lacks ilvG | 6 |
| V. harveyi BB170 | BB120 luxN::Tn5 (negative for AI-1 sensor, positive for AI-2 sensor) | 58 |
| Plasmids | ||
| R1drd19 | Ampr Kmr Cmr Smr IncFII finO | 20 |
| pCM18 | Emr pTRKL2-PCP25RBSII-gfp3*-To-T1 | 25 |
Ampr, Kmr, Cmr, Smr, and Emr denote ampicillin, kanamycin, chloramphenicol, streptomycin, and erythromycin resistance, respectively.
ATCC, American Type Culture Collection.
The knockout deletions in all the strains were confirmed by Baba et al. (2) by amplifying the regions flanking the deleted gene with two specific primers corresponding to the kanamycin gene (K1, 5′-CAGTCATAGCCGAATAGCCT, and K2, 5′-CGGTGCCCTGAATGAACTGC) by PCR. For example, to confirm the cpxA deletion, forward primer 5′-GCCAATAAAATCCTGTTAGTTGA was used with K1 and reverse primer 5′-GCCCGATATCCGGTTGATGTATA was used with K2. For rseA, forward primer 5′-GCCAGCGAGCAGTTAACGGACCA and reverse primer 5′-CCTTGCGCTGCCCCGAACTTAAT were used. For ompA, forward primer 5′-GCCTACACTTCAGGCTATGCACA and reverse primer 5′-GCCAAATATCAACAACTTGAAAA were used. For bssR, forward primer 5′-CCAACCCGGCTACCCCACAAATC and reverse primer 5′-CCATTGCGTGGGCTAACTTTAAG were used.
Conjugation.
Plasmid R1drd19 was conjugated (65) into the 40 isogenic mutants by using donor strain E. coli BW25113 cysB/R1drd19. The recipient colonies were selected on M9C glu plates containing 30 μg of chloramphenicol/ml, and the presence of R1drd19 was confirmed using four antibiotics (100 μg of ampicillin/ml, 50 μg of kanamycin/ml, 30 μg of chloramphenicol/ml, and 100 μg of streptomycin/ml) (20).
Crystal violet biofilm assay.
The crystal violet biofilm assay was based on that of Pratt and Kolter (44) but was modified to achieve consistent biofilm formation upon the addition of the conjugative plasmid. E. coli strains were grown in LB medium for 16 h, and then the overnight cultures were inoculated into fresh LB medium; when the turbidity at 600 nm reached 1.5, these cultures were diluted in LB medium to a turbidity at 600 nm of 0.05 and added to polystyrene 96-well plates, which were incubated at 37°C for 7 h without shaking. Each biofilm assay data point represented the average from 10 wells for each of 3 to 20 independent cultures.
Flow cell biofilm experiment and image analysis.
Strains were cultured overnight in LB medium with erythromycin to maintain pCM18 and chloramphenicol to maintain R1drd19. All the flow cells (14) were inoculated at a turbidity at 600 nm of 0.05 at 37°C for 2 h at a flow rate of 13 ml/h, and then fresh LB medium with 300 μg of erythromycin/ml was added at 13 ml/h. After 24 h, a TCS SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) was used to view the flow cell biofilms by imaging approximately eight random positions; for each position, 25 images were taken. Imaris confocal software (Bitplane, Zurich, Switzerland) was applied to process the images. Those 200-color confocal flow chamber images were converted to gray scale by using Image Converter (Neomesh Microsystems, Wainuiomata, Wellington, New Zealand). COMSTAT confocal software (27) was used to determine the biofilm parameters.
Growth rate measurement.
Strains and the mutants carrying the conjugative plasmid R1drd19 were grown in LB medium with appropriate antibiotics, and the increase in the turbidity at 600 nm from 0.08 to 0.6 was measured as function of time. Two independent cultures were used to determine each growth rate.
RNA isolation and DNA microarray analyses.
To study the impact of R1drd19 on E. coli wild-type biofilm formation, E. coli BW25113 biofilms with and without R1drd19 were developed on glass wool (Corning Glass Works, Corning, NY) in LB medium for 7, 15, and 24 h. Similarly, E. coli ATCC 25404 biofilms with and without R1drd19 were developed on glass wool in LB medium for 24 h, and E. coli MG1655 biofilms with and without R1drd19 were developed on glass wool in M9C glu for 24 h. Different media for the strains were chosen because these media were those in which R1drd19 influenced biofilm formation to the largest extent or allowed us to see the effects under different growth conditions. Biofilm cells were removed by sonicating the glass wool cultures in 200 ml of sterile 0.85% NaCl solution at 0°C, and then the total RNA was isolated as described previously (49). The E. coli GeneChip antisense genome array (catalog no. 900381; Affymetrix) was used to analyze the complete E. coli transcriptome as described previously (22). According to the manufacturer's guidelines, each array contains probes for more than 4,200 open reading frames. Each open reading frame is covered by 15 probe pairs consisting of a perfect match probe and a mismatch probe. The expression of each gene is evaluated by comparing the intensity for the perfect match probe and that for the mismatch probe in each of the 15 probe pairs, leading to reliable gene expression profiles (http://www.affymetrix.com/products/arrays/specific/ecoli_antisense.affx). The total signal intensity was scaled automatically in the software to an average value of 500 relative light units. Genes were identified as differentially expressed if the P value was less than 0.05 and if the expression ratio was greater than 2 to 2.5, since the standard deviations for the gene expression ratios in the data were 1.3 to 3.3 (48). The annotations of gene functions were obtained from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/) (18); The Institute for Genomic Research, University of California at San Diego; and the UNAM database (http://ecocyc.org/) (34).
Motility assay.
Cell motility (56) was examined by inoculating 16-h overnight cultures into fresh LB medium, growing the bacteria until the turbidity at 600 nm reached around 1, and inoculating motility agar (1% tryptone, 0.25% NaCl, and 0.3% agar) plates with these exponentially growing cells by using a toothpick. Motility halos were quantified using at least three plates for each set of culture conditions and two independent cultures for each strain.
Aggregation assay.
The aggregation assay was modified slightly from that described previously (51); E. coli strains were cultured for 16 h overnight in LB medium, and these overnight cultures were inoculated into fresh LB medium to yield exponentially growing cells at a turbidity at 600 nm of 1.5. The cells were washed with LB medium to remove antibiotics and concentrated in 3 ml of LB medium to a turbidity at 600 nm of 2.5. The cultures were placed in 14-ml sterile tubes, and the tubes were incubated quiescently at 37°C for 7 h. The turbidity was measured 5 mm underneath the surface to determine the cell concentration, which is an indirect determination of cell aggregation. Each data point was the average of measurements for two tubes for each independent culture, and two independent cultures were analyzed for each strain.
AI-2 assay.
V. harveyi BB170 was inoculated into AB medium and cultured at 30°C at 250 rpm for 16 h. E. coli overnight cultures in LB medium with antibiotics (grown for 16 h at 30°C) were inoculated into fresh LB medium without antibiotics, and 1.5-ml samples of the cell culture were taken at intervals and quickly centrifuged at 16,000 × g for 5 min. The samples were filter sterilized and stored at 0°C. Overnight cultures of V. harveyi BB170 were diluted 5,000-fold in 50 ml of AB medium. Aliquots (1.8 ml) of the diluted V. harveyi BB170 cultures and 0.2-ml aliquots of E. coli supernatants were mixed together and incubated at 30°C at 250 rpm for 4 h, as at the 4-h time point V. harveyi BB170 has the lowest level of bioluminescence. The luminescence levels of the cultures (0.1 ml each) were measured with a luminometer (Turner Design 20/20) after the mixture was preheated at 37°C for 2 min. For each set of conditions, two independent cultures were analyzed.
Microarray data accession numbers.
The expression data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession numbers GSM147162 to GSM147165 and GSM153383 to GSM153388 (18).
RESULTS
Conjugative plasmid R1drd19 promotes E. coli biofilm formation.
The addition of R1drd19 had no affect on the growth rate of the E. coli BW25113 wild-type strain in LB medium (specific growth rate, 1.53 ± 0.00 h−1 for the wild type versus 1.56 ± 0.05 h−1 for the BW25113 strain with R1drd19) (Table 2). However, in 96-well plates, adding R1drd19 increased BW25113 biofilm formation in LB medium by 1.9- ± 0.6-fold at 7 h and increased ATCC 25404 biofilm formation in LB medium by 3.5- ± 0.5-fold at 24 h.
TABLE 2.
Specific growth rates of E. coli strains in LB medium and flow cell COMSTAT analysis of biofilms formed in LB medium at 37°C after 24 ha
| Strain | Growth rate (h−1) | Biomass (μm3/μm2) | Substratum coverage (%) | Mean biofilm thickness (μm) | Roughness coefficient |
|---|---|---|---|---|---|
| BW25113 wild type | 1.53 ± 0.00 | 3 ± 3 | 3 ± 2 | 4 ± 4 | 1.7 ± 0.2 |
| BW25113/R1drd19 | 1.56 ± 0.05 | 11 ± 4 | 39 ± 16 | 14 ± 6 | 0.7 ± 0.3 |
| rseA/R1drd19 | 1.22 ± 0.02 | 2.7 ± 0.5 | 4 ± 3 | 5 ± 1 | 1.4 ± 0.1 |
| bssR/R1drd19 | 1.36 ± 0.00 | 3 ± 3 | 4 ± 5 | 5 ± 4 | 1.4 ± 0.4 |
| cpxA/R1drd19 | 1.24 ± 0.05 | 5 ± 2 | 22 ± 7 | 6 ± 2 | 1.3 ± 0.2 |
| ompA/R1drd19 | 0.98 ± 0.03 | 3 ± 1 | 5 ± 5 | 4 ± 1 | 1.6 ± 0.2 |
| rseA | 1.6 ± 0.1 | 9 ± 8 | 29 ± 9 | 11 ± 7 | 1.1 ± 0.1 |
| bssR | 1.37 ± 0.04 | 12 ± 9 | 47 ± 26 | 15 ± 9 | 0.9 ± 0.4 |
| cpxA | 1.22 ± 0.02 | 2.5 ± 0.5 | 23 ± 6 | 5 ± 1 | 1.11 ± 0.09 |
| ompA | 1.15 ± 0.00 | 3 ± 3 | 18 ± 10 | 4 ± 3 | 1.4 ± 0.2 |
One standard deviation is shown.
To corroborate the results of the 96-well biofilm assay, continuous-flow cells were used to study the E. coli BW25113 biofilm architecture with and without R1drd19 in LB medium at 37°C after 24 h. COMSTAT analysis (Table 2) indicated that upon the addition of R1drd19, the biofilm biomass of BW25113 increased 3.7-fold and the mean thickness of BW25113 biofilms increased 3.5-fold. Hence, R1drd19 increases biofilm formation considerably without affecting planktonic growth.
Profiles of gene expression upon the addition of R1drd19.
To gain insight into the genetic basis of the increased biofilm formation upon the addition of conjugative plasmid R1drd19, gene expression profiles of the biofilm cells of BW25113 in LB medium at 7, 15, and 24 h, MG1655 in M9C glu at 24 h, and ATCC 25404 in LB medium at 24 h were determined. The specific media were chosen to maximize the effects of R1drd19 on biofilm formation by each strain, as well as to demonstrate the effects of the conjugative plasmid in both minimal and rich media. Multiple strains were used so that a general effect of R1drd19 on biofilm formation could be discerned, and the microarray analyses were conducted with BW25113 at multiple time points to get a temporal response. The five sets of microarray data indicating the most induced and repressed genes are summarized in Table S1 in the supplemental material. The genes differently expressed upon the addition of R1drd19 included genes involved in amino acid transport and metabolism; carbohydrate transport and metabolism; cell motility; cell wall and membrane biogenesis; defense mechanisms; energy production and conversion; inorganic-ion transport and metabolism; lipid transport and metabolism; posttranslational modification; protein turnover; chaperoning; replication; recombination and repair; secondary-metabolite biosynthesis, transport, and catabolism; signal transduction mechanisms; and transcription; phage and phage-related genes; and genes with unknown functions.
The gene expression profiles for different strains and medium backgrounds varied. Adding R1drd19 to BW25113 at 7 h induced 451 genes (10% of the genome) more than 2.0-fold and repressed 291 genes (7% of the genome) more than 2.0-fold. Adding R1drd19 to BW25113 at 15 h induced 112 genes (2.5% of the genome) and repressed 10 genes (0.2% of the genome) more than 2.0-fold. Adding R1drd19 to BW25113 at 24 h induced 30 genes (0.7% of the genome) more than 2.0-fold and repressed 275 genes (6% of the genome) more than 2.0-fold. Adding R1drd19 to MG1655 at 24 h induced 39 genes (0.9% of the genome) more than 2.5-fold and repressed 50 genes (1.1% of the genome) more than 2.5-fold. Adding R1drd19 to ATCC 25404 at 24 h induced 53 genes (1.2% of the genome) more than 2.0-fold and repressed 9 genes (0.2% of the genome) more than 2.0-fold. As shown by the five microarray data sets, the only group of genes consistently induced in all data sets was the group of replication, recombination, and repair genes insA_1, insA_2, and insA_5. The genes oppABCDF, encoding oligopeptide ABC transporters, were induced up to fourfold in the BW25113 7- and 15-h microarrays. Also, energy production and conversion genes (sdhABCD, sucABCD, nuoABCEFGHIJKLM, and atpEFH) were induced two- to sevenfold in the BW25113 7- and 15-h microarrays. Within any single microarray data set, the group of genes with the most consistent differential expression upon the addition of R1drd19 was that of the 23 e-14 phage genes in MG1655 in M9C glu at 24 h, which were induced 3- to 104-fold. The psp operon was also induced consistently in BW25113 in LB medium at 15 h. The selection of genes (corresponding to 40 isogenic mutants) for further study was based primarily on two criteria: (i) the genes were significantly induced or repressed upon the addition of R1drd19 to BW25113 (hence, their study was facilitated by the use of the Keio collection of single-gene knockouts for this strain), or (ii) they were located just upstream or downstream of those with marked differential expression.
Biofilm formation by isogenic mutants.
Based on the microarray results, biofilm formation in the presence of R1drd19 was tested for 40 related isogenic knockout mutants of BW25113 in LB medium at 7 h by using the crystal violet assay (Fig. 1). To aid in their analysis, the isogenic mutants were categorized into five classes: cells with class I mutations (those in rseA, bssR, cpxA, and ompA) exhibited no difference from the wild-type strain in biofilm formation and no increase upon the addition of R1drd19 (Fig. 1A); cells with class II mutations (those in gatC, yagI, ompC, cspA, pspD, pspB, ymgB, gadC, pspC, ymgA, slp, cpxP, cpxR, cstA, rseC, ompF, and yqjD) showed increased biofilm formation compared to the wild-type strain, and the addition of R1drd19 decreased biofilm formation relative to that by the mutant without R1drd19 (Fig. 1B); cells with class III mutations (those in gadA) showed increased biofilm formation compared to the wild-type strain and increased biofilm formation upon the addition of R1drd19 (Fig. 1C); cells with class IV mutations (those in aceB, glgS, glpD, csgG, hdeD, pspA, gadB, tnaA, and crl) showed increased biofilm formation compared to the wild-type strain but had little change in biofilm formation upon the addition of R1drd19 (Fig. 1D); and cells with class V mutations (those in flhC, nmpC, flhD, pspE, icdA, atpF, atpH, rseB, and sodB) had no difference from the wild-type strain in biofilm formation but exhibited increased biofilm formation upon the addition of R1drd19 (Fig. 1E). It appears that the deletion of class I or II genes blocked the effects of R1drd19 on BW25113, which indicates that these genes are key genes involved in the R1drd19-related induction of biofilm formation.
FIG. 1.
Biofilm formation by E. coli BW25113 mutants of classes I through V in LB medium after 7 h at 37°C in 96-well plates. (A) Class I mutations produce no difference from the wild-type strain in biofilm formation and do not increase biofilm formation upon the addition of R1drd19. (B) Class II mutations increase biofilm formation compared to that by the wild-type strain but decrease biofilm formation upon the addition of R1drd19. (C) Class III mutations increase biofilm formation compared to that by the wild-type strain and increase biofilm formation upon the addition of R1drd19. (D) Class IV mutations increase biofilm formation compared to that by the wild-type strain but produce no difference in biofilm formation upon the addition of R1drd19. (E) Class V mutations produce no difference from the wild-type strain in biofilm formation but increase biofilm formation upon the addition of R1drd19. OD540nm, optical density at 540 nm.
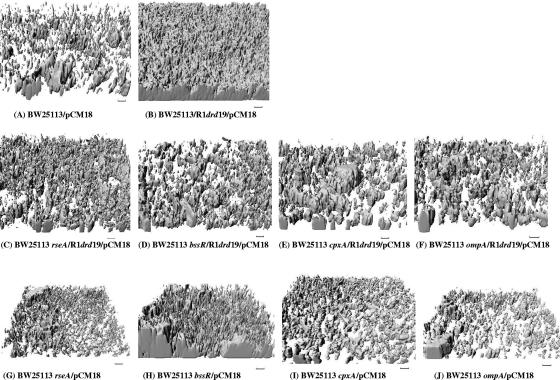
To corroborate the 96-well biofilm results for the class I mutants, we also conducted flow cell experiments in the presence (rseA/R1drd19, bssR/R1drd19, cpxA/R1drd19, and ompA/R1drd19 strains) and absence of R1drd19 in LB medium at 37°C after 24 h (Fig. 2C to J). Flow cell experiments were also conducted with rseA, bssR, cpxA, and ompA mutant strains as negative controls. COMSTAT analysis (Table 2) indicated that there was no increase in biomass or mean thickness upon the addition of R1drd19 to cells with the rseA, bssR, cpxA, and ompA mutations compared to that for the wild-type strain. Hence, the biofilm biomasses for rseA/R1drd19, bssR/R1drd19, cpxA/R1drd19, and ompA/R1drd19 strains decreased dramatically compared to that for BW25113/R1drd19 (4.1-, 3.7-, 2.2-, and 3.5-fold, respectively), as did the mean biofilm thicknesses (2.8-, 2.8-, 2.3-, and 13-fold, respectively) (Table 2). In contrast to the wild-type strain, the four class I mutants showed decreased or unaltered biomasses, substratum coverage levels, and mean biofilm thicknesses upon the addition of R1drd19 (Table 2). Therefore, the class I mutations (those in rseA, bssR, cpxA, and ompA) prevent R1drd19 from increasing biofilm formation as it does with the wild-type strain, and there was good agreement between the results from the 96-well and the flow cell biofilm experiments. Furthermore, the normalization of biofilm formation levels by cell growth did not affect the classification of the class I genes (data not shown).
FIG. 2.
Imaris images of flow cells in E. coli biofilms in LB medium after 24 h at 37°C. Scale bars represent 10 μm.
R1drd19 increases aggregation through class I genes.
To study the role of aggregation in biofilm formation by E. coli with R1drd19, we tested the aggregation of the four E. coli BW25113 class I mutants and the 17 class II mutants that we identified in relation to R1drd19 and biofilms. For the wild-type strain in LB medium, the addition of R1drd19 increases aggregation 3- ± 1-fold. Except for the cstA mutation and cpxR, which is the partner of class I gene cpxA and is part of the same two-component system, class II mutations have apparently inverse effects on biofilm enhancement by R1drd19 and aggregation stimulated by the presence of the plasmid in that these mutations alone had no significant effect on the aggregation of the wild-type strain but they increased aggregation 4- to 47-fold relative to that of the wild-type strain upon the addition of R1drd19 (Table 3). Class I mutations also had no effect on aggregation compared to that of the wild-type strain; however, unlike class II mutations, class I mutations did not increase aggregation upon the addition of R1drd19 except for the rseA mutant, which showed an extraordinary increase in aggregation (267-fold) upon the addition of R1drd19 (Table 3). Since the addition of R1drd19 increased the aggregation of the wild-type strain but not the aggregation of three of the four class I mutants, R1drd19 enhancement of the aggregation of the wild-type strain requires class I genes bssR, cpxA, and ompA. In contrast, the class II proteins repress aggregation since their inactivation results in increased aggregation. Also, this increase in aggregation is inversely proportional to the level of biofilm formation, as shown in Fig. 1B. Therefore, R1drd19 increases aggregation using class I proteins, and class II proteins repress aggregation.
TABLE 3.
Motility and aggregation assay results for E. coli BW25113 class I and class II mutants
| Mutant | Classification | Change (n-fold) relative to wild-type measurementa in:
|
|||
|---|---|---|---|---|---|
| Motility
|
Aggregation
|
||||
| Without R1drd19 | With R1drd19 | Without R1drd19 | With R1drd19 | ||
| cpxA | Class I | 0.4 ± 0.1 | 0.39 ± 0.09 | 1.04 ± 0.06 | 1.1 ± 0.1 |
| rseA | Class I | 2.8 ± 0.6 | 2.2 ± 0.5 | 1.13 ± 0.05 | 267 ± 43 |
| bssR | Class I | 1.6 ± 0.4 | 1.7 ± 0.4 | 1.04 ± 0.04 | 1.14 ± 0.06 |
| ompA | Class I | 1.0 ± 0.1 | 1.1 ± 0.1 | 1.01 ± 0.05 | 1.3 ± 0.1 |
| ymgA | Class II | 7 ± 1 | 2.0 ± 0.4 | 1.01 ± 0.03 | 47 ± 24 |
| slp | Class II | 1.2 ± 0.1 | 0.59 ± 0.07 | 1.00 ± 0.03 | 37 ± 6 |
| cstA | Class II | 4.4 ± 0.4 | 3.2 ± 0.3 | 3.3 ± 0.5 | 9 ± 3 |
| gatC | Class II | 3.9 ± 0.8 | 3.6 ± 0.7 | 1.11 ± 0.02 | 8.5 ± 0.8 |
| yqjD | Class II | 2.2 ± 0.2 | 1.6 ± 0.2 | 0.94 ± 0.03 | 10.2 ± 0.6 |
| cpxR | Class II | 4.4 ± 0.2 | 2.8 ± 0.1 | 1.38 ± 0.06 | 1.19 ± 0.07 |
| cpxP | Class II | 4.1 ± 0.7 | 3.6 ± 0.6 | 1.37 ± 0.02 | 4.6 ± 0.5 |
| yagI | Class II | 1.6 ± 0.3 | 1.4 ± 0.3 | 1.01 ± 0.03 | 11 ± 1 |
| ompC | Class II | 2.1 ± 0.4 | 1.9 ± 0.5 | 1.10 ± 0.05 | 13.2 ± 0.5 |
| cspA | Class II | 3.2 ± 0.6 | 2.9 ± 0.7 | 1.32 ± 0.07 | 9.3 ± 0.5 |
| pspD | Class II | 3.9 ± 0.7 | 3.4 ± 0.6 | 1.08 ± 0.04 | 17 ± 3 |
| pspB | Class II | 1.0 ± 0.4 | 2.1 ± 0.4 | 1.2 ± 0.2 | 13 ± 3 |
| ymgB | Class II | 1.7 ± 0.6 | 2.0 ± 0.2 | 1.16 ± 0.02 | 9 ± 1 |
| gadC | Class II | 3.6 ± 0.8 | 2.1 ± 0.5 | 1.1 ± 0.1 | 12 ± 2 |
| pspC | Class II | 2.8 ± 0.5 | 2.1 ± 0.4 | 1.04 ± 0.04 | 4 ± 1 |
| rseC | Class II | 3.0 ± 0.5 | 2.4 ± 0.4 | 1.23 ± 0.08 | 15 ± 2 |
| ompF | Class II | 4.3 ± 0.8 | 3.6 ± 0.7 | 1.5 ± 0.3 | 19 ± 2 |
Motility was determined after 8 h at 37°C, and aggregation was determined after 7 h in LB medium at 37°C. Results are expressed as the ratio of the measurement for the indicated mutant to the measurement for the wild type. Data are the averages of results for two independent cultures, and one standard deviation is shown.
R1drd19 increases biofilm formation by decreasing motility through class I and class II genes.
The addition of R1drd19 decreased BW25113 wild-type motility by 24% ± 7%. The motility of wild-type BW25113 was not substantial, corresponding to a diameter of only 0.6 ± 0.1 cm after 8 h. We also conducted motility assays for the class I and II mutants (Table 3). Of the 17 class II mutants, 88% exhibited an increase in motility of 1.6- to 7.0-fold compared to that of the wild type, and except for the slp mutant, all of them were more motile than the wild-type strain upon the addition of R1drd19, although for 88% of them, the increase in motility relative to that of the wild type was less than the increase without R1drd19 (Table 3). Hence, one of the reasons for the failure of the conjugative plasmid to increase biofilm formation appears to be the enhanced motility that occurs with the class II mutations.
Supporting this idea, the deletion of the class I genes prevented R1drd19 from decreasing motility as it did for the wild-type strain (Table 3) and motility was increased for the rseA, bssR, and ompA mutants. One of the class I genes, cpxA, encodes the upstream protein CpxA in the CpxR-P regulation pathway that down-regulates motility genes (10) (see Fig. 4). cpxA, cpxR, and cpxP in BW25113 in LB medium were induced 1.3-, 2.5-, and 6.1-fold, respectively, by the addition of R1drd19 at 7 h (see Table S1 in the supplemental material), which indicates the activation of the two-component system that led to 16 motility genes' being repressed by the addition of R1drd19 to the wild-type strain (see Table S1 in the supplemental material). Therefore, it appears that R1drd19 increases wild-type biofilm formation because it initiates the pathway which represses cell motility through the class I genes.
FIG. 4.
Hypothesized mechanism of E. coli biofilm formation with R1drd19. Envelope stress caused by conjugative pili initiates the network. OmpA is the potential outer membrane protein that receives the signal from conjugative pili and then translates the signal to the sensor, CpxA, of the two-component system in which CpxR is the response regulator. Phosphorylated CpxR (CpxR-P) then regulates biofilm-related gene expression. RseA-σE is another system involved in the envelope stress response system and, to some extent, overlaps with the CpxAR system. In this pathway, RseA is the sensor to detect the envelope stress caused by conjugative pili. MicA is a negative antisense regulator of OmpA synthesis, and this sRNA is induced by the overexpression of σE. OM stands for outer membrane, and IM stands for inner membrane.
R1drd19 increases biofilm formation by increasing the quorum-sensing signal AI-2.
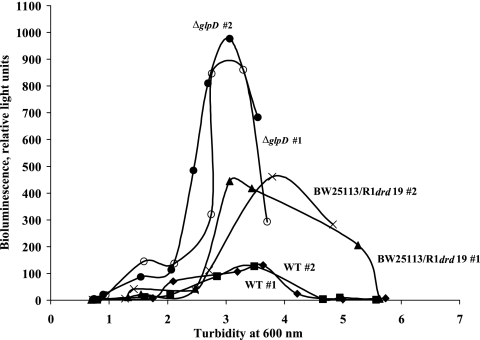
According to the microarray data for ATCC 25404 in LB medium at 24 h (see Table S1 in the supplemental material), the addition of R1drd19 repressed glpDKF 3.0- to 5.7-fold; the glpD mutation represses lsr transcription, which results in the accumulation of extracellular AI-2 (64). Therefore, we studied the effect of R1drd19 on extracellular AI-2 concentrations. As expected, extracellular AI-2 concentrations in both the wild-type strain BW25113 and BW25113/R1drd19 accumulated in the stationary phase (61) and decreased at the end of the stationary phase (Fig. 3). However, the addition of R1drd19 increased extracellular AI-2 concentrations 3.4-fold at a turbidity of 3.5 (Fig. 3). As a positive control, AI-2 concentrations for the glpD mutant were assayed, and the glpD mutation increased AI-2 by 7.1-fold, as expected (64). Therefore, adding R1drd19 increases the cell quorum-sensing signal AI-2, probably by repressing glpD, which leads to an increase of biofilm formation by E. coli, as has been seen with the direct addition of AI-2 (22).
FIG. 3.
Extracellular AI-2 concentrations demonstrated by V. harveyi bioluminescence for E. coli BW25113 (wild type [WT]), BW25113/R1drd19, and the BW25113 glpD strain (ΔglpD). Results for two replicates (#1 and #2) are shown.
DISCUSSION
In this study, we showed that E. coli biofilm formation is induced by the addition of R1drd19 to different strains (E. coli BW25113, MG1655, and ATCC 25404) in rich and minimal media. Using a whole-transcriptome approach, we discovered that the addition of this conjugative plasmid consistently affected the expression of class I genes (rseA, bssR, the cpx operon, and ompA) in different E. coli strains. We also discovered that mutations in these genes prevented the addition of R1drd19 from increasing biofilm formation as it does in the wild-type strain.
We hypothesize that the pili formed by the conjugative plasmid lead to the presence of unassembled or misfolded proteins in the membrane that increase E. coli K-12 biofilm formation through the associated stress response (Fig. 4), much as we have previously shown acid, heat, hydrogen peroxide, and cadmium stress to increase E. coli biofilm formation (67). The envelope stress response system responds to pili (11) and regulates genes involved in biofilm formation. RseA and CpxA are the sensors in the two different envelope stress response systems, the RseA-σE envelope stress response system and the CpxAR two-component system, respectively. Although the idea is highly speculative, the class I protein OmpA may also sense the signal from pili in the outer membrane and act as the activator of the CpxAR two-component system. Therefore, three of the four class I genes are involved in early regulation of the biofilm pathway by sensing the signal from conjugative pili which initiates the envelope stress response system and regulates biofilm-related genes, including adherence genes and motility genes. Upon the deletion of rseA, cpxA, and ompA, E. coli is not able to sense the signal upon the addition of R1drd19 and, thus, this conjugative plasmid fails to increase biofilm formation.
rseA encodes the envelope stress and anti-σE factor (1) and was induced 2.3-fold in BW25113 in the 7-h microarray analysis. This gene was differentially expressed in our other microarray studies (it was repressed 2.5-fold in BW25113 in LB medium at 24 h and repressed 2.3-fold in MG1655 in M9C glu at 24 h) (see Table S1 in the supplemental material). It appears that conjugation promotes RseA binding to σE, which blocks σE from binding RNA polymerase and thus possibly represses certain biofilm-related genes. Furthermore, rseA is induced by cold shock (42) and is down-regulated by OmpC, which is up-regulated in biofilm cells compared to that in stationary cultures (54). Here, the cold shock genes cspACDG in the BW25113 strains were induced 2.5- to 3.5-fold by R1drd19 (see Table S1 in the supplemental material). Hence, we also speculate that cold shock proteins are possibly involved in the mechanism of biofilm formation increase upon the addition of R1drd19 through the RseA system.
The relationship between Cpx, stress, and biofilm formation is more clear. Cpx expression in relation to envelope stress results from the expression of protein folding catalysts (DsbA, PpiA, and PpiD) and degrading factors like DegP, which cleaves RseA and releases σE (17). At the transcriptional level, Cpx represses genes for adherence, taxis, and motility whereas it activates genes involved in folding factor/protein degradation, the production of outer membrane proteins, and multidrug resistance (16). cpxAR and cpxP in BW25113 in LB medium were induced (1.3-, 2.5-, and 6.1-fold, respectively) upon R1drd19 addition at 7 h, and cpxR and cpxP in BW25113 in LB medium were induced 3.5-fold at 15 h. Possibly in association with cpxAR and cpxP induction, 16 cell motility genes in BW25113 in LB medium were repressed 1.7- to 4.9-fold upon the addition of R1drd19 at 7 h. Curli genes csgBEFG were also repressed 2.8-, 3.7-, 2.3-, and 6.5-fold, respectively. Hence, our DNA microarray data show that at the transcriptional level, the genes related to motility were repressed by the addition of R1drd19 and that CpxR is potentially the regulator involved in this mechanism (see Table S1 in the supplemental material).
The function of OmpA upon R1drd19 addition is less clear, even though we found that it is necessary for R1drd19 to increase E. coli biofilm formation. ompACF, genes encoding outer membrane proteins, were induced 3.0-, 2.8-, and 2.5-fold, respectively, upon R1drd19 addition to BW25113 in LB medium at 7 h. ompA was also induced 2.8- and 3.0-fold, respectively, upon R1drd19 addition to BW25113 in LB medium at 15 h and R1drd19 addition to ATCC 25404 in LB medium at 24 h. We predict that OmpA, similar to another outer membrane protein, NlpE, is positioned early in the stress response system. Adding R1drd19 induces ompA gene expression; consequently, overproduced OmpA activates the two-component system CpxAR to mediate biofilm formation. In addition, OmpA enhances E. coli swarming without significantly affecting swimming (30); usually, swarming and biofilm formation are correlated (55), so the addition of R1drd19 increases OmpA, which may then facilitate biofilm formation through its link to swarming.
Two of five sets of microarray data showed bssR to be repressed (see Table S1 in the supplemental material). In LB medium, the deletion of bssR repressed mtr, which encodes the protein importing indole, and induced acrEF, which encode proteins involved in the export of indole (15). DNA microarrays indicated that BssR regulates genes involved in biofilm formation (15), and 130 of these genes were affected by quorum sensing via AI-2. However, how R1drd19 interacts with BssR is not clear. It is possible that R1drd19 increases biofilm formation through indole regulated by BssR.
In support of the hypothesis of the link between the cell envelope stress response system and R1drd19 addition, we found that the genes that encode murein were induced upon the addition of R1drd19 to the E. coli strains: murE, glmS, and yeaF in BW25113 in LB medium at 7 h were induced 2.5- to 3.3-fold (see Table S1 in the supplemental material). Murein contributes to the mechanical stability of the E. coli cell wall (29). We postulate that the murein genes were induced due to the response to the cell envelope stress upon the addition of R1drd19. lpp, the gene encoding murein lipoprotein, was also induced 2.8- and 2.1-fold upon the addition of R1drd19 to BW25113 in LB medium at 7 and 15 h and 2.3-fold upon the addition of R1drd19 to ATCC 25404 in LB medium at 24 h (see Table S1 in the supplemental material). bolA, a possible regulator of murein genes (53), was also induced 2.0-fold in BW25113 in LB medium at 15 h but was repressed 2.0-fold in ATCC 25404 in LB medium at 24 h upon the addition of R1drd19. In addition, the operon encoding oligopeptide permeases, oppABCDF (28), was induced 2.8- to 4.0-fold upon the addition of R1drd19 to BW25113 in LB medium at 7 h; these permeases assist murein recycling (23). Hence, the induction of the murein genes upon R1drd19 addition corroborates the conclusion that the cell experiences envelope stress.
The conjugative plasmid may also increase cell persistence by inducing persistence genes in E. coli. Persisters are responsible for the high-level resistance of biofilms to various antimicrobials (57). Our microarray data indicated that the conjugative plasmid had a significant effect on persistence-gene transcription. In MG1655, 23 e-14 prophage genes (b1137 to b1159) were induced 3- to 104-fold by the addition of R1drd19; we also identified CP 4-6 prophage genes that were induced (b0275 in all the BW25113 microarrays and yafXZ in the BW25113 15-h microarray). Both e-14 prophage and CP 4-6 prophage genes have been reported previously to be induced in persister cells (37). Another operon associated with cell persistence, pspABCDE (33), was also induced upon R1drd19 addition (see Table S1 in the supplemental material). The psp genes in this operon were induced 3.5- to 6.5-fold in BW25113 in LB medium at 15 h and 1.6- to 2.5-fold in ATCC 25404 in LB medium at 24 h and repressed 3.5- to 4.3-fold in MG1655 in M9C glu at 24 h upon the addition of R1drd19. Therefore, the addition of the conjugative plasmid may influence cell persistence.
By investigating the differentially expressed genes of the host rather than the R1 plasmid itself, it was discovered here that cell envelope stress is one of the key reasons for the increase in biofilm formation upon the addition of a conjugative plasmid. Understanding how biofilms form when they are influenced by a conjugative plasmid is important since these plasmids enhance biofilm formation while overriding the importance of flagella, type I fimbriae, Ag43, and curli (46).
Supplementary Material
Acknowledgments
This research was supported by the NIH (grant no. 5R01EB003872-05) and the Army Research Office (grant no. W911NF-06-1-0408).
Footnotes
Published ahead of print on 14 March 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma E-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor, E., D. Walthers, L. J. Kenney, and M. Goulian. 2005. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins OmpF and OmpC. J. Bacteriol. 187:5723-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, M., R. Eferl, G. Zellnig, K. Teferle, A. Dijkstra, G. Koraimann, and G. Högenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J.-M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, E. A., J. L. Tupy, T. M. Gruber, S. Wang, M. M. Sharp, C. A. Gross, and S. A. Darst. 2003. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 11:1067-1078. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Las Peñas, A., L. Connolly, and C. A. Gross. 1997. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 10.De Wulf, P., O. Kwon, and E. C. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Wulf, P., A. M. McGuire, X. Liu, and E. C. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 12.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dionisio, F., I. C. Conceição, A. C. R. Marques, L. Fernandes, and I. Gordo. 2005. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1:250-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 15.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorel, C., P. Lejeune, and A. Rodrigue. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157:306-314. [DOI] [PubMed] [Google Scholar]
- 17.Duguay, A. R., and T. J. Silhavy. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694:121-134. [DOI] [PubMed] [Google Scholar]
- 18.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egler, M., C. Grosse, G. Grass, and D. H. Nies. 2005. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 21.González Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. [DOI] [PubMed] [Google Scholar]
- 22.González Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock, V., and P. Klemm. 2007. Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 75:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 26.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbφll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 28.Higgins, C. F., and M. M. Hardie. 1983. Periplasmic protein associated with the oligopeptide permeases of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 155:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue, T., R. Shingaki, S. Hirose, K. Waki, H. Mori, and K. Fukui. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 189:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junker, L. M., J. E. Peters, and A. G. Hay. 2006. Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 152:2233-2245. [DOI] [PubMed] [Google Scholar]
- 33.Kaldalu, N., R. Mei, and K. Lewis. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koraimann, G., and G. Högenauer. 1989. A stable core region of the tra operon mRNA of plasmid R1-19. Nucleic Acids Res. 17:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koraimann, G., C. Koraimann, V. Koronakis, S. Schlager, and G. Högenauer. 1991. Repression and derepression of conjugation of plasmid R1 by wild-type and mutated finP antisense RNA. Mol. Microbiol. 5:77-87. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay, S., M. Tasab, A. Rickard, M. Kertesz, and P. Gilbert. 2007. Persister cells: mechanisms towards biofilm recalcitrance, abstr. S10:3, p. 44. Fourth ASM Conf. Biofilms, Quebec, Canada. American Society for Microbiology, Washington, DC.
- 38.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nancharaiah, Y. V., P. Wattiau, S. Wuertz, S. Bathe, S. V. Mohan, P. A. Wilderer, and M. Hausner. 2003. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl. Environ. Microbiol. 69:4846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Dehò. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154:573-580. [DOI] [PubMed] [Google Scholar]
- 43.Pölzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Högenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 44.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 45.Raivio, T. L., and T. J. Silhavy. 1999. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 46.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 47.Reisner, A., B. M. Höller, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 88:630-642. [DOI] [PubMed] [Google Scholar]
- 49.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez, R. L., and R. C. Tait. 1983. Recombinant DNA techniques: an introduction. Benjamin/Cummings Publishing, Menlo Park, CA.
- 51.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 187:1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 53.Santos, J. M., M. Lobo, A. P. A. Matos, M. A. De Pedro, and C. M. Arraiano. 2002. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45:1729-1740. [DOI] [PubMed] [Google Scholar]
- 54.Schembri, M. A., K. Kjærgaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 55.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 56.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 57.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatum, E. L., and J. Lederberg. 1947. Gene recombination in the bacterium Escherichia coli. J. Bacteriol. 53:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udekwu, K. I., and E. G. H. Wagner. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, L., Y. Hashimoto, C. Tsao, J. Valdes, and W. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Will, W. R., and L. S. Frost. 2006. Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J. Bacteriol. 188:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood, T. K., A. F. G. Barrios, M. Herzberg, and J. Lee. 2006. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 72:361-367. [DOI] [PubMed] [Google Scholar]
- 64.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yee, D. C., J. A. Maynard, and T. K. Wood. 1998. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl. Environ. Microbiol. 64:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshioka, Y., H. Ohtsubo, and E. Ohtsubo. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J. Bacteriol. 169:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, X., R. García Contreras, and T. K. Wood. 2007. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 189:3051-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.