Abstract
We recently described a new method to activate antibiotic production in bacteria by introducing a mutation conferring resistance to a drug such as streptomycin, rifampin, paromomycin, or gentamicin. This method, however, enhanced antibiotic production by only up to an order of magnitude. Working with Streptomyces coelicolor A3(2), we established a method for the dramatic activation of antibiotic production by the sequential introduction of multiple drug resistance mutations. Septuple and octuple mutants, C7 and C8, thus obtained by screening for resistance to seven or eight drugs, produced huge amounts (1.63 g/liter) of the polyketide antibiotic actinorhodin, 180-fold higher than the level produced by the wild type. This dramatic overproduction was due to the acquisition of mutant ribosomes, with aberrant protein and ppGpp synthesis activity, as demonstrated by in vitro protein synthesis assays and by the abolition of antibiotic overproduction with relA disruption. This new approach, called “ribosome engineering,” requires less time, cost, and labor than other methods and may be widely utilized for bacterial strain improvement.
Strain improvement is important in applied microbiological research, especially in the production of clinically important antibiotics as well as antibiotics important in veterinary medicine and agriculture. Current methods of antibiotic production, ranging from classical random approaches to metabolic engineering, are either costly or labor-intensive. We recently described a new method to increase antibiotic production in bacteria by modulating ribosomal components (ribosomal proteins or rRNA), i.e., by the introduction of mutations conferring drug resistance, since many antibiotics target the ribosome (13, 29, 31). This new approach, called “ribosome engineering” (28), has several advantages. These advantages include the ability to screen for drug resistance mutations by simple selection on drug-containing plates, even if the mutation frequency is extremely low (e.g., <10−10), and the ability to select for mutations without prior genetic information. Thus, this method requires no induced mutagenesis. This approach has been used to enhance the production of salinomycin in an industrial strain of Streptomyces albus (33); to activate the synthesis of dormant antibiotics (15, 19); to improve chemical tolerance in Pseudomonas putida (12), a valuable bacterium for waste processing; and to enhance the synthesis of enzymes such as α-amylase in Bacillus subtilis (22). Interestingly, the introduction of several drug resistance mutations had a cumulative effect on antibiotic production. This was shown by the sequential introduction of three drug resistance mutations into Streptomyces coelicolor A3(2) (str, gen, and rif, which confer resistance to streptomycin [Sm], gentamicin [Gen], and rifampin [Rif], respectively) and three rounds of selection, with the resulting triple mutant, SGR, showing hierarchical increments of antibiotic production (14).
S. coelicolor A3(2), the genetically best-characterized strain of Streptomyces, produces at least four distinct classes of antibiotics (21), including the blue-pigmented polyketide antibiotic actinorhodin (Act), thus providing an easily tractable system for the methodological study of strain improvement. Based on our previous results using single mutations or multiple mutations, we demonstrate here the efficacy of septuple and octuple drug resistance mutations for the dramatic activation of antibiotic production. The mechanisms underlying this remarkable activation were also studied.
MATERIALS AND METHODS
Drugs and chemicals.
Geneticin, paromomycin (Par), fusidic acid (FA), lincomycin (Lin), and thiostrepton (Tsp) were purchased from Sigma; Sm was purchased from Nacalai Tesque, Inc. (Japan); and Gen, Rif, tetracycline (Tet), and hygromycin were purchased from Wako Pure Chemicals (Japan). The standards guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) were prepared enzymatically in our laboratory, each with a purity of >95%.
Bacterial strains and preparation of multiply drug-resistant mutants.
S. coelicolor A3(2) wild-type strain 1147, triple mutant strain SGR (14), and its multiply drug-resistant derivatives are listed in Table 1. All of the mutations arose spontaneously, and no induced mutagenesis was required. For each experiment, strains were inoculated onto GYM (31) or SFM (21) plates and incubated for approximately 7 to 10 days for sporulation. Fresh spore suspensions were inoculated into 100 ml medium in a 500-ml flask and incubated on a rotary shaker (200 rpm) at 30°C except where indicated.
TABLE 1.
S. coelicolor A3(2) and its drug-resistant derivatives used in this study
| Strain | Genotype or descriptionf | Mutation introduced at each stepa | Reference or source |
|---|---|---|---|
| 1147 | Prototrophic wild type | —a | 21 |
| K88E | str | K88E in ribosomal protein S12 | 8 |
| SG | str gen | ND | 14 |
| SGR | str gen rif | H437Y in RNA polymerase β-subunit | 14 |
| SGRP | str gen rif par | Insertion of a glycine residue at position 92 of S12 | This study |
| C5 | str gen rif par gnt | —b | This study |
| C6 | str gen rif par gnt fus | —c | This study |
| C7 | str gen rif par gnt fus tsp | —d | This study |
| C8 | str gen rif par gnt fus tsp lin | —e | This study |
| C8relA2 | str gen rif par gnt fus tsp lin relA | Insertional disruption of relA gene by hygromycin resistance cassette (hyg) | This study |
No mutation; wild type.
Mutation not determined.
Mutation not detected in elongation factor G.
Mutation not detected in ribosomal protein L11.
Mutation not detected in ribosomal proteins S7, L14, and L15.
str, gen, rif, par, gnt, fus, tsp, and lin confer resistance to streptomycin, gentamicin, rifampin, paromomycin, Geneticin, fusidic acid, thiostrepton, and lincomycin, respectively.
Triple mutant strain SGR (14) was used as the starter strain to select for multiple-drug-resistant mutants, with general screening procedures performed as described previously (28). The procedure used to prepare multiply drug-resistant mutants is illustrated in Fig. 1A. Spore suspensions (usually >109 spores) were spread and incubated for 7 days. Antibiotic-resistant clones were first screened for the production of Act on plate cultures, with candidates having the deepest blue color being assessed for Act production assays using liquid media. If necessary, mutations were mapped by DNA sequencing. In each step, one or two colonies with the highest Act production were used for the next round of screening. SGR spore suspensions were spread onto GYM plates containing various concentrations of Par, usually approximately 3- to 30-fold of the parental strain's MIC to select for Par resistance, followed by similar steps of screening for resistance to Geneticin, FA, Tsp, and Lin, finally yielding octuple mutants with resistance to eight different drugs.
FIG. 1.
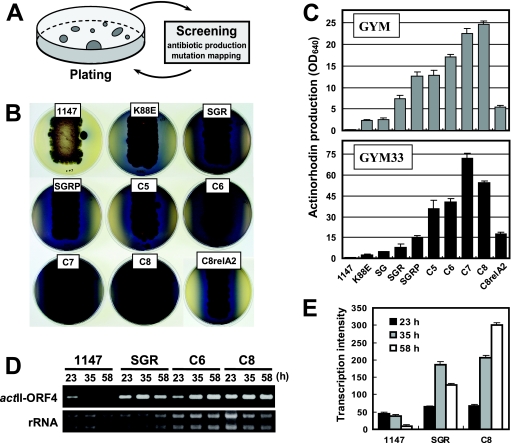
Act production and actII-ORF4 transcription in drug-resistant mutants. (A) Illustration of the procedure used for the sequential introduction of drug resistance mutations to generate multiple-drug-resistant mutants. (B) Act production on agar plates. Strains were inoculated onto GYM plates, which were incubated for 8 days. The reverse sides of the plates are shown to illustrate the blue antibiotic Act. (C) Act production in liquid culture. Strains were inoculated into liquid GYM or GYM33 medium and incubated on a rotary shaker (200 rpm) for 8 days. Act production by each strain was measured as the maximum OD640 value. (D) Transcriptional analysis of actII-ORF4 by RT-PCR. Total RNA preparation and RT-PCR were performed as described in Materials and Methods. Eight microliters of each PCR product was loaded for electrophoresis. The gel profile of rRNA (4 μg per lane) is presented as a reference. (E) Transcriptional analysis of actII-ORF4 by real-time qPCR. Total RNA preparation and real-time qPCR were performed as described in Materials and Methods. Each transcriptional assay was normalized to that of hrdB. The error bars indicate the standard deviations of the means of triplicate samples.
Antibiotic production assay.
GYM, R2YE (21), R5 (21), and R3 (31) media were used to screen for the production of the blue antibiotic Act on agar plates by directly assessing the density of the blue color. For Act production in liquid media, culture samples (0.5 ml) were mixed with equal volumes of 2 M KOH, vortexed, and centrifuged at 3,000 × g for 5 min. The Act concentration in the supernatants was determined by measuring the absorbance at 640 nm (optical density at 640 nm [OD640]; ɛ = 25,320 for the pure compound) (21). Red in liquid GYM medium and calcium-dependent antibiotic (CDA) on plates were assayed as described previously (21). For antibiotic production, three flasks were always used for each strain, with production confirmed by at least two separate experiments.
Determination of MICs.
To determine MICs, spore solutions were dotted onto GYM plates containing various concentrations of a drug and incubated at 30°C for 48 h, with the minimum drug concentration able to fully inhibit growth defined as the MIC.
Mutation analysis.
Primers used to amplify the candidate DNA fragments are listed in Table S1 in the supplemental material. PCR amplification was carried out with ExTaq (Takara). Purified PCR products were directly sequenced with BigDye Terminator cycle sequencing kits (Perkin-Elmer Applied Biosystems, Foster City, CA). The sequence data were aligned using the GENETIX program (Software Development Co., Tokyo, Japan).
Disruption of relA.
S. coelicolor A3(2) is characterized by high-frequency conjugation due to the presence of the sex factor plasmid SCP1 (21). To knock out the relA gene in the octuple mutant C8, conjugation was performed using the relA null mutant M570 (5), in which most of the relA gene (corresponding to amino acid residues 167 to 683 of 847 residues) was replaced by a hygromycin resistance gene (hyg). Strains C8 and M570 were mix cultured on a R2YE plate, and the spores that formed after 5 days were spread onto a selection plate containing 5 μg/ml of hygromycin and all eight drugs. The conjugant (C8relA2) thus obtained was assayed by PCR for the absence of the internal segment of relA and the presence of hyg.
Analysis of gene transcription.
Total RNAs were purified from cells grown on GYM plates covered with cellophane for the indicated times using Isogen reagent (Nippon Gene) according to the manufacturer's protocol. After treatment with RNase-free DNase I (amplification grade; Invitrogen), 1 μg of each of the total RNAs was used as a template for reverse transcription (RT) (20 μl) with a ThermScript RT-PCR kit (Invitrogen). Primers used for RT-PCR are listed in Table S1 in the supplemental material. The amount of RT products used as a PCR template and numbers of PCR cycles were optimized for each gene. In a 50-μl PCR mixture, 1 and 2 μl of reverse transcript products were used for actII-ORF4 and hrdB (encoding the major sigma factor), respectively, which were amplified with 26 cycles, and 4 μl was used for relA for 28 cycles.
Real-time quantitative PCR (qPCR) analysis of gene transcription was conducted using the 7300 real-time PCR system and Sybr green PCR master mix (Applied Biosystems) as described previously by Kasai et al. (20), except that random hexamers included in the ThermScript RT-PCR kit (Invitrogen) were used in the RT reaction and except that the annealing temperature of qPCR was 60°C. The transcription of hrdB, a gene encoding the principal sigma factor of RNA polymerase, was used as the internal control. Each transcriptional assay was normalized to the corresponding transcriptional level of hrdB. Primers used for real-time qPCR are listed in Table S1 in the supplemental material.
Preparation of ribosomes and the S-150 fraction.
Ribosomes and the S-150 fraction (i.e., the supernatant following centrifugation at 150,000 × g for 3 h) were prepared from cells harvested at various growth phases in YEME medium (21), as described previously (11), except that cells were first washed twice with a high-salt buffer containing 1 M (instead of 30 mM) ammonium acetate before washing with the standard buffer and except that 2 mM (instead of 1 mM) phenylmethylsulfonyl fluoride was included in all buffers.
In vitro protein synthesis.
Cell-free synthesis of green fluorescent protein (GFP) was performed as described previously (11). In brief, 20 A260 units/ml of ribosome or 0.5 mg/ml of the S-150 fraction was incubated at 30°C for 15 min in a 100-μl total volume of a mixture containing 1.2 mM ATP, 0.8 mM GTP, 0.64 mM cyclic AMP, 15 μg Escherichia coli total tRNAs, and 0.4 mM each of the 20 natural l-amino acids. GFP synthesis was initiated by the addition of 100 μg gfp mRNA, which had been synthesized in vitro from a plasmid gfp gene using T7 RNA polymerase (Takara). Aliquots (10 μl) were withdrawn every 30 min, electrophoresed on native 10% polyacrylamide gels, and subjected to fluorescence analysis using a FluoroImager (Molecular Dynamics).
Assay of ppGpp.
Intracellular ppGpp was extracted from cells cultured on GYM and on modified R5 (16) plates covered with cellophane with 1 M formic acid and assayed by high-performance liquid chromatography (27). For nutritional shift down, exponentially growing cells in chemically defined (CD) medium (27) supplemented with 3% Casamino Acids (vitamin free; Difco) were rapidly transferred into fresh CD medium without Casamino Acids. Cells were harvested 0, 15, 30, and 60 min after cell transfer for the extraction of ppGpp; pppGpp was not detectable in these samples.
RESULTS AND DISCUSSION
Construction of multiply drug-resistant mutants.
Starting with triple mutant strain SGR (14), we screened for Par resistance by spreading SGR spores onto GYM plates containing various concentrations of Par, as illustrated in Fig. 1A. About 20% of the colonies that developed within 7 days due to spontaneous mutations showed increased Act production compared with that of parental strain SGR. Since we previously found that a Par resistance mutation (P91S) in ribosomal protein S12 effectively resulted in Act overproduction (29), we sequenced the S12-encoding gene, rpsL, in SGR and dozens of Act-overproducing colonies. As expected, we found that in addition to the rpsL-encoded mutation K88E present in the SGR mutant, all colonies tested possessed a second amino acid substitution in S12, including the P91S mutation. To our surprise, we found that one colony contained a novel Par resistance mutation consisting of an inserted glycine residue (G, encoded by ggg) at position 92 of S12 (92::G), although it appeared at a relatively low frequency. More interestingly, this new insertion mutation plus K88E rendered cells able to produce even more Act than P91S combined with K88E. The introduction of 92::G (and P91S) (not shown) increased the sensitivities of mutants to many antibiotics, including Geneticin, FA, Lin, and Tet (Table 2). Consequently, a quadruple mutant, SGRP, containing 92::G was used for further selection.
TABLE 2.
Levels of resistance of S. coelicolor A3(2) strains to various drugs
| Strain | MIC (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sm | Gen | Rif | Par | Geneticin | FA | Tsp | Lin | Tet | |
| 1147 | 1 | 0.1 | 10 | 0.1 | 0.5 | 90 | 1 | 50 | 10 |
| K88E | 100 | 0.1 | 30 | 0.3 | 0.8 | 90 | 1 | 50 | 10 |
| SG | 100 | 0.3 | 30 | 0.3 | 0.8 | 90 | 1 | 50 | 10 |
| SGR | 100 | 0.3 | 300 | 0.3 | 0.5 | 70 | 1 | 50 | 10 |
| SGRP | 100 | 0.3 | 300 | 5 | 0.3 | 30 | 1 | 8 | 5 |
| C5 | 100 | 0.4 | 300 | 5 | 0.5 | 30 | 0.5 | 10 | 5 |
| C6 | 100 | 0.4 | 300 | 10 | 0.8 | 70 | 1 | 8 | 5 |
| C7 | 100 | 0.3 | 300 | 10 | 0.3 | 70 | 50 | 8 | 2 |
| C8 | 100 | 0.3 | 300 | 10 | 0.3 | 90 | 50 | 200 | 3 |
The MICs were determined after incubation on GYM plate at 30°C for 48 h.
Subsequent sequential screenings were performed to isolate strains that were resistant to Geneticin, FA, Tsp, and Lin, generating quintuple, sextuple, septuple, and octuple mutants (C5, C6, C7, and C8, respectively) (Table 1). During these screenings, we found that as many as 5% to 15% of the mutant strains produced more Act than the corresponding parental strain. The period required for all screenings was 8 months. In general, as the number of mutations increased, the growth rate slowed (see Fig. 3A), a finding consistent with the opinion that drug resistance was often obtained at the cost of growth fitness (1).
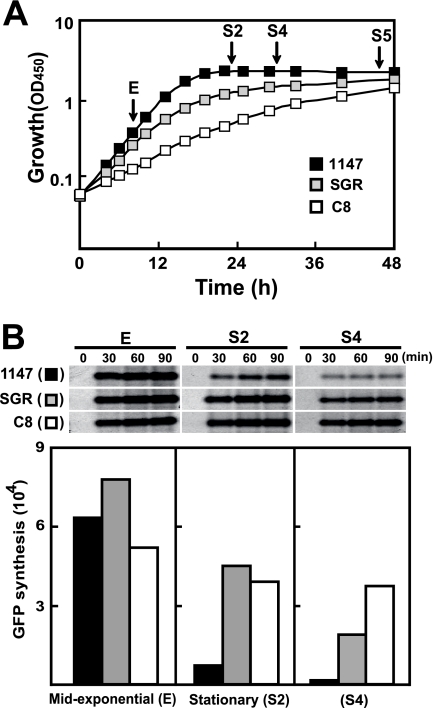
FIG. 3.
Growth patterns and in vitro protein synthesis activities of wild-type strain 1147, triple mutant strain SGR, and octuple mutant strain C8. (A) Growth profiles. Spores were inoculated into liquid YEME medium, with absorbance at 450 nm (OD450) monitored for the growth curve. An OD450 of 0.06 was defined as the zero time point approximately 16 to 24 h after inoculation. Arrows indicate the time points at which cells were harvested for preparation of ribosomes and the S-150 fraction. (B) In vitro GFP synthesis was assayed using ribosomes from 1147 (▪), SGR (□), and C8 (□), with nascent gfp mRNA as the template. Top, fluorographs. Bottom, fluorescence intensities of bands after 90 min of reaction, as determined by scanning the fluorographs.
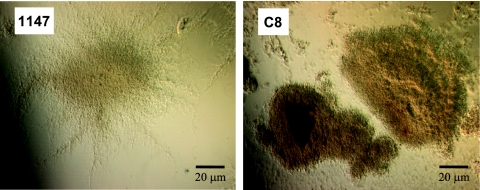
Octuple mutant strain C8 was somewhat temperature sensitive; wild-type strain 1147 grew at 40°C, but C8 did not grow at 39°C. In addition, C8 exhibited extremely slow growth in a chemically defined medium. In liquid culture, mutant strain C8 showed a tight “core” pellet with short-fragmented mycelia, whereas the wild type (1147) possessed a core surrounded by many naturally stretching, branched mycelia (Fig. 2). Importantly, mutant strain C8 was genetically stable; no obvious changes in morphology and Act productivity were found even after 16 sequential inoculations onto GYM agar plates.
FIG. 2.
Morphological appearance of wild-type strain 1147 and octuple mutant strain C8 during exponential growth phase in liquid R2YE medium. Bars represent 20 μm.
Antibiotic overproduction and mutation analysis.
The relationship between Act production and the number of mutations is shown in Fig. 1B and C. We found that C7 produced 0.51 g/liter and that C8 produced 0.55 g/liter Act, 88- and 95-fold higher, respectively, than the 5.8 mg/liter produced by wild-type 1147 in liquid GYM medium. It was noteworthy that low temperatures could further enhance Act production by these multiply drug-resistant mutants. When strains were incubated at 27°C, C7 produced 0.63 g/liter Act, and C8 produced 0.90 g/liter Act, or 24% and 64% higher, respectively, than at 30°C. Increased Act production was more pronounced when mutants were cultivated in a dense medium, GYM33 (GYM medium supplemented with 3% starch and 3% soybean powder). In this medium, C7 and C8 produced 1.63 g/liter and 1.22 g/liter Act, respectively, or 180- and 136-fold more than the 9 mg/liter produced by wild-type strain 1147 (Fig. 1C).
Transcriptional analysis by RT-PCR showed that the expression of the actII-ORF4 gene, encoding the positive regulator of Act biosynthesis (7), was greatly enhanced in drug-resistant mutants, especially in the late phase of mutants (C6 and C8) (Fig. 1D). This was confirmed by real-time qPCR, in which the expression level of actII-ORF4 in the C8 mutant was sevenfold higher than that in wild-type strain 1147 (Fig. 1E). These results indicate that drug resistance mutations exerted their effects on antibiotic production at the transcriptional level. In addition to Act, these multiply drug-resistant mutants showed increased productivity of undecylprodigiosin (red) and CDA (data not shown), although the observed increase (20- to 30-fold) was not as dramatic as that of Act.
Structural studies on the mechanism of action of antibiotics have shown that many of these agents target ribosomal components (35, 36). For example, most FA resistance mutations are clustered in the fusA gene, which encodes elongation factor G (3); Tsp resistance is often due to mutations in or a deletion of the ribosomal protein L11 (27, 30), and Lin resistance is frequently due to mutations in the ribosomal proteins S7, L14, and L15 (17). Octuple mutant strain C8, however, did not have mutations in the fusA gene, the L11-encoding gene rplK, and the genes encoding S7, L14, and L15. Apparently, new types of drug resistance mutations tend to appear in these “unnatural ribosomes” that contain multiple drug resistance mutations.
Mutant ribosomes sustain a high level of protein synthesis even at the late growth phase.
Although some mutations were not identified, it is likely that ribosomal properties and concomitant protein translation were modified largely by drug resistance mutations, because all drugs used in this study, except rifampin, target ribosomal components. We therefore assessed the in vitro protein synthesis activities of the mutants using nascent GFP mRNA as a template. Ribosomal and S-150 fractions were prepared from wild-type 1147, triple mutant strain SGR, and octuple mutant strain C8 cells grown in YEME medium to mid-exponential phase (E), early stationary phase (S2), and late stationary phase (S4) (Fig. 3A). GFP was abundantly synthesized by ribosomes prepared from cells of all strains in the E phase. Ribosomes prepared from wild-type cells in S2 synthesized much less GFP, and ribosomes prepared from wild-type cells in S4 lost their ability to synthesize protein. By contrast, ribosomes prepared from drug-resistant mutant strains SGR and C8 in both S2 and S4 showed high levels of protein synthesis activity (Fig. 3B). Of note, ribosomes prepared from the octuple mutant C8 during exponential and stationary phases maintained relatively constant levels of protein synthesis activity, decreasing only about 25% upon entering stationary phase (Fig. 3B). Furthermore, ribosomes prepared from cells at an extremely late growth phase (S5) (Fig. 3A) had over 60% protein synthesis activity compared with ribosomes from E-phase cells (data not shown). Thus, the ability of late-growth-phase cells to sustain a high level of protein synthesis activity appears to be a key feature of drug-resistant mutants, as we demonstrated previously (11), an activity especially important for the expression of stationary-phase-specific genes, including those for secondary metabolism.
Mutant cells acquire an increased ability to accumulate ppGpp.
Bacterial cells exert “stringent control” over a wide variety of genes and enzymes when they encounter adverse environmental conditions, such as the limited availability of an essential nutrient. ppGpp is a key mediator of this response and is therefore called a bacterial alarmone (4). Experiments using knockout mutants of genes necessary for ppGpp synthesis or forcing ppGpp synthesis under nutrient-sufficient conditions have indicated that ppGpp plays a pivotal role in the onset of antibiotic production in bacteria, including Streptomyces spp. (5, 6a, 9, 26, 27) and B. subtilis (18).
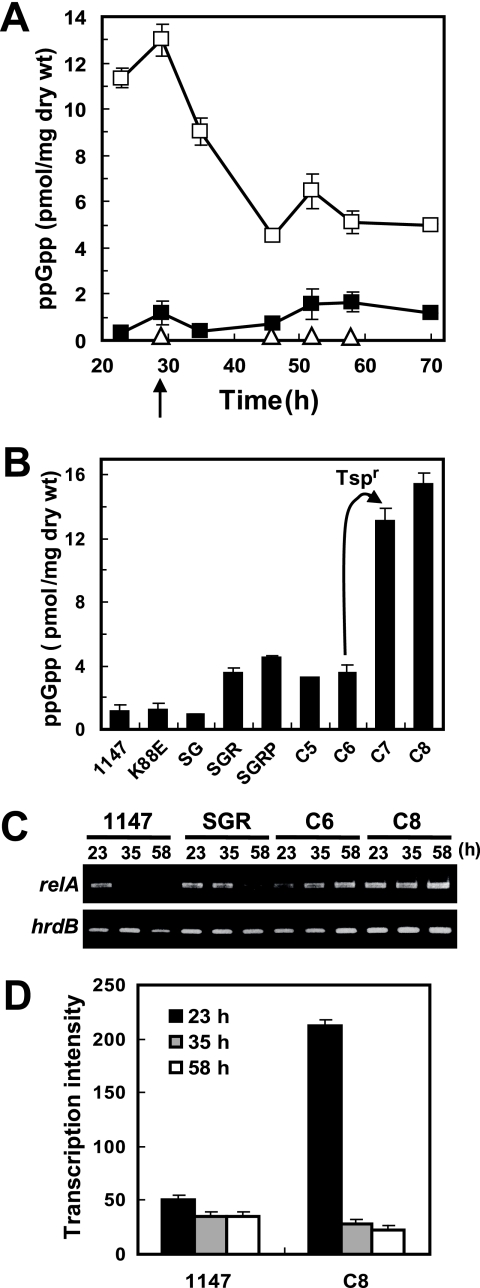
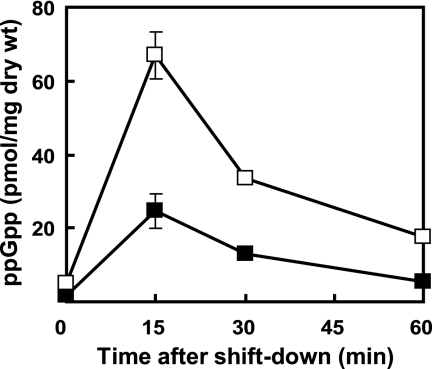
Strikingly, when grown on GYM agar, the octuple mutant C8 had a much higher intracellular level of ppGpp (4.5 to 13 pmol/mg [dry weight]) than wild-type strain 1147 (0.3 to 1.6 pmol/mg [dry weight]) (Fig. 4A). The high level of ppGpp in C8 was especially pronounced during the early growth phase (20 to 30 h), when its ppGpp level was 10- to 30-fold higher than that in the wild type. Similar results were obtained in liquid culture (see Fig. S1 in the supplemental material) and on modified R5 plates (data not shown). The ability to accumulate a high level of ppGpp was conferred mainly upon the introduction of Tsp resistance, as measured by intracellular ppGpp levels of each drug-resistant mutant at 29 h (Fig. 4B). To clearly demonstrate the intrinsic role of mutant ribosomes, the ability of the mutant to synthesize ppGpp was analyzed in shift-down experiments. In both wild-type and mutant strains, ppGpp synthesis was activated immediately after the depletion of amino acids, peaking 15 min after nutritional shift down. As expected, we found that C8 synthesized threefold more ppGpp than did wild-type strain 1147 (Fig. 5), indicating that C8 mutant ribosomes synthesized and accumulated higher levels of ppGpp.
FIG. 4.
Accumulation of intracellular ppGpp and transcriptional analysis of the relA gene in drug-resistant mutants. (A) Intracellular ppGpp levels of wild-type strain 1147, octuple mutant strain C8, and relA deletion mutant strain C8relA2 during growth on GYM plates. Cells were harvested at the indicated time points for ppGpp extraction. ▪, 1147; □, octuple mutant strain C8; ▵, relA deletion mutant strain C8relA2. (B) Intracellular ppGpp levels of each drug-resistant mutant at 29 h (corresponding to the arrow in A). The increase in the ppGpp level upon the introduction of Tsp resistance is indicated by an arrow. (C) Transcriptional analysis of relA in drug-resistant mutants by RT-PCR using the total RNA samples as described in the legend of Fig. 1D. Transcription of hrdB was utilized as an internal control. (D) Transcriptional analysis of relA by real-time qPCR. Total RNA preparation and real-time qPCR were performed as described in Materials and Methods. Each transcriptional assay was normalized to that of hrdB. The error bars indicate the standard deviations of the means of triplicate or more samples.
FIG. 5.
Intracellular ppGpp accumulation after nutritional shift down. After growth to mid-exponential phase in CD medium plus 3% Casamino Acids, cells were rapidly transferred into fresh CD medium without Casamino Acids, and ppGpp was extracted with 1 M formic acid at the indicated time points. The error bars indicate the standard deviations of the means of triplicate samples. ▪, wild-type strain 1147; □, octuple mutant strain C8.
Although the detailed molecular mechanisms underlying these phenomena are at present not known, it is important that the expression of the relA gene, which encodes ppGpp synthetase, was elevated remarkably at the transcriptional level in the mutants, as shown by RT-PCR analysis (Fig. 4C) and as confirmed by real-time qPCR analysis, which displayed a 4.4-fold-elevated level of transcription of the relA gene in mutant strain C8 (Fig. 4D). It was also confirmed that these multiply drug-resistant mutants have no mutations in the relA gene. Several kinds of drug resistance mutations (including rif and fus mutations) were previously reported to disturb ppGpp synthesis (23, 24) but show a reduction in the ppGpp level. Our finding is therefore the first to demonstrate the existence of mutations (especially a tsp mutation) that cause an increase in ppGpp levels.
relA disruption abolishes a mutant's antibiotic overproduction.
To confirm the important role of ppGpp in the dramatic activation of Act production in multiply drug-resistant mutants, the relA gene was knocked out in octuple mutant strain C8 by conjugation with relA null mutant strain M570 (5), which is entirely deficient in ppGpp synthesis. This conjugant, C8relA2, showed a complete inability to accumulate ppGpp throughout all growth phases (Fig. 4A) as well as a >60% decrease in Act production (Fig. 1B and C), indicating the critical role of elevated ppGpp levels in the antibiotic overproduction observed in mutant strain C8. The impaired growth of mutant strain C8 was restored only partially by introducing the relA mutation (data not shown).
Recent analyses of the ppGpp system in plant cells (6, 32, 34) and X-ray structural analysis of the RNA polymerase-ppGpp complex (2) have enhanced our understanding of the physiological significance of the stringent control mediated by ppGpp. Analysis of the effect of ppGpp accumulation on changes in the S. coelicolor A3(2) transcriptome suggested a global role for ppGpp in cellular function (10). The induction of ppGpp synthesis activated the transcription of genes for Act and CDA biosynthesis, a finding in good agreement with the results shown here.
Concluding remarks.
We have developed a more rapid and cost-effective and less labor-intensive method (i.e., ribosome engineering) for the dramatic activation of antibiotic production by constructing multiply drug-resistant mutants. Our results demonstrate that this dramatic overproduction of antibiotics was due to mutant ribosomes with aberrant protein and ppGpp synthesis activities. These findings indicate that ribosomes can be good targets for strain improvement in bacteria, although it is still unclear why ribosomal mutations markedly enhance relA transcription (Fig. 4C and D). We reported similar results for metK (coding for S-adenosylmethionine synthetase), the expression of which was activated more than 30-fold by an rsmG mutation in S. coelicolor, which results in the failure to methylate 16S rRNA at position G518 (25). Clarification of these peculiar phenomena at the molecular level may open new horizons for the study of still-unknown ribosomal functions. Although in the present study, we used mainly three strains (1147, SGR, and C8) as representatives to be characterized in some detail, more convincing evidence should come from a thorough analysis of each successive step in mutagenesis, since ribosomal alterations and altered ppGpp synthesis abilities do not necessarily occur together or occur in each successive mutant strain.
The present findings are in agreement with our proposal (11, 28) that the cell's capacity to synthesize protein at a late growth phase is crucial for accelerating the initiation of onset of secondary metabolism and for the production of abundant biosynthetic enzymes. The principal regulator of Act production in S. coelicolor appears to be the availability of the pathway-specific transcriptional regulatory protein ActII-ORF4, a threshold concentration of which is required for the efficient transcription of its cognate biosynthetic structural genes (7). Although we do not yet know how the drug resistance mutations mediated preferential gene transcription work (Fig. 1D and E), it is conceivable that the expression of pathway-specific regulatory genes (e.g., actII-ORF4 and redD) is governed by higher-order regulatory proteins and that the expression of the latter presumptive regulatory proteins may be significantly affected under conditions associated with enhanced protein synthesis during the stationary phase in the mutants.
Supplementary Material
Acknowledgments
This work was supported by grants to K.O. from the Effective Promotion of Joint Research of Special Coordination Funds (Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government).
The relA null mutant M570 was a generous gift from Mervyn Bibb of the John Innes Institute.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299-310. [DOI] [PubMed] [Google Scholar]
- 3.Besier, S., A. Ludwig, V. Brade, and T. A. Wichelhaus. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 47:463-469. [DOI] [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 5.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Givens, R. M., M. H. Lin, D. J. Taylor, U. Mechold, J. O. Berry, and V. J. Hernandez. 2004. Inducible expression, enzymatic activity, and origin of higher plant homologues of bacterial RelA/SpoT stress proteins in Nicotiana tabacum. J. Biol. Chem. 279:7495-7504. [DOI] [PubMed] [Google Scholar]
- 6a.Gomez-Escribano, J. P., P. Liras, A. Pisabarro, and J. F. Martin. 2006. An rplkΔ29-PALG-32 mutation leads to reduced expression of the regulatory genes ccaR and claR and very low transcription of the ceaS2 gene for clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol. Microbiol. 61:758-770. [DOI] [PubMed] [Google Scholar]
- 7.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 8.Hesketh, A., and K. Ochi. 1997. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsl (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J. Antibiot. (Tokyo) 50:532-535. [DOI] [PubMed] [Google Scholar]
- 9.Hesketh, A., J. Sun, and M. Bibb. 2001. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 39:136-144. [DOI] [PubMed] [Google Scholar]
- 10.Hesketh, A., W. J. Chen, J. Ryding, S. Chang, and M. Bibb. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosaka, T., J. Xu, and K. Ochi. 2006. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol. Microbiol. 61:883-897. [DOI] [PubMed] [Google Scholar]
- 12.Hosokawa, K., N. H. Park, T. Inaoka, Y. Itoh, and K. Ochi. 2002. Streptomycin-resistant (rpsL) or rifampicin-resistant (rpoB) mutation in Pseudomonas putida KH146-2 confers enhanced tolerance to organic chemicals. Environ. Microbiol. 4:703-712. [DOI] [PubMed] [Google Scholar]
- 13.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, H., and K. Ochi. 2001. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 67:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, H., Q. Zhang, and K. Ochi. 2002. Activation of antibiotic biosynthesis by specific mutations in the rpoB gene (encoding the RNA polymerase β subunit) of Streptomyces lividans. J. Bacteriol. 184:3984-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hummel, H., W. Piepersberg, and A. Bock. 1979. Analysis of lincomycin resistance in Escherichia coli. Mol. Gen. Genet. 169:345-347. [DOI] [PubMed] [Google Scholar]
- 18.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 19.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 279:3885-3892. [DOI] [PubMed] [Google Scholar]
- 20.Kasai, K., T. Nishizawa, K. Takahashi, T. Hosaka, H. Aoki, and K. Ochi. 2006. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 188:7111-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 22.Kurosawa, K., T. Hosaka, N. Tamehiro, T. Inaoka, and K. Ochi. 2006. Improvement of α-amylase production by modulation of ribosomal component protein S12 in Bacillus subtilis 168. Appl. Environ. Microbiol. 72:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little, R., J. Ryals, and H. Bremer. 1983. rpoB mutation in Escherichia coli alters control of ribosome synthesis by guanosine tetraphosphate. J. Bacteriol. 154:787-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacVanin, M., U. Johanson, M. Ehrenberg, and D. Hughes. 2000. Fusidic acid-resistant EF-G perturbs the accumulation of ppGpp. Mol. Microbiol. 37:98-107. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura, K., T. Hosaka, S. Tokuyama, S. Okamoto, and K. Ochi. 2007. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 189:3876-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochi, K. 1987. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 169:3608-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochi, K. 1990. A relaxed (rel) mutant of Streptomyces coelicolor A3(2) with a missing ribosomal protein lacks the ability to accumulate ppGpp, A-factor and prodigiosin. J. Gen. Microbiol. 136:2405-2412. [DOI] [PubMed] [Google Scholar]
- 28.Ochi, K., S. Okamoto, Y. Tozawa, T. Inaoka, T. Hosaka, J. Xu, and A. Kurosawa. 2004. Ribosome engineering and secondary metabolite production. Adv. Appl. Microbiol. 56:155-184. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto-Hosoya, Y., T. A. Sato, and K. Ochi. 2000. Resistance to paromomycin is conferred by rpsL mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2). J. Antibiot. (Tokyo) 53:1424-1427. [DOI] [PubMed] [Google Scholar]
- 30.Porse, B. T., I. Leviev, A. S. Mankin, and R. A. Garrett. 1998. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 276:391-404. [DOI] [PubMed] [Google Scholar]
- 31.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, K., K. Kasai, and K. Ochi. 2004. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. USA 101:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamehiro, N., T. Hosaka, J. Xu, H. Hu, N. Otake, and K. Ochi. 2003. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol. 69:6412-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Biezen, E. A., J. Sun, M. J. Coleman, M. J. Bibb, and J. D. Jones. 2000. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. USA 97:3747-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, D. N. 2004. Antibiotics and the inhibition of ribosome function, p. 449-527. In D. N. Wilson and K. H. Niehaus (ed.), Protein synthesis and ribosome structure: translating the genome. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
- 36.Wirmer, J., and E. Westhof. 2006. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 415:180-202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.