Abstract
Members of the rhodophytan order Cyanidiales are unique among phototrophs in their ability to live in extreme environments that combine low pH levels (∼0.2 to 4.0) and moderately high temperatures of 40 to 56°C. These unicellular algae occur in far-flung volcanic areas throughout the earth. Three genera (Cyanidium, Galdieria, and Cyanidioschyzon) are recognized. The phylogenetic diversity of culture isolates of the Cyanidiales from habitats throughout Yellowstone National Park (YNP), three areas in Japan, and seven regions in New Zealand was examined by using the chloroplast RuBisCO large subunit gene (rbcL) and the 18S rRNA gene. Based on the nucleotide sequences of both genes, the YNP isolates fall into two groups, one with high identity to Galdieria sulphuraria (type II) and another that is by far the most common and extensively distributed Yellowstone type (type IA). The latter is a spherical, walled cell that reproduces by internal divisions, with a subsequent release of smaller daughter cells. This type, nevertheless, shows a 99 to 100% identity to Cyanidioschyzon merolae (type IB), which lacks a wall, divides by “fission”-like cytokinesis into two daughter cells, and has less than 5% of the cell volume of type IA. The evolutionary and taxonomic ramifications of this disparity are discussed. Although the 18S rRNA and rbcL genes did not reveal diversity among the numerous isolates of type IA, chloroplast short sequence repeats did show some variation by location within YNP. In contrast, Japanese and New Zealand strains showed considerable diversity when we examined only the sequences of 18S and rbcL genes. Most exhibited identities closer to Galdieria maxima than to other strains, but these identities were commonly as low as 91 to 93%. Some of these Japanese and New Zealand strains probably represent undescribed species that diverged after long-term geographic isolation.
The Cyanidiales are an order of asexual, unicellular red algae that are able to grow in low-pH environments and at moderately high temperatures throughout the globe (4, 31, 32). These algae are not red, but blue-green, due to their predominant pigments, c-phycocyanin and chlorophyll a. Many members of this order are known to grow at temperatures as high as 56°C and at pH levels from 0.2 to 4.0. No other photosynthetic microorganisms are known to inhabit this combination of conditions, and these algae often form well-developed mats in acidic geothermal locations. Surprisingly though, little is known about the ecology, biodiversity, and geographical distribution of these organisms. In these acidic habitats, no prokaryotic phototrophs are known to exist below a pH level of ∼4 and the number of species reaching levels below pH 5 is small (37).
The order Cyanidiales consists of three recognized genera, Cyanidium, Galdieria, and Cyanidioschyzon (6, 13, 16), referred to colloquially as “cyanidia” in this work. The genera Cyanidium and Cyanidioschyzon are thought to include a single species each, Cyanidium caldarium and Cyanidioschyzon merolae, respectively, whereas the genus Galdieria has been classified into four species, Galdieria sulphuraria, Galdieria maxima, Galdieria partita, and Galdieria daedala, based on cell morphology. The last three species have recently been considered to be strains of G. sulphuraria, an opinion based on the similarities of sequences of the gene encoding the large subunit of ribulose bisphosphate carboxylase (rbcL) (7). The three genera have been distinguished morphologically, based on reproductive patterns, chloroplast and mitochondrion shapes, the presence or absence of a vacuole, cell size, and the presence or absence of a cell wall. Strains of the genus Cyanidioschyzon are smaller (usually 1 or 2 μm in breadth), with an oval, club-like shape. They reproduce by binary cytokinesis and lack rigid cell walls. Galdieria and Cyanidium have more spherical shapes, possess rigid cell walls, and reproduce through the formation of 4 to 32 small daughter cells within a mother cell. Chloroplasts vary from a multilobed shape in G. sulphuraria to a spherical shape in Cyanidium. Variation is also seen in mitochondrial shape. Some types may also be distinguished by physiological characteristics. G. sulphuraria has the unique ability to grow heterotrophically on at least 50 different carbon sources, including several sugars, sugar alcohols, amino acids, and trichloroacetic acid cycle intermediates (2, 15, 26). This character has been viewed as a plausible explanation for its ability to grow in darkness or semidarkness in soil, gravel, and as endolithic populations, presumably by using organic compounds released from other microorganisms in the community (10, 15, 26). Studies have also shown that G. maxima also can grow heterotrophically, although poorly (15). Up to this point, only G. sulphuraria lacks the ability to use nitrate as the sole nitrogen source (12).
Thermoacidic environments are scattered disparately throughout the earth, but cluster mainly in volcanic areas that have constant geothermal activity and low pH levels, due primarily to the presence of sulfuric acid as a result of the biological and abiological oxidation of elemental sulfur or hydrogen sulfide. Since the Cyanidiales tested thus far cannot tolerate desiccation, the geographical isolation of thermoacidic sites may have led to allopatric speciation events within this group over time (3, 4, 13). One geothermal region with many acidic habitats is Yellowstone National Park (YNP), an approximately 9,000-km2 area with numerous hydrothermal locations that vary in pH, temperature, exposure to high solar radiation, water availability, and concentrations of soluble metals and metalloids (25). This wide spectrum of physicochemical factors could exert selection pressures, potentially providing opportunities for distinct ecotypes to arise even in island-like features within YNP. Indeed, evidence of YNP population “endemism” or habitat and temperature specialization has previously been found for the thermophilic cyanobacterium Mastigocladus within YNP (22). The presence of the archaean Sulfolobus (25), as well as temperature specialization within YNP for the thermophilic cyanobacterium and other observations, establishes precedence for our efforts summarized herein, where both genetic and morphological criteria were used to examine the diversity of a large number of axenic cyanidial isolates obtained from many disparate sites within YNP and from larger spatial scales that include Japan and New Zealand. The data are consistent with the suggestion that geographical isolation has led to speciation within the Cyanidiales on a global scale and to the evolution of site-specific ecotypes within YNP.
MATERIALS AND METHODS
Sample collection and cultivation.
Samples were collected from various thermoacidic areas within YNP (Wyoming) during four summer field seasons from 2001 to 2005 (Fig. 1) and also from three acidic geothermal areas in Japan in the summer of 2003 and seven New Zealand sites in 2005. The global positioning system locations for nearly all sites are recorded in Table 1. Each isolate's CCMEE (Culture Collection of Microorganisms from Extreme Environments) number, general location of its collection site, type of environment, temperature, pH, designation of type (I to VI), and GenBank accession number are found in Table 2. The samples were kept in the dark after collection and refrigerated when possible until they were brought back to the laboratory. We have determined that cyanidial specimens remain intact at room temperature in the dark for about 2 months without any apparent loss of viability. Clonal cultures of the samples were established after serial dilution to extinction in modified Allen's medium at pH 2.0 [2.64 g liter−1 (NH4)2SO4, 0.54 g/liter KH2PO4, 0.24 g/liter NaCl, 0.5 g/liter MgSO4·7H2O, 0.14 g/liter CaCl2·2H2O, 1.0 ml FeCl3 solution (0.29 g/liter)] (23) and 0.5 ml of trace element solution (5) by using sterile Falcon round-bottom polystyrene tubes with ribbed caps that allow gas exchange (Fisher Scientific, Pittsburgh, PA). Maximum dilution enrichments were plated on the same medium as above at pH 2.5 solidified with 7 g liter−1 Agargel (Sigma-Aldrich, St. Louis, MO). Single colonies were subcultured twice from plates and placed in sterile tubes with liquid medium. Dilution tubes and plates were all incubated at 40 to 42°C under constant illumination of cool-white fluorescent lamps at about 20 to 50 μmol·m−2·s−1. For types that were more difficult to isolate (e.g., morphotypic C. merolae), single cells from a streak of separated cells on Agargel blocks were picked with watchmaker's forceps under a magnification of ×40 to ×60 with an AO 570 dissecting microscope. All cultures are numbered (Table 2) and are in the CCMEE, University of Oregon (http://cultures.uoregon.edu). Slow-growing cultures (under an even lower light intensity) were maintained by transfer in liquid medium once per 6 weeks and were also conserved at −80°C using dense, stationary-phase cultures in original medium.
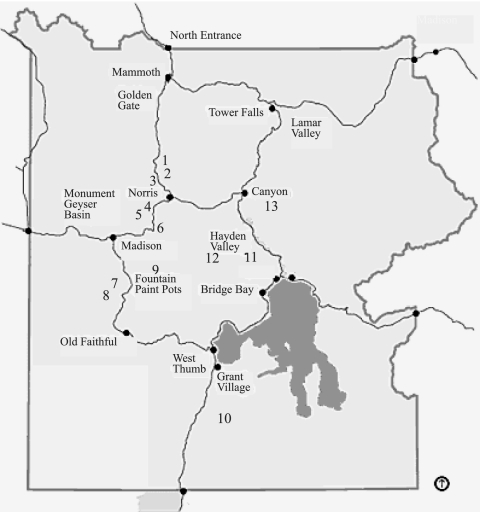
FIG. 1.
Map of YNP, with numbers indicating from which sampling site the majority of cyanidial cultures were isolated. These sites are also indicated by number in Tables 1, 5, and 6. Site numbers and sample sites are as follows: 1, Lemonade Creek; 2, Obsidian Creek; 3, Nymph Creek; 4, Norris Geyser Basin; 5, Sylvan Springs; 6, Artists Paint Pots; 7, River Group; 8, Imperial Spring and Geyser and Fairy Falls Trailhead; 9, Rabbit Creek source; 10, Heart Lake Trail; 11, Mud Volcano; 12, Highland Creek; 13, Sour Creek, Clear Lake area. The distance “as the crow flies” is approximately 65 km from Mammoth to Grant Village.
TABLE 1.
Global positioning system coordinates for all sampling sites in Yellowstone National Park, Japan, and New Zealand
| Sampling sitea | Coordinates |
|---|---|
| Star Plunge, Thermopolis, WY | 43°39′06"N, 108°12′09"W |
| Heart Lake Trailhead (10) | 44°18′32"N, 110°31′58"W |
| Fairy Falls Trailhead (8) | 44°30′5"N, 110°49′54"W |
| Rabbit Creek Source (9) | 44°31′17"N, 110°48′39"W |
| Imperial Spring (8) | 44°31′55"N, 110°53′17"W |
| River Group (7) | 44°33′32"N, 110°50′02"W |
| Highland Creek (12) | 44°36′55"N, 110°36′52"W |
| Mud Volcano (11) | 44°37′24"N, 110°26′01"W |
| Artists Paint Pots (6) | 44°41′35"N, 110°44′18"W |
| Sylvan Springs/Sylvan Crust (5) | 44°41′59"N, 110°46′01"W |
| Sour Creek (13) | 44°42′39"N, 110°28′45"W |
| Crystal Lake (13) | 44°42′39"N, 110°28′45"W |
| Cyanidium Creek, Norris Basin (4) | 44°43′16"N, 110°42′28"W |
| Dragon Spring, Norris Basin (4) | 44°43′54"N, 110°42′41"W |
| “Extreme site,” Norris Basin (4) | 44°44′02"N, 110°42′39"W |
| Nymph Creek (3) | 44°44′59"N, 110°43′40"W |
| Obsidian Creek (2) | 44°46′53"N, 110°44′28"W |
| Lemonade Creek (1) | 44°47′59"N, 110°43′43"W |
| Owakudani, Japan | 35°10′60"N, 139°1′60"E |
| Kusatsu, Japan | 36°37′21.5"N, 138°35′47"E |
| Nakabusa, Japan | 36°24′N, 137°45′E |
| White Island, NZ | 37°31′0"S, 177°4′0"E |
| Waiotapu, NZ | 38°21′0"S, 176°22′0"E |
| Whaka, NZ | 38°10′S, 176°15′E |
| Rotorua, NZ | 38°07′S, 176°15′E |
| Waimangu, NZ | 38°16′60"S, 176°22′60"E |
| Craters of the Moon, NZ | 38°39′32"S, E176°03′55"E |
Numbers in parentheses correspond to the site locations on the map in Fig. 1.
TABLE 2.
Identification, location, location code, environment, temperature, pH, and GenBank accession number of each strain used in this study
| Strain IDa | Location | Location code | Environment | Temp (°C) | pH | Type | Accession no.b |
|---|---|---|---|---|---|---|---|
| 5562 | Artists Paint Pots, YNP | APP | Acid steam hole | 44 | 3.9 | IA | EF6750891, EF6751173 |
| 5570 | Crystal Lake, YNP | CRY | Steam | Unknown | Unknown | IA | EF6751143 |
| 5564 | Cyanidium Creek, YNP | CC | Acid stream | 56 | 3.3 | IA | EF6750991 |
| 5566 | Cyanidium Creek, YNP | CC | Acid stream | 56 | 3.3 | IA | EF6751083 |
| 5508 | Dragon Spring, Norris Basin, YNP | DRG | Acid stream | 50 | 1.9 | IA | EF6750851, EF6751352 |
| 5639 | Extreme site B, Norris Basin, YNP | EXTB | Acid soil | 55 | 0.5 | IA | EF6751272 |
| 5640 | Extreme site B, Norris Basin, YNP | EXTB | Acid soil | 55 | 0.5 | IA | EF6751372 |
| 5506 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | IA | EF6750871, EF6751462 |
| 5507 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | IA | EF6751602 |
| 5511 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | II | EF6751742 |
| 5572 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | II | EF6750731, EF6751822 |
| 5573 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | II | EF6750921, EF6751712 |
| 5574 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | II | EF6750981 |
| 5631 | Extreme site, Norris Basin, YNP | EXT | Acid soil | 55 | 1 | IA | EF6751402 |
| 5633 | Fairy Falls Trailhead, YNP | FF | Steam | Unknown | Unknown | IA | EF6751193 |
| 5636 | Fairy Falls Trailhead, YNP | FF | Steam | Unknown | Unknown | IA | EF6751123 |
| 5651 | Heart Lake Trail, YNP | HRT | Acid soil | Unknown | Unknown | IA | EF6750771, EF671213 |
| 5652 | Heart Lake Trail, YNP | HRT | Acid soil | Unknown | Unknown | IA | EF6750801, EF6751223 |
| 5654 | Heart Lake Trail, YNP | HRT | Acid soil | Unknown | Unknown | IA | EF6750711 |
| 5625 | Highland Creek, YNP | HLD | Acid stream | 46 | 2.5-3 | IA | EF6750781, EF6751582 |
| 5624 | Imperial Spring, YNP | IMP | Steam | Unknown | Unknown | IA | EF6750821 |
| 5576 | Lemonade Creek, YNP | LEM | Acid stream | 54 | 2.2 | IA | EF6750841, EF6751302 |
| 5578 | Lemonade Creek, YNP | LEM | Acid stream | 54 | 2.2 | IA | EF6751203 |
| 5620 | UTEX2393 | Unknown | Unknown | II | EF6750901, EF6751702 | ||
| 5580 | Mud Volcano, YNP | MV | Acid stream | 43 | 2.2 | IA | EF6750881, EF6751163 |
| 5584 | Nymph Creek, YNP | NYM | Acid stream | 42 | 3 | IA | EF6751001, EF6751113, EF6751642 |
| 5585 | Nymph Creek, YNP | NYM | Acid stream | 42 | 3 | IA | EF6750791, EF6751522 |
| 5587.1 | Nymph Creek, YNP | NYM | Acid stream | 42 | 3 | II | tbd1,2 |
| 5593 | Obsidian Creek, YNP | OBS | Acid stream | 47 | 1.9 | IA | EF6751242 |
| 5647 | Obsidian Creek, YNP | OBS | Acid stream | 47 | 1.9 | IA | EF6751073 |
| 5602 | Rabbit Creek Source, YNP | RC | Acid pool edge | 52 | <4 | IA | EF6750861, EF6751153 |
| 5605 | River Group, YNP | RG | Acid pool edge | 52 | 3.9 | IA | EF6750761 |
| 5614 | Sour Creek, YNP | SOUR | Acid stream | 41 | 1.9 | IA | EF6750811, EF6751103 |
| 5615 | Sour Creek, YNP | SOUR | Acid stream | 41 | 1.9 | IA | EF6751133 |
| 5617 | Star Plunge, Thermopolis, WY | STAR | Steam | Unknown | Unknown | IA | EF6750751 |
| 5610 | Sylvan Crust, YNP | SYLCR | Acid crust | 40 | 4 | IB | EF6750831, EF6751093, EF6751252 |
| 5611 | Sylvan Crust, YNP | SYLCR | Acid crust | 40 | 4 | IA | EF6750741 |
| 5612 | Sylvan Crust, YNP | SYLCR | Acid crust | 40 | 4 | IA | EF6751183 |
| 5609 | Sylvan Springs, YNP | SYL | Acid stream | 48 | 2.2 | IA | EF6751233, EF6751442 |
| 5664 | Kusatsu, Japan | KUS | Acid stream | 45 | 1.9 | IIIB | EF6751452 |
| 5665 | Kusatsu, Japan | KUS | Acid pool | 49 | 2 | IIIA | EF6751292 |
| 5667 | Kusatsu, Japan | KUS | Acid stream | 45 | 1.9 | IIIB | EF6751512 |
| 5676 | Kusatsu, Japan | KUS | Acid pool | 49 | 2 | IIIB | EF6751432 |
| 5677 | Kusatsu, Japan | KUS | Acid stream | 45 | 1.9 | IIIA | EF6751322 |
| 5678 | Kusatsu, Japan | KUS | Acid stream | 45 | 1.9 | IIIB | EF6750961, EF6751682 |
| 5679 | Kusatsu, Japan | KUS | Acid stream | 41 | 2 | IIIB | EF6751632 |
| 5680 | Kusatsu, Japan | KUS | Acid pool | 40 | 1.9 | IIIB | EF6750721, EF6751672 |
| 5681 | Kusatsu, Japan | KUS | Acid stream | 41 | 2 | IIIB | EF6751572 |
| 5660 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6751562 |
| 5661 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6751502 |
| 5662 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6751542 |
| 5663 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6751592 |
| 5674 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6751532 |
| 5675 | Nakabusa, Japan | NAK | Steam | Unknown | Unknown | IIIB | EF6750941, EF6751552 |
| 5657 | Owakudani, Japan | OWA | Acid pool edge | >45 | 2.5 | IIIA | EF6750911, EF6751392 |
| 5658 | Owakudani, Japan | OWA | Acid pool edge | >45 | 2.5 | IIIB | EF6751622 |
| 5659 | Owakudani, Japan | OWA | Acid pool edge | >45 | 2.5 | IIIA | EF6750971, EF6751382 |
| 5668 | Owakudani, Japan | OWA | Acid stream | 30 | 2.5 | IIIA | EF6751362 |
| 5669 | Owakudani, Japan | OWA | Acid stream | 30 | 2.5 | IIIA | EF6751262 |
| 5670 | Owakudani, Japan | OWA | Acid stream | 30 | 2.5 | IIIA | EF6751482 |
| 5671 | Owakudani, Japan | OWA | Acid stream | 30 | 2.5 | IIIA | EF6751332 |
| 5672 | Owakudani, Japan | OWA | Acid pool edge | 42-55 | 3 | IIIA | EF6750951, EF6751312 |
| 5673 | Owakudani, Japan | OWA | Acid pool edge | 42-55 | 3 | IIIB | EF6751412 |
| 5706 | Craters of the Moon, New Zealand | CR | Thermal soil | Unknown | Unknown | IV | EF6751061, EF6751772 |
| 5712 | Craters of the Moon, New Zealand | CR | Acid steam hole | Unknown | Unknown | IV | EF6751782 |
| 5716 | Craters of the Moon, New Zealand | CR | Acid steam hole | Unknown | Unknown | V | EF6751041, EF6751422 |
| 5717 | Rotorua, New Zealand | RT | Acid crust | 32 | 3-3.5 | IV | EF6751762 |
| 5705 | Rotowhero, New Zealand | ROTO | Acid lake | 32 | 3-3.5 | V | EF6751662 |
| 5702 | Waimangu, New Zealand | WM | Steam | Unknown | Unknown | V | EF6751692 |
| 5703 | Waimangu, New Zealand | WM | Steam | Unknown | Unknown | V | EF6751652 |
| 5704 | Waimangu, New Zealand | WM | Steam | Unknown | Unknown | V | EF6751031, EF6751282 |
| 5714 | Waimangu, New Zealand | WMC | Acid crust | 44 | Unknown | IV | EF6751802 |
| 5707 | Waiotopu, New Zealand | WTP | Acid steam | 31 | 2.25 | IV | EF6751812 |
| 5713 | Waiotopu, New Zealand | WTP | Acid lake | 31 | 2.25 | IV | EF6751472 |
| 5719 | Waiotopu, New Zealand | WTP | Steam | Unknown | Unknown | IV | EF6751752 |
| 5708 | Whaka, New Zealand | WKA | Acid stream | 30 | 4 | IV | EF6751722 |
| 5709 | Whaka, New Zealand | WKA | Acid stream | 30 | 4 | V | EF6751011, EF6751612 |
| 5715 | Whaka, New Zealand | WKA | Acid stream | 30 | 4 | V | EF6751492 |
| 5718 | Whaka, New Zealand | WKA | Steam | Unknown | Unknown | IV | EF6751792 |
| 5710 | White Island, New Zealand | WI | Acid stream | 40 | 1-1.5 | IV | EF6751832 |
| 5720 | White Island, New Zealand | WI | Acid stream | 45 | 2.5-3 | IV | EF6751021, EF6751342 |
| 5711 | White Island, New Zealand | WI | Acid stream | Unknown | Unknown | V | EF6751051, EF6751732 |
ID, identification number.
Numbers in superscripts refer to the gene to which each accession number relates. 1, 18S rDNA; 2, rbcL; 3, SSR9.
DNA extractions.
DNA was extracted from culture isolates via modifications of the method of Moré et al. (24). Algal cells were combined with 0.1-mm-diameter silica/zirconium beads (BioSpec Products, Bartlesville, OK), 600 μl of 20 mM Na-phosphate buffer (pH 8.0), and 300 μl of a lysis solution consisting of 10% sodium dodecyl sulfate, 0.5 M Tris-HCl (pH 8.0), and 0.1 M NaCl and then disrupted in a Bio101 bead beater (Savant, Farmingdale, NY) for 3 min on high speed. The tubes were then centrifuged at 12,000 × g, the supernatant was removed, and then 7.5 M ammonium acetate (2:5 ratio of ammonium acetate to supernatant) was added to the supernatant. The tubes were centrifuged at 12,000 × g, the supernatant was removed, and isopropanol was added to the supernatant to precipitate the DNA at room temperature. The tubes were centrifuged for 60 min at 4°C at 12,000 × g, and the resultant nucleic acid pellet was washed with −20°C 70% ethanol and centrifuged again for 5 min. The pellet was air dried and resuspended in 100 μl nuclease-free water (Promega, Madison, WI).
PCR amplification.
All primers for PCR studies are listed in Table 3. The 18S rRNA gene was amplified from 37 culture isolates from YNP, New Zealand, and Japan. The PCR mixture contained 50 ng of template DNA, 200 μM deoxynucleoside triphosphates (dNTPs), 1× PCR buffer, 2.5 mM MgCl2, 25 pmol primers CdmF and CdmR, and 5 U of Taq polymerase (Promega, Madison, WI). Cycling conditions were 95°C for 5 min, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 74°C for 1 min, and a final extension at 74°C for 10 min. PCR amplification of the rbcL gene was performed on 73 isolates. The reaction mixture contained 50 ng DNA, 200 μM dNTPs, 1× PCR buffer, 2.5 mM MgCl2, 25 pmol rbcL-1F and rbcL-1R, and 2.5 U Taq polymerase (Promega, Madison, WI). PCR products for both rbcL and 18S ribosomal DNA (rDNA) were sequenced bidirectionally by Macrogen, Inc. (Seoul, South Korea).
TABLE 3.
Primers used in this study
| Primer | Primer sequence | Target | Reference or source |
|---|---|---|---|
| CdmF | GTCAGAGGTGAAATTCTTGGATTTA | 18S rRNA | 13 |
| CdmR | AAGGGCAGGGACGTAATCAACG | 18S rRNA | 13 |
| RbcL1F | AACCTTTCATGCGTTGGAGAGA | rbcL | This study |
| RbcL1R | CCTGCATGAATACCACCAGAAGC | rbcL | This study |
| SSR9F | CTAACTGAATCCTATCTTGCTC | Chloroplast | 17 |
| SSR9R | GTGCAATTCTCTATTTGATGC | Chloroplast | 17 |
PCR amplification of a chloroplast short sequence repeat region (cpSSR) for select type I isolates was performed using primers SSR9F and SSR9R (positions 65600 to 67092 of the chloroplast genome of Cyanidioschyzon merolae strain 10D). Cycling conditions were a hot start at 96°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and a final extension for 10 min at 72°C. The reaction mixture contained 50 ng of template DNA, 200 μM dNTPs, 1× Q solution (Qiagen, Valencia, CA), 10× PCR buffer, 2.5 mM MgCl2, 25 pmol of each primer, and 5 U of Taq polymerase (Promega, Madison, WI). PCR products for 17 isolates were sequenced bidirectionally by Macrogen, Inc. (Seoul, South Korea). Due to the inability of the SSR primers to amplify YNP type II isolates, all YNP isolates were screened by using the SSR primers to determine to which type they belong. The major tree topologies were the same for each analysis.
Phylogenetic analyses.
All sequences for the 18S rDNA (487 nucleotides), rbcL (516 nucleotides), and cpSSR clones were edited using Sequencher (Gene Codes Corporation, Ann Arbor, MI) and aligned using ClustalX (www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html). Phylogenetic analysis for the rbcL, 18S rDNA, and cpSSR data sets was carried out using maximum parsimony methods with PAUP (http://paup.csit.fsu.edu/about.html). For maximum parsimony methods, 1,000 bootstrap replicates were included in a heuristic search, starting with a random tree and the tree bisection-reconnection branch-swapping algorithm. Percent variation calculations were made by comparing all isolates to the nearest BLAST relative. For Bayesian analysis (MrBayes, version 3.0b4), Metropolis-coupled Markov chain Monte Carlo analysis was performed with a random starting tree and four chains, of which three were hot and one was cold. Trees were sampled every 100 generations for 500,000 total generations by using a general time-reversible model and a gamma distribution.
Accession numbers for the 18S rDNA genes of previously sequenced rhodophytes used here for phylogenetic comparisons are AB090849 for G. sulphuraria L1B, AB090837 for G. sulphuraria A3D, AF441363 for G. sulphuraria ISG, AB090835 for G. sulphuraria FF1B, AF441359 for G. sulphuraria RT-27, AF441371 for G. sulphuraria MSh, AF441360 for G. sulphuraria AZ, AB091229 for G. sulphuraria 107.79, AF441361 for G. sulphuraria 136, AF441375 for G. sulphuraria ELS, AB090829 for G. sulphuraria FF3A, AB090851 for G. sulphuraria 86C, AF441373 for G. sulphuraria J, AF441365 for G. sulphuraria 21.92, AB090840 for G. sulphuraria 1C, AB090832 for G. maxima IPPAS P507, AB090844 for G. partita J5D, AF441372 for G. partita K, AB090839 for G. daedala IPPAS P508, AF441374 for C. caldarium 182, AF441366 for Cyanidium sp., AB158485 for C. merolae 10D, AF441376 for C. merolae 199, AB090833 for C. merolae 61D, AF168623 for Porphyridium aerugineum, and D88387 for Bangia atropurpurea.
Accession numbers for the rbcL gene sequences used for phylogenetic comparisons are AY541305 for G. sulphuraria DBV015NAFG, AY119768 for G. sulphuraria DBV009VTNE, AY541307 for G. sulphuraria DBV021MEVU, AY119769 for G. sulphuraria DBV063AGCS, AY119767 for G. sulphuraria SAG108.79, AY541302 for G. daedala IPPAS P508, AB018008 for G. partita, AY391370 for G. maxima IPPAS P507, AY119766 for C. merolae DBV001NAPS, AY541296 for C. merolae DBV202NAMN, AY541299 for C. merolae DBV182APAS, AY541298 for C. merolae DBV182JAVA, D63675 for C. merolae 10D, AY541297 for C. caldarium DBV019SIPE, AY541301 for Cyanidium sp. Monte Rotaro 20, AY541300 for Cyanidium sp. Monte Rotaro 19, AY119771 for Bangia fuscopurpurea, and AY119772 for Bangiopsis subsimplex. All accession numbers for the isolates used in the present study are listed in Table 2.
RESULTS
Phylogenetic analysis of rbcL and 18S rDNA.
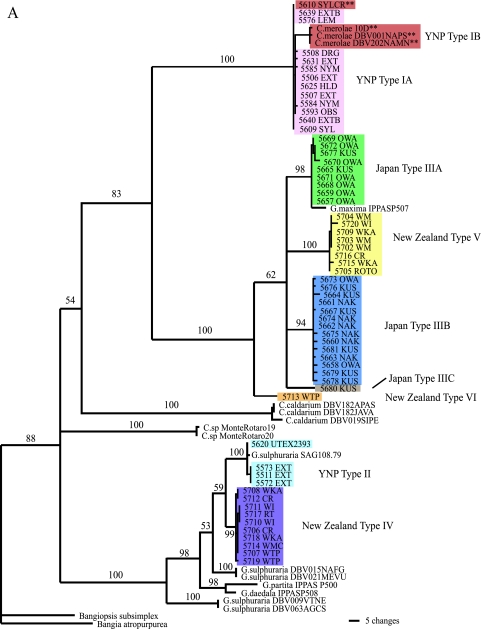
Phylogenetic analysis of 18 YNP chloroplast rbcL sequences resolved two major groups of Cyanidiales with strong support ( Fig. 2A). Members of the largest group of isolates (referred to here as type IA) were about 99% identical at the rbcL locus to the corresponding site in C. merolae strain 10D (originally from an Italian site), with little within-group variation (Table 4). Using 18S rDNA sequences, we found that the type IA YNP isolates were also about 99% identical to C. merolae strain 10D (Table 4 and Fig. 2B). In both analyses, the YNP type IA isolates clearly grouped with C. merolae strain 10D and were quite distinct from G. maxima, G. sulphuraria, and C. caldarium.
FIG. 2.
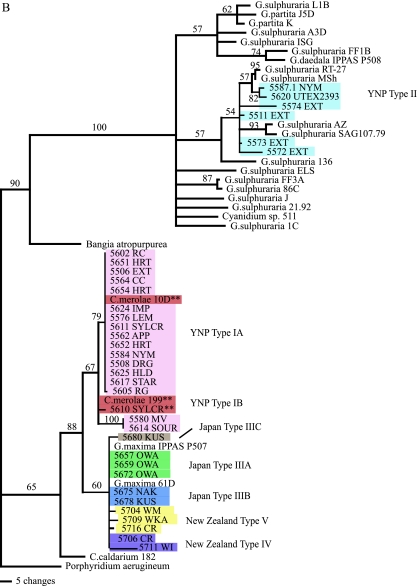
Maximum parsimony phylogenetic analysis of the cyanidial cultures described in this study. (A) Tree based on rbcL gene sequences using PAUP. The double asterisks represent those cultures with morphologies similar to that of C. merolae. Colors distinguish the various types. The reference strains (e.g., Cyanidium sp. Monte Rotaro 19 and Monte Rotaro 20 and most of the G. sulphuraria clusters [DVB 015, 021, 009, and 063]) may be found in reference 6. (B) Tree based on 18S rDNA gene sequences using PAUP. For both trees, branch lengths are proportional to the number of changes on a given branch, and bootstraps are given for each node above 50%. The double asterisks represent those cultures with morphologies similar to that of C. merolae. Colors of the various cultures correspond to the types indicated in panel A.
TABLE 4.
Nearest BLAST relative for each type of isolate from YNP, Japan, and New Zealand for both rbcL and 18S rDNAa
| Isolate source | Type | Nearest BLAST relativea
|
% Variation within groupb | |
|---|---|---|---|---|
| rbcL | 18S rDNA | |||
| YNP | IA/IB | C. merolae 10D (99) | C. merolae 10D (99) | 0.38/1.9 |
| II | G. sulphuraria UTEX 2393 (99) | G. sulphuraria SAG 107.79 (98) | 0.38/4 | |
| Japan | IIIA | G. maxima IPPAS507 (99) | G. maxima IPPAS P507 (100) | 1/0 |
| IIIB | G. maxima IPPAS507 (96) | G. maxima IPPAS P507 (100) | 1/0 | |
| IIIC | G. maxima IPPAS507 (93) | G. maxima IPPAS P507 (99) | 0 | |
| New Zealand | IV | G. sulphuraria SAG 108.79 (96) | G. maxima IPPAS P507 (99) | 0.8/1 |
| V | G. maxima IPPAS P507 (93) | G. maxima IPPAS P507 (99) | 1/1 | |
| VI | G. maxima IPPAS P507 (91) | G. maxima IPPAS P507 (99) | 0 | |
The numbers in the parentheses indicate the percent identity to the nearest GenBank relative.
Results are shown as percent variation for rbcL/percent variation for 18S rDNA.
Four sequenced isolates from YNP were referred to as type II and were closely related to G. sulphuraria based on both the rbcL and 18S rDNA analyses (Fig. 2A and B). These cultures share 99% sequence identity at the rbcL locus to a culture of G. sulphuraria (UTEX 2393) isolated from California and 98% sequence identity at the 18S level to G. sulphuraria (culture SAG 107.79) (Table 4). As with the type IA strains, there was little variation among the rbcL alleles of the YNP type II strains (<0.38% divergence), although variation among the 18S rRNA genes was higher at 4% (Table 4).
An analysis of the rbcL sequences also showed two major clades among the isolates from Japan, with their closest identities to G. maxima IPPAS P507 (Fig. 2A), which was originally isolated from Kunashir Island of the Kurile Group of Russia near the northeastern coast of Japan. Clade type IIIA cultures were isolated exclusively from Owakudani, a volcanic area in Hakone National Park, Japan, and showed 99% rbcL sequence identity to the G. maxima IPPAS P507 strain, with little intragroup variation (Table 4). The second clade (type IIIB), with only 96% identity to G. maxima IPPAS P507, was made up of isolates from the Kusatsu and Nakabusa hot spring sites in Japan (Table 4). We also found a single isolate from Kusatsu (CCMEE 5680) sharing only 93% identity to G. maxima IPPAS P507 (Fig. 2A). In contrast to the differences found among the rbcL sequences and their geographically distinct locations, the 18S rDNA sequences varied little among all of the Japanese isolates; in a comparison of 18S rRNA sequences, all 24 Japanese strains were nearly 99% identical to the G. maxima strain IPPAS P507 (Fig. 2B).
The New Zealand strains fell into two main groups based on rbcL sequences. Members of the first group, type IV (Fig. 2A and Table 4), were 96% identical to G. sulphuraria SAG 108.79, whereas the closest database match for the type V isolates was G. maxima IPPAS P507 (Fig. 2A and Table 4), albeit at only 93% identity. Intragroup variation ranged from 0.8% to 1.0% (Table 4). There was also a single isolate, referred to as type VI, that was only 91% identical to G. maxima IPPAS P507, its closest match (Fig. 2A). By sequencing the 18S rDNA for a few selected New Zealand isolates, we established that they were all 99% identical to G. maxima strains IPPAS P507 and 61D and did not show the variation revealed by rbcL sequences (Fig. 2B). The 18S sequence is more highly conserved than that of the rbcL gene.
Morphology.
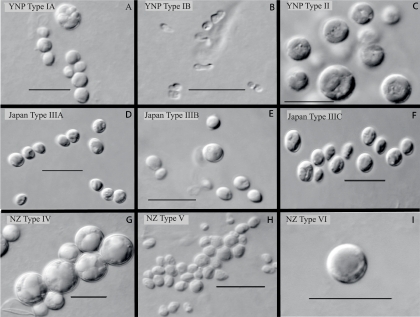
The isolates varied in cell morphology. In some cases, the morphological characteristics corresponded to separate clades identified in the rbcL-based phylogenetic analysis. For example, the G. maxima-like Japanese type IIIA isolates and the sister clades comprising the New Zealand type V and Japanese type IIIB isolates (Fig. 2A) were fairly similar in shape and size at maturity (Fig. 3D, E, and H). Likewise, the G. sulphuraria-like YNP type II and the G. sulphuraria-like New Zealand type IV isolates were also similar in both characteristics (Fig. 3C and G). In a similar fashion, different rbcL-based clades also reflected clear differences in cell size. For example, compare the G. sulphuraria-like New Zealand type IV with the New Zealand type V (Fig. 2A and 3G and H). As another example, the Japanese type IIIC cell was somewhat oval (Fig. 3F), distinguishing it from all other organisms isolated in this study, and this concurs with the fact that its rbcL identity match with any strain in the database was poor (Table 4). It should be remembered, however, that Galdieria-like cells grow to a somewhat indefinite size (depending on culture conditions) before giving rise (by an internal cell division) to 4 to 32 small daughter cells that then grow to a larger size. Therefore, the selected morphotypes (as shown in Fig. 3) cannot be totally relied on to show stable characteristics of each type.
FIG. 3.
The photomicrographs are selected examples of cultures of the various types included in Fig. 1A. Only type 1B illustrates the morphology of a typical C. merolae cell. The spherical cells in different images illustrate the typical morphology of Galdieria. However, the images of Japan types IIIB and IIIC and New Zealand (NZ) type V indicate cells that are somewhat oblong. The type of division for these three strains remains undetermined. CCMEE culture numbers for the types indicated in the panel are as follows: 5508 (A), 5610 (B), 5573 (C), 5665 (D), 5664 (E), 5680 (F), 5706 (G), 5704 (H), and 5713 (I).
The most interesting deviation from sequence and morphology match is the lack of morphological agreement among the YNP type IA and IB isolates. All of the YNP type IA isolates were clearly identified as being C. merolae by both 18S rDNA and rbcL criteria (Fig. 2A and B), but the isolates did not share the distinctive morphology of C. merolae, which is considerably smaller, lacks a rigid wall, and is pear shaped, as shown in Fig. 3B. Instead, the type IA isolates are spherical with a rigid wall and divide internally into four to eight walled daughter cells within the mother cell. This type IA matched the cell morphology and size of Galdieria rather than those of Cyanidioschyzon (Fig. 3, compare panels A and B). These cultures reproduced with absolute fidelity to the Galdieria morphotype, with no sign of Cyanidioschyzon-like cells. The few isolates that did possess the currently accepted morphology of C. merolae (type IB, e.g., culture CCMEE 5610 4A) also reproduced with complete fidelity to the C. merolae morphotype. In other work, we further checked the possibility that Galdieria morphotypes were contaminated with a few C. merolae type cells that might more readily rupture and release their DNA because of the lack of a cell wall, leading to the preferential amplification of their DNA. A culture of a Galdieria sulphuraria type II from YNP (CCMEE 5572), possessing a rigid cell wall, was mixed in various proportions with a “true” Cyanidioschyzon merolae morphotype (CCMEE 5610 4a). Clone libraries (18S rDNA) of the graded series of dual mixtures indicated that the Galdieria morphotype amplified as readily as did the C. merolae morphotype (D. J. Skorupa, R. W. Castenholz, and T. R. McDermott, unpublished data). In another check, 12 new cultures of CCMEE 5508 (a Galdieria morphotype type IA from YNP) were grown from single cells isolated manually under a dissecting scope. All 18S sequences were identical to that of C. merolae 10D from Italy (D. J. Skorupa, R. W. Castenholz, and T. R. McDermott, unpublished data).
cpSSR typing.
Recently, we found that cpSSRs (or microsatellites) could be used to identify genetic differences among six cultured Galdieria-like type IA morphotypes that shared identical 18S rRNA gene sequences with C. merolae (17). Therefore, in the current study, we employed an approach similar to the above method to more closely examine these strains for evidence of genotypic variation. Variation in cpSSR repeat number translated into different amplicon lengths that could easily be distinguished by agarose gel electrophoresis, resulting in a classification of type IA isolates into five major variants based on the number of repeats found in one cpSSR, previously referred to as cpSSR9 (17) (Table 5). Variant 1 of cpSSR9 contained 2.4 repeats of a 136-bp repeat unit sequence, variant 2 contained 3.4 repeats, variant 3 contained 4.4 repeats, and variant 4 had 5.4 repeats. The DNA from some type IA isolates did not amplify with the SSR9 primer set (but would amplify 18S rDNA and rbcL, as described above), and so for the purposes of comparison here, the isolates are referred to variant 5 (Table 5). When summarized as a function of distribution within YNP, cpSSR9 variant 2 isolates were found at all sampled sites except at the Imperial Spring steam vent (Fig. 1 and Table 6). SSR9 variants 3 and 5 were both isolated from nearly half of the sites, although they occurred together in only one, that being in Artists Paint Pots (Fig. 1 and Table 6).
TABLE 5.
Selected strains from YNP types IA and IB categorized into SSR variants and their locations in YNP
| Culture no. or strain | Locationa | SSR variant | No. of repeats | % Similarity of repeats |
|---|---|---|---|---|
| 5651 | Heart Lake10 | 1 | 2.4 | 92 |
| 5652 | Heart Lake10 | 1 | 2.4 | 92 |
| C. merolae 10D | Naples, Italy | 2 | 3.4 | 90 |
| 5566 | Cyanidium Creek4 | 2 | 3.5 | 89 |
| 5570 | Crystal Lake4 | 2 | 3.4 | 84 |
| 5584 | Nymph Creek3 | 2 | 3.5 | 89 |
| 5610 | Sylvan Crust5 | 2 | 3.4 | 89 |
| 5614 | Sour Creek13 | 2 | 3.4 | 84 |
| 5636 | Fairy Falls Trail8 | 2 | 3.4 | 85 |
| 5647 | Obsidian Creek2 | 2 | 3.4 | 84 |
| 5562 | Artists Paint Pots6 | 3 | 4.4 | 87 |
| 5578 | Lemonade Creek1 | 3 | 4.4 | 87 |
| 5580 | Mud Volcano11 | 3 | 4.4 | 94 |
| 5602 | Rabbit Creek9 | 3 | 4.4 | 94 |
| 5612 | Sylvan Crust5 | 3 | 4.4 | 88 |
| 5615 | Sour Creek13 | 3 | 4.4 | 83 |
| 5633 | Fairy Falls Trail8 | 3 | 4.4 | 87 |
| 5609 | Sylvan Springs5 | 4 | 5.4 | 90 |
| 5561 | Artists Paint Pots6 | 5 | 0 | 0 |
The superscript numbers correspond with location numbers on the map in Fig. 1.
TABLE 6.
Distribution of SSR variants from sites throughout the park
| Locationa | Distribution for variantb:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Artists Paint Pots6 | + | + | + | ||
| Clear Lake13 | + | + | |||
| Cyanidium Creek4 | + | + | |||
| Dragon Spring4 | + | ||||
| Extreme site4 | + | + | |||
| Fairy Falls Trail8 | + | + | |||
| Heart Lake10 | + | + | + | ||
| Highland Creek12 | + | + | |||
| Imperial Spring8 | + | ||||
| Lemonade Creek1 | + | + | |||
| Mud Volcano11 | + | + | |||
| Nymph Creek3 | + | + | + | ||
| Obsidian Creek2 | + | + | |||
| Rabbit Creek9 | + | + | |||
| River Group7 | + | + | |||
| Sour Creek13 | + | ||||
| Star Plunge (Thermopolis, WY) | + | ||||
| Sylvan Creek5 | + | + | |||
| Sylvan Crust5 | + | + | |||
The superscript numbers correspond with location numbers on the map in Fig. 1.
By using agarose gel electrophoresis, all culture isolates from type IA in YNP were screened to determine their SSR variant groups. A positive (+) result for a SSR9 variant group represents the presence of a culture isolate that was determined to belong to that variant group.
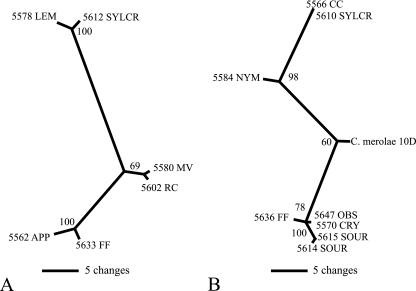
We then sought to evaluate cpSSR9 sequence variance as an additional phylogenetic tool. Not all cpSSR9 varietal amplicons could be used for this purpose, due to problems with ambiguous alignments encountered within a cpSSR9 repeat unit. However, selected isolates of variants 2 and 3 could be used for this purpose, and by using maximum parsimony, the cpSSR9 sequences of these isolates were compared phylogenetically (Fig. 4). For comparisons of the strains of variants 2 and 3, groupings and separations were readily apparent. This result showed that in addition to simply the fractional repeat differences, various isolated strains could be differentiated by using cpSSR loci for type IA strains that could not be differentiated using rbcL or 18S rDNA.
FIG. 4.
Unrooted maximum parsimony trees of SSR9 variant 2 (A) and variant 3 (B) of selected YNP type IA and IIIA culture isolates. Bootstrap values are given for all nodes exceeding 50%. Branch lengths are proportional to the number of changes on a given branch.
DISCUSSION
The extensive sampling, isolation, and phylogenetic characterization of cyanidial algae from acidic geothermal sites throughout YNP and from sites in Japan and New Zealand yielded several novel observations. The phylogenetic diversity of these isolates was based on both the rbcL and the 18S rRNA genes, which considerably expands a previous survey conducted by Gross et al. (13), who used 18S rDNA as the basis of their biogeographic comparisons of a limited number of isolates from various culture collections (usually one from each site), but none from New Zealand. It also extends a more recent study by Ciniglia et al. (6) who used rbcL to describe organism/population variation at sites within the Phlegrean Fields in the Pisciarelli hydrothermal system located near Naples, Italy. The current study provided more extensive evidence of biogeographical patterning within the Cyanidiales similar to that found within the thermophilic cyanobacteria (21, 27).
Genotypic comparisons within YNP.
Comparisons of 18S rDNA and rbcL sequences from the YNP isolates were almost boringly predictable, falling into only two types. The type II YNP isolates were closely related to G. sulphuraria based on both rbcL and 18S rDNA analyses (Fig. 2A and B). Indeed, our isolates grew heterotrophically in the dark on glucose and were not able to use nitrate as the sole nitrogen source (data not shown), traits that are consistent with the G. sulphuraria strains isolated near Naples, Italy, and elsewhere (12). A second and much more intriguing group of YNP isolates was that for the organisms we refer to as type I, which comprised 95% of our more than 120 culture isolates from YNP and were made up of two different subtypes (IA and IB). Both of these classes have more than 99% rbcL and 18S rDNA sequence identity to C. merolae. However, only four of these isolates (type IB) have morphologies consistent with that currently recognized for C. merolae (20). All of our type I strains (type IA) have morphologies and cell division characteristics consistent with those of Galdieria (Fig. 3, compare panels A and B). The isolation of this type of organism in the current study serves to verify the previous reports of similar identity uncertainty by Ferris et al. (10) and Lehr et al. (17). More importantly, the extensive isolation of type IA morphophylotypes from throughout YNP in the current study also serves to demonstrate that these previous reports do not represent fortuitous observations. Rather, this Galdieria-like organism appears to represent the predominant member of the Cyanidiales within YNP. There is precedence for rDNA sequences commonly used for species determination to be the same in two distinctly different phenotypes. In the case of two photosynthetic dinoflagellates (18), the sequenced DNA fragments (internal transcribed spacers 1 and 2, 5.8S, the large subunit [D1/D2], and the small subunit) were identical in both organisms, but numerous stable morphological, physiological, and ecological characteristics of these two organisms were quite unlike.
As the rbcL and 18S rDNA phylotypes were found to be inadequate for describing genotypic diversity among the type IA isolates from YNP, we turned to cpSSRs in an attempt to identify evidence of some genetic diversity among these organisms. SSRs have been useful for resolving subspecies- or ecotype-level relationships in other eukaryotic organisms (29) as well as strain-level diversity among bacteria (8, 34). As in a previous study using cpSSRs for the purpose of seasonal monitoring of type IA cyanidial populations in a single YNP spring (17), we also showed the utility of SSRs for differentiating a small number of type IA culture isolates. In the current study, we were able to further separate YNP type IA isolates based on differences in cpSSR9 frequencies and sequences. An assessment of isolation sites of these variants revealed that there was an almost complete lack of cohabitation between variants 3 and 5 (Table 6), although there was nothing obvious in the within-YNP geographical distribution patterns for these particular cpSSR9 variants that would suggest migration constraints within YNP (compare Table 6 and Fig. 4 with Fig. 1). While at this juncture there is no information that directly links cpSSR sequence variation with important adaptive traits, it is possible that such clustering and the apparent separation of variants 3 and 5 may correspond to variations in the physicochemical characteristics that may occur among acid thermal environments in YNP (1, 4). For example, some habitats in the Norris Geyser Basin contain elevated levels of mercury (28) and we have measured exceptionally high total mercury concentrations in the soils at the extreme site B (250 μg·g−1 soil [analysis provided by Ed Nater, University of Minnesota]). In assessing the otherwise limited information available for the study sites, there was no apparent relationship between cpSSR9 variant types and temperature measured at sampling times (data not shown). Site distribution separation for variants 3 and 5 did not appear to be related to pH, although the variants were both more frequently encountered at sites where pH values were >2.0. There was also some evidence of pH effect for some of the other SSR9 variants. SSR9 variant 2 was found at all sites in YNP and at all pHs, although it was a more frequently encountered isolate (relative to the other cpSSR9 variants) as site pH values increased from 2.0 to 4.0.
Numerous cultures from samples collected at most sites and all cpSSR9 types were capable of growing well on the isolation medium. Nevertheless, a cultivation bias might fail to detect other SSR9 variants and it is possible that not all SSR variants were cultured from each sampling site. Thus, the apparent distribution patterns summarized in Table 6 may merely be coincidental and without additional genetic and physiologic information it is not yet possible to functionally associate SSR variants with environmental conditions. In the future, we hope to use the cpSSR9 locus, other appropriate loci, more detailed physiological characterization, and detailed chemical analyses of the sampling sites to further investigate possible “speciation” events that may have given rise to ecotypes within type IA in YNP.
A recent study of acid hot springs in Italy showed a high level of rbcL variation in the clone libraries and cultures from the Pisciarelli area within the Phlegrean Field complex and other areas of Italy (6). Two different clades of G. sulphuraria were found, one corresponding to isolates from endolithic sites and one from aquatic environments. In addition to G. sulphuraria, clone libraries and culturing revealed C. caldarium type organisms as well as C. merolae types. Some of the Cyanidium phylotypes were nonthermophilic forms from the Monte Rotaro area of Italy (Fig. 2A). The rbcL data from the Italian study support the biogeographic and habitat niche separation of different cyanidial clades. The Italian cyanidial lineages, however, are distinct from the Japan, New Zealand, and Yellowstone rbcL clades.
A recent report describing photosynthetic organisms in a single acidic siliceous crust in the Norris Geyser Basin (YNP) demonstrated that the majority of the 16S rDNA PCR clones exhibited their closest identities to the C. caldarium chloroplast phylotype (98 or 99%) (36). However, the clone libraries did not detect the presence of Galdieria sp. or C. merolae in this one endolithic sample. In contrast, we did not detect any C. caldarium phylotypes through extensive culturing attempts from collections in YNP, Japan, and New Zealand, nor has C. caldarium been detected in 18S rDNA molecular surveys of Dragon Spring in Norris Geyser Basin (17) or in Nymph Creek (10), both in YNP. The paper by Walker et al. (36) claims to have identified a cyanidial sequence closest to C. caldarium from an endolithic sample in the Norris Geyser Basin, but we believe this claim is in error (unpublished data). It is possible that no C. caldarium phylotype exists in YNP. In earlier studies by Brock (4), all cyanidia were referred to as C. caldarium. However, it is now clear from those results, which included a demonstration of dark heterotrophic growth, that G. sulphuraria was the organism being characterized.
Evolutionary hypotheses and questions.
Seckbach (32, 33) has hypothesized an evolutionary order from C. merolae (the simplest morphologically) to C. caldarium to G. sulphuraria by comparing morphological and physiological characteristics. However, a recent study comparing the complete genome of C. merolae strain 10D with expressed sequence tags and high-throughput genomic sequence reads of G. sulphuraria (covering 70% of the genome of G. sulphuraria strain 074W) suggests an alternative hypothesis (2). It appears that approximately 30% of the G. sulphuraria genome has no relation to that of C. merolae. Both organisms have similar varieties of sugar kinases, but only G. sulphuraria has a high number of monosaccharide transporters. The lack of transport proteins helps explain the obligate photoautotrophy of C. merolae, and the presence of such proteins in G. sulphuraria explains its ability to grow on numerous carbon sources. The same genomic comparison (2) also established that C. merolae lacks genes coding for enzymes involved in the synthesis, modification, and degradation of the cell wall and the synthesis of red algal floridosides (for osmoregulation) and also displayed less cellular compartmentalization. Others have suggested that the cell wall architecture of G. sulphuraria may allow it to tolerate drier environments, while C. merolae may be found mainly in aquatic niches that are osmotically stable and, thus, do not require a rigid wall (2, 6). These cell structural differences suggest that the cyanidia may have evolved from a common ancestor possessing a cell wall and heterotrophic capability as in G. sulphuraria (2). In such an evolutionary scenario, G. sulphuraria would have retained the ability to grow heterotrophically in moist, darker sites (e.g., moist acid soils and gravels), where it would more likely encounter organic carbon sources. The ancestor of C. merolae may have colonized osmotically stable, inorganic habitats and lost the ability to synthesize a cell wall or use organic compounds (2).
Potentially, the YNP type IA isolates represent an interesting piece of this evolutionary puzzle. With their cell walls and spherical shapes, they resemble a Galdieria-type organism, yet genetically, they are nearly identical to C. merolae, at least when viewed using the rbcL and 18S loci. Our culture isolates of both type IA and type IB are from acid streams as well as from moist soil environments. In preliminary screens of a sampling of these organisms, none grew heterotrophically on glucose, but otherwise they varied considerably with respect to growth rate, ability to use nitrate as a sole nitrogen source, and their cardinal temperatures (data not shown). And while such differences are certainly noteworthy with respect to the characterization of these organisms, the extent of their genetic differences is as yet unknown and, thus, it would be difficult to reclassify these various strains into separate ecotypes or other taxonomic entities at this juncture.
The finding of this abundant type IA organism in YNP (a possible intermediate between a typical spherical G. sulphuraria-type cell and a typical Cyanidioschyzon) may help in understanding the evolutionary events that may have allowed these two types of organisms to diverge into different habitats. Natural genetic exchange is so far unknown in the Cyanidiales. If this lack is, in fact, true, the evolutionary history of these organisms may be easier to unravel. Minoda et al. (23) have successfully accomplished limited gene transformation in C. merolae through electroporation, presumably facilitated by the lack of a cell wall in this member of the Cyanidiales. According to Yoon et al. (39), the cyanidial group probably evolved as the earliest branch of the red algae in the Precambrian era, earlier than 1.3 billion years before the present. A few members of the Cyanidiales live in nonthermal acidic habitats (14, 31). It is therefore possible that primordial cyanidia evolved into thermophilic types later, through periodic selection and adaptive divergence, thus escaping from the competition within a less extreme acidic environment. The same evolutionary path may have been followed even earlier, i.e., an escape from extensive competition in a neutral environment to a nonthermal acidic environment where, even today, relatively few species of algae occur.
Global distribution of cyanidia.
On a global scale, phylogenetic analyses based on the rbcL gene identified two different Japanese clades most closely related to G. maxima, which was originally isolated from the Kurile Islands off the northeast coast of Hokkaido (Fig. 2A). However, when we compared these organisms based upon their 18S rDNA sequence data, the clade structures partially collapsed (Fig. 2B). A similar discrepancy between rbcL and 18S phylogenies was observed with the isolates from New Zealand. Organisms more closely clustered with G. sulphuraria, based on rbcL comparisons (e.g., New Zealand type IV [5722 WI and 5706 CR]), were more closely related to G. maxima when viewed from the 18S rDNA perspective (Fig. 2, compare panels A and B). The discrepancy between the chloroplast rbcL and nuclear 18S rDNA sequence data may indicate a difference in the time scale of chloroplast evolutionary changes (or at least of the rbcL gene) within the cyanidia of Japan and New Zealand that are independent of the nuclear DNA as characterized by 18S rDNA, which shows little divergence. There is precedence for rates of chloroplast DNA evolution differing from those of nuclear DNA (11, 35). This precedence also hints at lateral transfer of the rbcL gene. The ancestor of the New Zealand type IV strains could have obtained its plastid rbcL gene from a strain somewhat related to G. sulphuraria, while the more conserved, nuclear 18S rDNA sequence indicates phylogenetic identity closest to G. maxima (Table 4). Although no natural mechanism of gene transfer is known in the Cyanidiales, this does not preclude the possibility of its existence (9). The low rbcL sequence identity for both the Japanese type IIIB (96%) and type IIIC (94%) to G. maxima strain IPPAS P507 suggests the possibility of many more ecotypes or species in existence that are somewhat distantly or closely related to G. maxima than were previously known (Table 4). Some of these new taxa may be distinguished by differences in plastid characteristics rather than by nuclear or cytoplasmic dissimilarities. We anticipate further characterization of these isolates by using physiological differences and the sequences of more variable genes and proteins.
The geographically specific rbcL-based and even 18S-based clades of G. sulphuraria and G. maxima lineages suggest the possibility that unique cyanidial types evolved during long-term isolation over great geographical distances between western North America, Japan, and New Zealand (19). Because of the distances and the great ages of the volcanic hot springs in all three regions and because of the fidelity of specific 18S and rbcL genotypes to only one region, allopatric divergence and speciation must be regarded as the most likely basis of the genetic differences among these geographical provinces. This might not be the case if it were not for the fact that the ranges in chemistry of the acidic habitats sampled in all three areas are very similar. It is known from the literature and from our tests (unpublished data) that cyanidia do not tolerate desiccation, and therefore dispersal of viable founder populations over great distances must be a very rare event (38). The most probable vector may be migratory birds that carry moist acidic mud on their feet or pass viable cells through their acidic digestive systems. The latter case has been demonstrated for unicellular green algae, diatoms, cyanobacteria, and others, even in species not inhabiting acid waters and without the aid of resting stages (30).
Even without multilocus analyses, the divergences of the rbcL and 18S genes suggest speciation events that may have taken place over thousands or millions of years. A caveat of the conclusions raised here is that more strains, species, or ecotypes probably exist in Japan and New Zealand and collecting and isolation attempts have not reached saturation. However, we are probably close to saturation in Yellowstone, since cyanidia are readily cultivated and isolates from almost all acidic areas in the park are in the culture collection and have been sequenced using the rbcL and 18S loci.
Our study of the cyanidia of acid hot springs in YNP, Japan, and New Zealand shows that we are only beginning to understand the diversity of this interesting group of primitive red algae in an environment quite atypical of those of all other members of this phylum.
Acknowledgments
Primary funding for this work was from the National Aeronautics and Space Administration (grants NNA05CP98A and NAG 5-8807) to R.W.C., who is a member of the NASA Astrobiology Institute. Additional minor support from the Montana Agricultural Experiment Station to T.R.M. (911310) and NSF grant MIP 0702177 are also acknowledged.
We thank John Varley and Christie Hendrix at the Yellowstone Center for Resources for general assistance and for obtaining research permits. We also thank Dana J. Skorupa for valuable assistance as well as the three unidentified reviewers of the original manuscript for their constructive input.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Ball, J. W., D. K. Nordstrom, J. M. Holloway, and P. L. Verplanck. 2002. Water-chemistry data for selected springs, geysers, and streams in Yellowstone National Park, Wyoming, 1999-2000. United States Geological Survey Open File Report 02-382. U.S. Geological Survey, Boulder, CO.
- 2.Barbier, G., C. Oesterhelt, M. D. Larson, R. G. Halgren, C. Wilkerson, R. M. Garavito, C. Benning, and A. P. Weber. 2005. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137:460-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock, T. D. 1967. Life at high temperatures. Evolutionary, ecological, and biochemical significance of organisms living in hot springs is discussed. Science 158:1012-1019. [DOI] [PubMed] [Google Scholar]
- 4.Brock, T. D. 1978. Thermophilic microorganisms and life at high temperatures. Springer-Verlag, New York, NY.
- 5.Castenholz, R. W. 1988. Culturing methods (cyanobacteria). Methods Enzymol. 167:68-93. [Google Scholar]
- 6.Ciniglia, C., H. S. Yoon, A. Pollio, G. Pinto, and D. Bhattacharya. 2004. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol. Ecol. 13:1827-1838. [DOI] [PubMed] [Google Scholar]
- 7.Cozzolino, S., P. Caputo, O. De Castro, A. Moretti, and G. Pinto. 2000. Molecular variation in Galdieria sulphuraria (Galdieri) Merolae and its bearing on taxonomy. Hydrobiologia 433:145-151. [Google Scholar]
- 8.de Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwiche, C. F., and J. D. Palmer. 1996. Rampant horizontal transfer and duplication of Rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13:873-882. [DOI] [PubMed] [Google Scholar]
- 10.Ferris, M. J., K. B. Sheehan, M. Kühl, K. Cooksey, B. Wigglesworth-Cooksey, R. Harvey, and J. M. Henson. 2005. Algal species and light microenvironment in a low-pH, geothermal microbial mat community. Appl. Environ. Microbiol. 71:7164-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glöckner, G., A. Rosenthal, and K. Valentin. 2000. The structure and gene repertoire of an ancient red algal plastid genome. J. Mol. Evol. 51:382-390. [DOI] [PubMed] [Google Scholar]
- 12.Gross, W. 1999. Revision of comparative traits for the acido- and thermophilic red algae Cyanidium and Galdieria, p. 437-446. In J. Seckbach (ed.), Enigmatic microorganisms and life in extreme environments. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Gross, W., I. Heilmann, D. Lenze, and C. Schnarrenberger. 2001. Biogeography of the Cyanidiaceae (Rhodophyta) based on 18S ribosomal RNA sequence data. Eur. J. Phycol. 36:275-280. [Google Scholar]
- 14.Gross, W., C. Oesterhelt, G. Tischendorf, and F. Lederer. 2002. Charaterization of a non-thermophilic strain of the red algal genus Galdieria isolated from Soos (Czech Rebublic). Eur. J. Phycol. 37:477-482. [Google Scholar]
- 15.Gross, W., and C. Schnarrenberger. 1995. Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 36:633-638. [Google Scholar]
- 16.Heilmann, I., and W. Gross. 2001. Genetic diversity of thermo-acidophilic red algae according to random amplified polymorphic DNA (RAPD) analysis. Nova Hedwigia 123:531-539. [Google Scholar]
- 17.Lehr, C. R., S. D. Frank, T. B. Norris, S. D'Imperio, A. V. Kalinin, J. A. Toplin, R. W. Castenholz, and T. R. McDermott. 2007. Cyanidia (Cyanidiales) population diversity and dynamics in an acid-sulfate chloride spring in Yellowstone National Park. J. Phycol. 43:3-14. [Google Scholar]
- 18.Logares, R., K. Rengefors, A. Kremp, K. Shalchian-Tabrizi, A. Boltovskoy, T. Tengs, A. Shurtleff, and D. Klaveness. 2007. Phenotypically different microalgal morphospecies with identical ribosomal DNA: a case of rapid adaptive evolution? Microb. Ecol. 53:549-561. [DOI] [PubMed] [Google Scholar]
- 19.Martiny, J. B., B. J. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki, M., O. Misumi, I. T. Shin, S. Maruyama, M. Takahara, S. Y. Miyagishima, T. Mori, K. Nishida, F. Yagisawa, Y. Yoshida, Y. Nishimura, S. Nakao, T. Kobayashi, Y. Momoyama, T. Higashiyama, A. Minoda, M. Sano, H. Nomoto, K. Oishi, H. Hayashi, F. Ohta, S. Nishizaka, S. Haga, S. Miura, T. Morishita, Y. Kabeya, K. Terasawa, Y. Suzuki, Y. Ishii, S. Asakawa, H. Takano, N. Ohta, H. Kuroiwa, K. Tanaka, N. Shimizu, S. Sugano, N. Sato, H. Nozaki, N. Ogasawara, Y. Kohara, and T. Kuroiwa. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653-657. [DOI] [PubMed] [Google Scholar]
- 21.Miller, S. R., R. W. Castenholz, and D. Pedersen. 2007. Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 73:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, S. R., M. D. Purugganan, and S. E. Curtis. 2006. Molecular population genetics and phenotypic diversification of two populations of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 72:2793-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minoda, A., R. Sakagami, F. Yagisawa, T. Kuroiwa, and K. Tanaka. 2004. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 45:667-671. [DOI] [PubMed] [Google Scholar]
- 24.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordstrom, D. K., J. W. Ball, and R. B. McClesey. 2005. Ground water to surface water: chemistry of thermal outflows in Yellowstone National Park, p. 73-94. In W. P. Inskeep (ed.), Geothermal biology and geochemistry in Yellowstone National Park. Thermal Biology Institute at Montana State University, Bozeman.
- 26.Oesterhelt, C., and W. Gross. 2002. Different sugar kinases are involved in the sugar sensing of Galdieria sulphuraria. Plant Physiol. 128:291-299. [PMC free article] [PubMed] [Google Scholar]
- 27.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 28.Phelps, D., and P. R. Buseck. 1980. Distribution of soil mercury and the development of soil mercury anomalies in the Yellowstone geothermal area, Wyoming. Econ. Geol. 75:730-741. [Google Scholar]
- 29.Powell, W., M. Morgante, C. Andre, J. W. McNicol, G. C. Machray, J. J. Doyle, S. V. Tingey, and J. A. Rafalski. 1995. Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr. Biol. 5:1023-1029. [DOI] [PubMed] [Google Scholar]
- 30.Proctor, V. W. 1959. Dispersal of fresh-water algae by migratory water birds. Science 130:623-624. [DOI] [PubMed] [Google Scholar]
- 31.Seckbach, J. 1999. The Cyanidiophyceae: hot spring acidophilic algae, p. 425-435. In J. Seckbach (ed.), Enigmatic microorganisms and life in extreme environments. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Seckbach, J. 1994. Evolutionary pathways and enigmatic algae: Cyanidium caldarium (Rhodophyta). Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 33.Seckbach, J. 1994. Methods for algal investigations, p. 333-336. In J. Seckbach (ed.), Evolutionary pathways and enigmatic algae: Cyanidium caldarium (Rhodophyta) and related cells. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 34.Versalovic, J., T. Koeuth, E. R. McCabe, and J. R. Lupski. 1991. Use of the polymerase chain reaction for physical mapping of Escherichia coli genes. J. Bacteriol. 173:5253-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogl, C., J. Badger, P. Kearney, M. Li, M. Clegg, and T. Jiang. 2003. Probabilistic analysis indicates discordant gene trees in chloroplast evolution. J. Mol. Evol. 56:330-340. [DOI] [PubMed] [Google Scholar]
- 36.Walker, J. J., J. R. Spear, and N. R. Pace. 2005. Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 434:1011-1014. [DOI] [PubMed] [Google Scholar]
- 37.Ward, D. M., and R. W. Castenholz (ed.). 2000. Cyanobacteria in geothermal habitats. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 38.Whitaker, R. J. 2006. Allopatric origins of microbial species. Philos. Trans. R. Soc. Lond. B 361:1975-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon, H. S., K. M. Muller, R. G. Sheath, F. D. Ott, and D. Bhattacharya. 2006. Defining the major lineages of red algae (Rhodophyta). J. Phycol. 42:482-492. [Google Scholar]