Abstract
Isolation and cultivation are a crucial step in elucidating the physiology, biogeochemistry, and ecosystem role of microorganisms. Many abundant marine bacteria, including the widespread Roseobacter clade-affiliated (RCA) cluster group, have not been cultured with traditional methods. Using novel techniques of cocultivation with algal cultures, we have accomplished successful isolation and propagation of a strain of the RCA cluster. Our experiments revealed that, in addition to growing on alga-excreted organic matter, additions of washed bacterial cells led to significant biomass decrease of dinoflagellate cultures as measured by in vivo fluorescence. Bacterial filtrate did not adversely affect the algal cultures, suggesting attachment-mediated activity. Using an RCA cluster-specific rRNA probe, we documented increasing attachment of these algicidal bacteria during a dinoflagellate bloom, with a maximum of 70% of the algal cells colonized just prior to bloom termination. Cross-correlation analyses between algal abundances and RCA bacterial colonization were statistically significant, in agreement with predator-prey models suggesting that RCA cluster bacteria caused algal bloom decline. Further investigation of molecular databases revealed that RCA cluster bacteria were numerically abundant during algal blooms sampled worldwide. Our findings suggest that the widespread RCA cluster bacteria may exert significant control over phytoplankton biomass and community structure in the oceans. We also suggest that coculture with phytoplankton may be a useful strategy to isolate and successfully grow previously uncultured but ecologically abundant marine heterotrophs.
It is now well established that most bacteria cannot grow as colonies on high-nutrient plates (60). Efforts to culture marine bacteria by dilution to extinction without solid substrates (10) have been quite successful, yielding, for example, the cultivation of several strains of the ubiquitous SAR11 group (45). Many other abundant groups, however, including the Roseobacter clade-affiliated (RCA) cluster, have been resistant to cultivation with such methods. The RCA cluster, discovered 3 years ago by Selje et al. (55), contains closely related (>98.5% similarity by 16S sequence) and abundant bacteria found worldwide in temperate and polar waters. These bacteria have been found attached to particles as well as free living and can comprise up to 10% of the coastal bacterial community (55). The ecosystem role of RCA cluster bacteria remains unknown, due in part to the unavailability of isolates from this group.
Based on the knowledge that most of the organic matter utilized by marine bacteria originates from phytoplankton, we investigated the possibility that novel, non-colony-forming bacteria such as the RCA cluster could be isolated from the surface and grown in the presence of algal cells. This type of algal-bacterial interaction would be expected to be dominant during algal blooms, when algal biomass is elevated, so we focused our efforts on samples collected from a bloom of the dinoflagellate Lingulodinium polyedrum. We further tested the idea that such bacteria can affect the growth of the algal cells (either positively or negatively), which would clarify their ecosystem role and biogeochemical consequences.
Bacteria that kill phytoplankton have previously been documented (see review in reference 33), though algicidal activity has not been previously associated with numerically dominant Alphaproteobacteria such as the RCA cluster. Surprisingly, there remains no conclusive evidence that algicidal bacteria kill phytoplankton in nature: all studies of this subject have been performed in the laboratory. The only attempt to directly enumerate algicidal bacteria in nature used a polyclonal antibody with unknown specificity (26) and examined only bacteria in the free-living fraction. Many algicidal bacteria require attachment to their host to kill it (33), and evidence of algicidal bacterial attachment in nature is lacking. Several studies have also indirectly suggested that algicidal bacteria may be involved in algal bloom decline, by utilizing a most-probable-number analysis using laboratory algal cultures (25). Such studies add diluted seawater bacteria (usually 0.6-μm filtrate) into algal cultures; the highest dilutions causing cell lysis allow indirect back-calculations of the number of putative algicidal bacteria in the original sample. Further evidence of algicidal bacteria killing phytoplankton in nature might entail quantification of infection rates in natural blooms. This has been carried out for eukaryotic pathogens (reviewed in reference 40) and algal viruses (reviewed in reference 7) but never with algicidal bacteria.
Using L. polyedrum as a model system, here we report evidence consistent with the following hypotheses: (i) RCA cluster bacteria can be isolated using algal cells and successfully propagated with additions of alga-derived organic matter, (ii) our RCA cluster strain significantly decreased the biomass of certain phytoplankton in the laboratory, and (iii) the dynamics of RCA cluster attachment to dinoflagellates in nature suggest that they play a direct role in bloom decline. We also examined previous microbial community structure analyses of algal blooms and suggest that RCA cluster bacteria play a major role in regulating algal bloom dynamics worldwide.
MATERIALS AND METHODS
Culture conditions.
All cultures, enrichments, and single-cell washes were carried out in borosilicate glass. Axenic (bacterium-free) L. polyedrum CCMP strain 1932 was grown statically in modified f/4 medium (21), with vitamin concentrations increased fourfold, at 18°C illuminated by cool white fluorescent tubes at 160 μE m−2 s−1on a 12-h/12-h light/dark cycle. Axenic status was routinely verified with DAPI (4′,6′-diamidino-2-phenylindole) staining and microscopic examination at 1,000× resolution, as well as PCR with bacterium-specific primers and fluorescent in situ hybridization (FISH) with bacterium-specific probes. Surface seawater was collected from the Scripps pier, La Jolla, CA, on several dates during an intense L. polyedrum bloom in summer 2005 and incubated in 2-liter flasks under the culture conditions mentioned above. On one occasion, one of these incubations crashed overnight (the algal cells formed a pellet at the bottom of the flask). We collected the bacterial fraction of this water by removing organisms larger than 0.6 μm by filtration and added it to the axenic culture (primary enrichment). After bacteria were allowed to colonize the algae for several days, single dinoflagellate cells from these incubations were collected with a pipette and washed several times in sterilized seawater (to remove unattached bacteria) before being added to new L. polyedrum cultures (secondary enrichments). We specifically targeted dinoflagellate cells that appeared moribund or unhealthy by collecting algal cells that were swimming slower or that had lost motility completely. Secondary enrichments were analyzed with bacterium-specific PCR-denaturing gradient gel electrophoresis (DGGE) (36) to check for single bands indicating potentially pure bacterial isolates. Algal biomass was monitored by measuring in vivo fluorescence with a TD700 fluorometer (Turner Designs) to quickly identify bacterial enrichments that decreased algal growth (compared to no-addition controls). Subsequent additions of pure RCA cluster strain LE17 were performed at late algal exponential phase (5,000 L. polyedrum cells ml−1), with 105 washed bacterial cells ml−1 added. We tested the ability of our bacterial strain to grow in both liquid and solid (1.5% agar, agarose, and Noble agar) media of the following constituents: f/4 medium (autoclaved seawater from a nonbloom period with added phosphate, nitrate, trace metals, and vitamins), both 0.22-μm-filtered and unfiltered axenic L. polyedrum cultures, and diluted ZoBell bacterial medium (38) consisting of seawater with 0.05 g peptone and 0.01 g yeast extract liter−1. Plates were monitored for the presence of colonies with a stereomicroscope for 3 weeks. Growth in liquid was monitored with a FACSort flow cytometer (BD Biosciences) for 11 days. Duplicate samples were fixed in 5% formalin for 30 min and frozen at −80°C until analysis. Samples were thawed on ice, stained for 15 min in the dark in 1× Sybr green II (30) (Invitrogen), and diluted 10- or 100-fold. Quantification of cells was based on green fluorescence and forward scatter. Unstained samples and stained axenic L. polyedrum cultures served as controls.
16S sequencing and phylogenetic analysis.
One-microliter secondary enrichment samples were incubated with Lyse-N-Go (Pierce Biotechnology) and amplified by PCR with primers 27F and 1492R (17). PCR products were purified with a QIAquick PCR purification kit (Qiagen) and directly sequenced with internal primers on a Megabase sequencer (Amersham). The sequences were imported into the Jan04 ARB database (31), aligned using the island hopping algorithm, checked manually, and added to the global tree using parsimony. Seventeen of the most closely related sequences (and two outgroups) were imported into PAUP*4.10b (62) and analyzed using maximum likelihood. Model parameters were optimized from the data using Modeltest (44), and 100 bootstrap replicate heuristic searches were performed.
Field sampling and probe design.
Surface samples during the summer 2005 L. polyedrum bloom were collected from the Scripps pier during the period between 17 June and 12 August. Water was collected with a surface sampler on a rope and fixed (within 10 min) in 2% (final volume) unbuffered formaldehyde. The abundance of L. polyedrum cells in 2-ml aliquots was quantified with light microscopic manual counts in 24-well plates with an inverted microscope (Olympus BX-71; 10× objective). Within 24 h, one-half of the fixed samples were filtered through 0.22-μm polycarbonate membranes and frozen at −20°C. For quantification of bacteria attached to L. polyedrum cells, the algal cells from the other half of the samples were allowed to gravity settle overnight at 4°C. These cells were washed in sterile phosphate-buffered saline (PBS) buffer and stored in 50% ethanol at −20°C until hybridization. Quantification of RCA cluster bacteria was accomplished using catalyzed reported deposition FISH (CARD-FISH) (41) with a 16S rRNA probe (see below). While previously published RCA cluster-specific probe RCA-825 (55) hybridized successfully against our isolate, it also exhibited a positive signal against Roseobacter strain Y3F (GenBank accession no. AF253467), which has a 1-bp difference at the probe binding site and is not a member of the RCA cluster. Increasing the hybridization stringency was not successful in removing the positive signal in strain Y3F without also removing it in strain LE17. Thus, we designed another RCA-specific probe, LE17-998 (5′-TCTCTGGTAGTAGCACAG) with helper probes RCA-980H (5′-GATGTCAAGGGTTGGTAA) and RCA-1016H (5′-CCCGAAGGGAACGTACCA). Using Roseobacter strain DG1128 (GenBank accession no. AY258100), which has two base pair differences at the probe binding site, we optimized hybridization conditions (35% formamide at 35°C hybridization temperature) to remove the positive signal in strain DG1128 while retaining the signal in strain LE17.
CARD-FISH.
Total RCA cluster bacteria during the 2005 bloom were enumerated using probe RCA-825, before we realized that the probe cross-reacted with a closely related Roseobacter isolate. Subsequently, RCA cluster bacteria attached to L. polyedrum were enumerated using probe LE17-998. Filtered samples (for total counts) were dipped in 0.1% low-melting-point agarose and air dried. Thawed liquid samples (for attached counts) were spotted on Teflon-coated well slides and air dried. The slides were dipped in 0.1% low-melting-point agarose and again air dried. Both types of samples were subsequently treated similarly, with a protocol slightly modified from the work of Pernthaler et al. (41). Samples were incubated in 1 mg ml−1 lysozyme (Sigma) in Tris-EDTA buffer at 37°C for 1 h. Slides or filters were then washed three times in MilliQ water, incubated for 10 min in 0.1 N HCl, washed again three times in 1× PBS (3 min), and finally dehydrated in an ethanol series (50%, 80%, and 95% for 3 min each). Samples were incubated for 2 h at 35°C in a hydrated chamber with hybridization buffer (35% formamide, 900 mM NaCl, 20 mM Tris, 0.01% sodium dodecyl sulfate, 20% Roche Diagnostic Boehringer blocking reagent) containing 1 μl probe for every 25 μl buffer (final concentration, 2 ng μl−1). Samples were subsequently washed for 20 min at 37°C in wash buffer (70 mM NaCl, 5 mM EDTA, 20 mM Tris, 0.01% sodium dodecyl sulfate), rinsed in MilliQ, and overlaid with TNT buffer (0.1 M Tris-HCl, pH 7.6, 0.15 M NaCl, 0.05% Tween 20) for 15 min. Samples were then incubated with 1 μl tyramide-Alexa Fluor 488 (Invitrogen) in 100 μl 1× PBS with 0.01% Boehringer blocking reagent and 0.003% H2O2 for 30 min at room temperature in the dark. They were then washed with TNT buffer at 55°C, rinsed in water, air dried, stained with DAPI, and mounted with Vectashield (Vectorlabs, CA). Negative (no probe) and positive (LE17) controls were always performed concurrently with all hybridizations. Bacteria were visualized on an Olympus BX51 epifluorescence microscope with a standard DAPI filter set and a fluorescein isothiocyanate-Texas Red dual band filter set (Chroma Technology Corp., VT). For attached RCA cluster counts, we examined at least 50 individual L. polyedrum cells for each sample and counted the total number of probe-positive bacterial cells on them. Our counts were most probably underestimates due to the inability to detect all probe-positive bacterial cells located behind the autofluorescent algal cells.
Statistical analyses.
We examined three indices of RCA cluster abundance over the course of the bloom: (i) total cells; (ii) colonization frequency, defined as the percentage of L. polyedrum cells colonized by at least one RCA cluster bacterium; and (iii) colonization intensity, defined as the mean number of RCA cluster bacteria colonizing the algal cells. It is important to note that the last index did not include algal cells with no detected RCA cluster colonizers, making it independent from the colonization frequency index. To examine the temporal relationships among the datasets, we performed cross-correlation analyses by lagging one data set with respect to the other before calculating correlations. Data were first log-transformed to satisfy the assumptions of normality. Due to uneven temporal sampling, bacterial data were manually lagged in both directions (0 to 3 days) relative to algal abundance data (in Microsoft Excel), and correlation analyses for each lag were performed with the statistical software JMP v.5.
Nucleotide sequence accession numbers.
The sequences of cultures LE17 and LE20 were deposited in GenBank under accession numbers EF661583 and EU391659, respectively.
RESULTS
Cultivation and phylogenetic analysis.
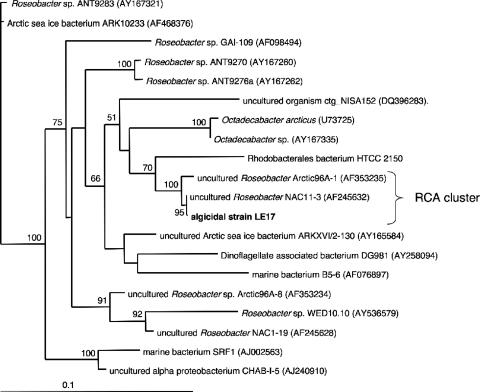
Incubation of seawater bacteria (0.6-μm filtrate) with L. polyedrum cultures followed by single algal cell micromanipulation into new algal cultures was successful in enriching for bacteria capable of attachment to algal cells. Several such enrichments displayed single bands (phylotypes) after DGGE (36) and were considered monospecific bacterial cultures (data not shown). Two such cultures (LE17 and LE20) were associated with lower in vivo fluorescence and were sequenced (GenBank accession numbers EF661583 and EU391659, respectively). Strain LE17 was revealed to be a member of the RCA cluster (99.9% sequence identity to clone NAC11-3 and 100% bootstrap support) (Fig. 1). RCA cluster sequences from the work of Selje et al. (55) were not included in this analysis as they do not cover the whole 16S gene (partial sequences were analyzed below). Although a close relative (HTCC2150, 95.5% 16S sequence similarity) has been isolated and its genome partially sequenced (GenBank accession number NZ_AAXZ0000000), LE17 is, to our knowledge, the first successfully isolated and serially transferred RCA cluster bacterium and remains in culture 2 years after isolation.
FIG. 1.
Maximum likelihood phylogenetic analysis of LE17 16S sequence within closely related Roseobacter sequences from the family Rhodobacteriaceae (GenBank accession numbers in parentheses). Numbers at nodes indicate full heuristic search maximum likelihood bootstrap values (100 replicates). Scale bar indicates percent sequence divergence. The model of molecular evolution (GTR+G+I) was chosen using MODELTEST (44).
RCA cluster dynamics in the laboratory.
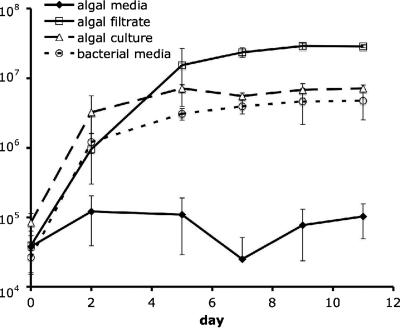
We investigated the growth characteristics of strain LE17 in various types of organic matter additions. LE17 grew to moderate abundances (3 × 107 cells ml−1) in coculture with L. polyedrum or in the presence of its filtrate. Incubations in diluted bacterial medium resulted in slower growth; those in nonbloom seawater amended with inorganic nutrients showed no measurable growth (Fig. 2). LE17 never formed visible colonies on solid media, which were identical to the liquid media tested above but with 1.5% agar, agarose, or Noble agar.
FIG. 2.
Abundance of RCA cluster strain LE17 over time in seawater with inorganic nutrient (f/4 algal medium), in L. polyedrum stationary culture filtrate, in log-phase L. polyedrum whole culture, and in 100×-diluted ZoBell bacterial medium (seawater with 0.05 g peptone and 5 mg yeast extract liter−1) measured with flow cytometry. Error bars represent standard deviations of triplicate incubations.
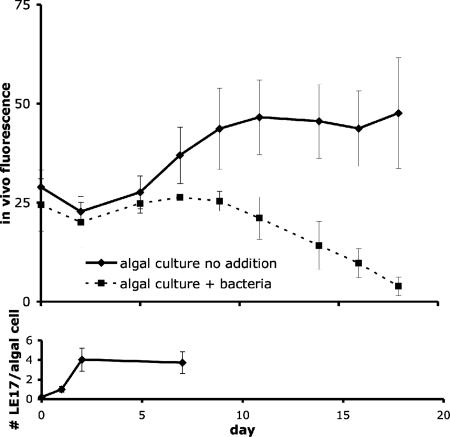
Since RCA cluster strain LE17 grew best in the presence of L. polyedrum, we tested whether algal growth was in turn influenced by the bacteria to determine if strain LE17 maintained a mutualistic, parasitic, or commensal relationship with the algae. Axenic L. polyedrum cultures incubated with RCA cluster strain LE17 exhibited a dramatic and repeatable decrease in fluorescence compared to no-addition controls (Fig. 3, top panel), leading us to describe strain LE17 as parasitic to algae (algicidal). Algicidal activity in laboratory cultures was likely mediated by attachment, with an average of four bacterial cells attached to each algal cell (Fig. 3, bottom panel). There was a lag of several days between initial bacterial colonization and the noted decrease in fluorescence. Samples collected before the decreased fluorescence and examined under the microscope revealed that RCA cluster-colonized dinoflagellates appeared to swim slower than those in axenic control cultures. This is consistent with our initial isolation approach targeting moribund L. polyedrum cells. Cell-free filtrates never displayed algicidal or motility-inhibiting activity (data not shown), leading us to conclude that the activity was likely mediated by attachment.
FIG. 3.
Abundance of L. polyedrum cells over time monitored by in vivo fluorescence with and without RCA cluster strain LE17 ± standard error of triplicate incubations (upper panel); colonization intensity quantified as number of RCA cluster bacteria per algal cell ± 95% confidence intervals measured with CARD-FISH (lower panel).
RCA cluster dynamics in nature.
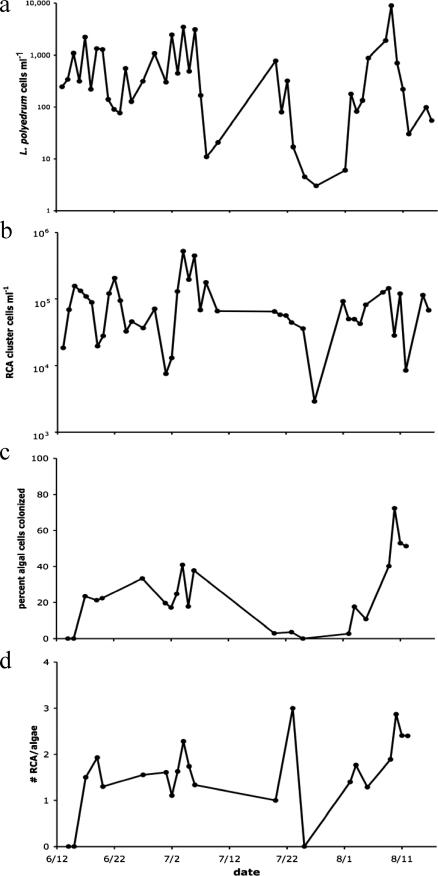
The summer 2005 L. polyedrum bloom consisted of two major peaks in abundance separated by more than a month, interspersed with a period of intermediate cell concentrations (Fig. 4a). In order to examine the population dynamics of RCA cluster bacteria during this bloom, we first quantified total free-living counts. Numbers of RCA cluster bacteria ranged from 3 × 103 to 5 × 105 cells ml−1 (Fig. 4b). Cross-correlation analyses between free-living RCA cluster bacteria and L. polyedrum abundances were not statistically significant, indicating no temporal relationship between the datasets (data not shown).
FIG. 4.
Population dynamics of L. polyedrum (a), total RCA cluster bacteria (b), RCA cluster bacterial attachment frequency (c), and RCA cluster attachment intensity (d) during the summer 2005 bloom at the Scripps pier, La Jolla, CA.
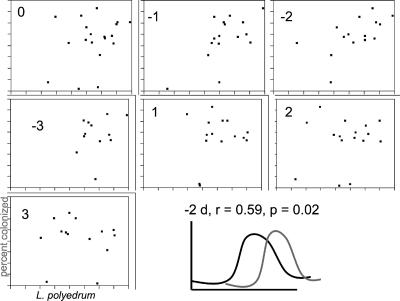
For the RCA cluster colonization data, a total of 2,179 L. polyedrum cells from 22 field samples were examined. Colonization frequency (the percentage of L. polyedrum cells colonized by at least one RCA cluster bacterium) appeared to increase with L. polyedrum abundance (Fig. 4c). The most intense colonization occurred at the end of the second stage of the bloom, with 51 to 72% of the algal cells colonized by RCA cluster bacteria on 10 August (before the final bloom crash beginning on 11 August). Cross-correlation of L. polyedrum abundances and RCA bacterial colonization frequency showed a significant positive correlation with a negative 2-day lag (dinoflagellate peaks leading the bacterial peaks), implying a potential response in bacterial colonization to high algal numbers (Fig. 5).
FIG. 5.
Cross-correlation analyses between log-transformed L. polyedrum abundances and RCA cluster colonization frequency during the summer 2005 bloom. The number inside each chart refers to the lag (in days). A lag of −2 days was statistically significant and positive. The last panel shows a graphical representation of the relationship between algal numbers (black line) and bacterial colonization frequency (gray line).
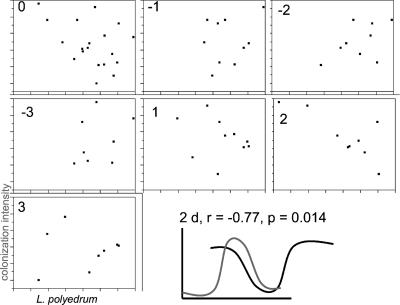
RCA cluster colonization intensity (measured by the average number of RCA cluster bacteria on each colonized algal cell) also seemed to increase with L. polyedrum abundances, in magnitudes similar to what we saw in laboratory cultures (up to an average of three RCA cluster bacteria per algal cell). Peaks in colonization occurred on 5 July, 23 July, and 10 August, again close to peaks in algal abundances (Fig. 4d). Cross-correlation analyses of L. polyedrum abundances and RCA cluster colonization intensity were statistically significant (and negative) with a positive 2-day lag (bacteria peaked first) (Fig. 6). This implies that peaks in RCA cluster colonization intensities preceded (by 2 days) crashes in L. polyedrum abundances, consistent with predator-prey or infection dynamics. These data are consistent with the hypothesis that RCA cluster bacteria killed L. polyedrum cells during the summer 2005 bloom in La Jolla.
FIG. 6.
Cross-correlation analyses between log-transformed L. polyedrum abundances and RCA cluster colonization intensity during the summer 2005 bloom. The number inside each chart refers to the lag (in days). A lag of +2 days was statistically significant and negative. The last panel shows a graphical representation of the relationship between algal numbers (black line) and bacterial colonization intensity (gray line).
We searched GenBank for previous studies that examined microbial diversity in marine phytoplankton blooms, both natural and mesocosm induced. Over 70% (11/15) of these studies reported a sequence >99.5% similar to that of LE17 (Table 1). These studies included blooms of dinoflagellates (13, 49, 51), prymnesiophytes (8, 19), a raphidophyte, diatoms (35, 47), and undescribed phytoplankton (42, 52). Although three of the sequences were relatively short (∼150 bp), they spanned variable regions of the 16S rRNA, were 100% identical to the LE17 sequence, and failed to match with any other organisms in the database. Thus, we are confident that these short sequences represent the same 16S phylotype as LE17.
TABLE 1.
Characteristics of RCA cluster sequences from phytoplankton blooms
| % Similarity to LE17 | Source | Clone name | GenBank accession no. | Reference | No. of base pairs |
|---|---|---|---|---|---|
| 99.5 | Chattonella bloom, coastal Japan | N7 | AB254272 | Unpublished | 151 |
| 100 | L. polyedrum bloom, California coast | ATT9b | AF125336 | 13 | 164 |
| 100 | Mesocosm bloom, California | MBE14 | AF191765 | 47 | 169 |
| 99.9 | Emiliania huxleyii bloom, Georgia | NAC11-3 | AF245632 | 19 | 1,414 |
| 100 | Phytoplankton microcosm, Spain | ST-11 | AY573528 | 42 | 492 |
| 100 | Mesocosm Phaeocystis bloom, North Sea | Band_4a | AY672827 | 8 | 505 |
| 100 | Dinoflagellate bloom, Catalan coast (Spain) | BH7 | DQ008454 | 51 | 512 |
| 100 | Spring phytoplankton bloom, Wadden Sea | GWS-a3-FLa | DQ080937 | Unpublished | 498 |
| 100 | Spring phytoplankton bloom (North Sea) | F089a | DQ289544 | 52 | 562 |
| 99.9 | Diatom bloom, Oregon coast | NH10_24 | DQ372848 | 35 | 1,426 |
| 99.6 | Bay of Fundy Alexandrium bloom | AFB-2a | AY353557 | 49 | 475 |
Clone representative of several RCA cluster clones.
Clone collected from the attached fraction.
We also exhaustively searched the GenBank nr database and the Global Ocean Survey (GOS) metagenomic database (data available on the CAMERA website [http://camera.calit2.net]) for RCA cluster sequences from temperate environments (not including the algal bloom samples mentioned above). Sequences >98.8% similar to the 16S of isolate LE17 were found in 35 previous molecular diversity studies of temperate coastal marine waters and in six coastal Atlantic Ocean samples from the GOS data set (50) (Table 2).
TABLE 2.
Characteristics of RCA cluster sequences found in nonbloom temperate environments
| % Similarity to LE17 | Source | Clone name | GenBank accession no. | No. of base pairs | Referencea |
|---|---|---|---|---|---|
| 100 | Saline lake, Japan | 1w | AB154432 | 524 | 28 |
| 100 | Japan coast | HB02-8b | AB265989 | 169 | 22 |
| 100 | Coast of Georgia | GAI-36 | AF007259 | 400 | 18 |
| 100 | Long Island Sound | pC2-12 | AF055225 | 298 | 16 |
| 100 | Coastal North Sea | OTU_E | AF207853 | 534 | 65 |
| 99.0 | Coastal Georgia | EC-I1 | AF287022 | 392 | 11 |
| 100 | Weser estuary, Germany | WM11-37b | AF497861 | 523 | 54 |
| 100 | English Channel | DGGE band 5 | AJ242822 | 515 | 15 |
| 99.0 | Mediterranean coast, France | S1-090-F-C-Nr1 | AJ508432 | 523 | Unpublished |
| 99.5 | North Sea, Germany | UNHYB_26b | AJ630678 | 450 | 53 |
| 100 | Oregon coast | HTCC152c | AY102029 | 647 | 10 |
| 99.7 | Marine sediment | s29 | AY171302 | 1,018 | 27 |
| 100 | Coast of Skagerrak Sea | SKA55c | AY317122 | 429 | 57 |
| 99.3 | Black Sea oxycline | BSBd6-20/40mb | AY360519 | 829 | 64 |
| 98.8 | Estuary, Portugal | RAN-63 | AY499446 | 592 | 23 |
| 99.1 | North Sea, United Kingdom | PEL-52 | AY550815 | 550 | 14 |
| 100 | Estuary, Massachusetts | PI_RT343b | AY580461 | 816 | 1 |
| 100 | Coastal salt pond, United States | SP_C23 | AY589480 | 783 | 56 |
| 99.8 | Estuary, Oregon | LS-F4 | AY628657 | 608 | 4 |
| 100 | Wadden Sea, Germany | Flo-37b | AY684343 | 465 | 20 |
| 100 | Salt marsh, Georgia | SIMO-662b | AY712199 | 451 | Unpublished |
| 100 | Coast of Scotland | FFW402 | AY828410 | 719 | Unpublished |
| 99.5 | Fjord, Norway | LUR12 | AY960287 | 570 | 48 |
| 99.7 | California coast | SPOTSAPR01_5m124b | DQ009296 | 1,400 | 6 |
| 99.9 | Eutrophic bay, Washington | PB1.27b | DQ071074 | 1,386 | 29 |
| 99.7 | Northeast Pacific, 2,500-m depth | CTD005-37B-02 | DQ513055 | 1,427 | 24 |
| 99.2 | Coast of Chile | Chili1-G4b | DQ669616 | 404 | 43 |
| 99.6 | O2 minimum, Chilean coast | ESP10-K27II-52 | DQ810325 | 1,301 | Unpublished |
| 99.6 | North Sea, Denmark | NS5 | DQ839250 | 539 | Unpublished |
| 99.9 | Coast of China | PV2-27 | EF215774 | 918 | Unpublished |
| 100 | North Atlantic, 1,700 m | 001733_3285_1268b | NAe | 60 | 59 |
| 99.5 | Coast, North Carolina | OM65b | U70682 | 643 | 46 |
| 99.6 | Northwest Mediterranean | T41_191 | DQ436604 | 751 | Unpublished |
| 100 | Plum Island Sound, United States | PIdgge30 | AY308694 | 132 | 12 |
| 99.6 | Coastal Mediterranean | Isolate a | EF018061 | 498 | Unpublished |
| 100 | Gulf of Mained | 1097156605048 | NA | 694 | 50 |
| 99.8 | Brown's Bank, Mained | 1097156701473 | NA | 1,226 | 50 |
| 99.9 | Bedford Basin, Nova Scotiad | 1097159073554 | NA | 935 | 50 |
| 100 | Newport Harbor, RId | 1097169034943b | NA | 1,470 | 50 |
| 98.7 | Cape May, NJd | 1097173026724 | NA | 946 | 50 |
| 100 | NAgs Head, NCd | 109720503350 | NA | 1,213 | 50 |
“Unpublished” indicates that sequences found in GenBank are not yet associated with a publication.
Representative sequence among several RCA cluster clones.
Reported dilution-to-extinction culture but unable to successfully propagate.
Sample from the GOS metagenomic analysis.
NA, not available.
DISCUSSION
Our results highlighting the physical interaction of bacteria from the Roseobacter group with phytoplankton, in particular dinoflagellates, are consistent with previous studies. Laboratory work has shown that cultured Roseobacter strains physically interact with dinoflagellates (34). Culture-independent field studies have shown that Roseobacter bacteria are often associated with algal blooms (reviewed in reference 9). Using this previous knowledge, we have isolated a member of the RCA cluster, one of the most common marine phylotypes in the ocean (37), and provided evidence to support the hypothesis that these bacteria affect algal bloom dynamics through pathogenesis.
Our finding that RCA cluster strain LE17 did not form visible colonies and yet required organic matter inputs is perhaps unexpected. It is contrary to a previous report that suggested that oligotrophic bacteria are unable to form colonies while copiotrophic bacteria are able to do so (58). Perhaps many marine bacteria, whether copiotrophic or oligotrophic, cannot form visible colonies on solid surfaces, consistent with the noted differences between cultured and uncultured phylogenetic diversity taken from the same seawater sample (3). In addition, our data documenting the inability of strain LE17 to grow in nonbloom seawater may shed some light on previous difficulties in growing RCA cluster bacteria (and some other abundant marine bacteria) with dilution-to-extinction methods. We suggest that adding organic matter to such dilutions may be a fruitful approach to grow copiotrophs that cannot form colonies. One additional puzzling characteristic of our strain is its inability to form colonies on L. polyedrum agar coupled with its ability to colonize L. polyedrum cells in liquid media. The most likely explanation is that our strain (and potentially many marine bacterial particle colonizers) can form colonies of a limited size, after which the drawbacks of colony formation outweigh the benefits. These colonies may simply be too small to be detected by stereomicroscope. Previous work has indeed shown that many marine bacteria make microcolonies on agar visible only with an epifluorescence microscope (57).
Our laboratory experiments further revealed that strain LE17 displayed a parasitic relationship with the dinoflagellate L. polyedrum. We have yet to resolve the exact mechanism of this interaction, although it appeared to be based on physical interaction between the organisms. Two perplexing aspects remain: (i) the number of LE17 cells attached to L. polyedrum cells was never particularly high, and (ii) we did not observe LE17 cells inside the L. polyedrum cells. The first finding raises the question of how a few attached bacteria on a large dinoflagellate can have any effect on its physiology. Our colonization intensities are consistent with previous work that showed that as few as two or three bacteria attached to a large cyanobacterial cell could affect its nitrogen fixation rates (39). The second aspect could cast a doubt on the ability of strain LE17 to be parasitic without being intracellular. However, many ectoparasites exist at all levels of biology, including bacterial ectoparasites on nematodes (63) and protist ectoparasites on phytoplankton (61). In addition, intracellular localization does not imply parasitism, as many dinoflagellates harbor intracellular bacterial symbionts and commensals (5, 32).
Based on the knowledge that RCA cluster strain LE17 required attachment to kill L. polyedrum in the laboratory, we proceeded to test the hypothesis that RCA cluster attachment dynamics during an L. polyedrum bloom were consistent with a pathogenic interaction. The null hypothesis that we attempted to reject was that attachment dynamics were random and showed no temporal interaction with algal numbers. In a Lotka-Volterra-type predator-prey system (here the bacteria are the predators and L. polyedrum the prey), peaks in the predator populations follow peaks in the prey. These peaks in predators cause subsequent decreases in the prey population, followed by decreases in the predator. Thus, increases in RCA cluster bacteria just after peaks in the L. polyedrum population and subsequent decreases in the L. polyedrum population would be consistent with our hypothesis that LE17 was a factor in bloom termination. Our colonization data demonstrated such dynamics with statistical robustness: increased RCA cluster colonization frequency occurred 2 days after L. polyedrum peaks, and L. polyedrum abundances were low 2 days after peaks in RCA cluster colonization intensity. The latter finding is most significant because it strongly suggests that L. polyedrum directly responded to RCA cluster colonization. An alternative and, we believe, less likely explanation is that RCA cluster colonization responded first to another factor that was ultimately responsible for bloom demise. However, for this to occur, this unknown factor would have to have a delay of more than 2 days before bloom decline: first a delay for RCA cluster attachment to increase in response to it, followed by an additional 2-day delay between RCA cluster attachment and noticeable bloom decline. We also propose that RCA cluster attachment may have acted in concert with another factor (such as nutrient limitation) to cause bloom decline. However, since LE17 can kill nutrient-replete L. polyedrum in the laboratory, we do not believe that another factor was necessarily involved.
We found 16S sequences from the same phylotype as strain LE17 (>99.5% similarity) in the majority of past phytoplankton bloom molecular diversity studies, suggesting that these bacteria may also play a role in those bloom dynamics. Unfortunately, the majority of these studies did not include a temporal component, and it is unknown whether RCA cluster colonization of algal cells increased at the end of the blooms. One exception is a study of an L. polyedrum bloom from 1997 in the same location as our study (13), in which the RCA cluster sequence (clone ATT9) was detected in the attached fraction at the end of the bloom and subsequently disappeared postbloom, consistent with our findings. To further validate the hypotheses that RCA cluster bacteria kill phytoplankton in general, more studies on RCA cluster attachment dynamics coupled with isolation of RCA cluster bacteria causing mortality of a range of phytoplankton taxa are needed.
We also found phylotypes >98.7% similar to LE17 from many other coastal temperate sites worldwide, suggesting that the success of the RCA cluster transcends episodic phytoplankton blooms. It may become abundant under any eutrophic condition, perhaps making it one of the most successful marine copiotrophic bacteria in the oceans. Indeed, based on the GOS metagenomic survey, this organism has been identified as one of the 20 most abundant phylotypes in surface marine waters (37, 50). In addition, the widespread distribution of the RCA cluster raises the possibility that these bacteria are also killing phytoplankton under nonbloom conditions, a hypothesis that remains to be tested. One additional intriguing aspect is that strain LE17 is unable to grow in nonbloom seawater and yet is widely distributed in coastal samples not taken during algal blooms. However, since there is no standard definition of “algal bloom,” we contend that many of the samples were probably obtained from eutrophic waters that contained high phytoplankton biomass but no apparent water discoloration. In addition, moderate phytoplankton biomass might be enough to support RCA cluster growth, particularly in microzones of high nutrients within the phycosphere of phytoplankton cells (2).
The first successful cultivation of a member of the RCA cluster and its recognition as an algicidal bacterium are noteworthy for several reasons. First, it demonstrates the usefulness of cocultivation with another organism(s) to isolate bacteria that will not grow on their own. While most efforts have focused on decreasing organic matter and using nonsolid media (10), here we have shown that organic matter enrichments to single bacterial cells may be a fruitful approach to isolating previously uncultured organisms. Second, the isolation of a strain of the RCA cluster will allow future biochemical, physiological, and genomic analyses of one of the most common bacterial phylotypes in the oceans. Third, it lends support to the hypothesis that bacterial killing of phytoplankton cells is not an artifact of laboratory incubations and that this phenomenon is an important mechanism shaping phytoplankton community structure in aquatic environments.
Acknowledgments
We thank R. Mueller and F. Lauro for discussions on a previous version of the manuscript and P. Von Dassow, M. A. Moran, and D. Green for phytoplankton or bacterial isolates. We are grateful to Y. Tanaka, J. Nguyen, and F. Malfatti for assistance in field sampling.
This work was funded by a NOAA ECOHAB award to P.J.S.F. and F.A. and an NSF grant to F.A.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Azam, F., and F. Malfatti. 2007. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5:782-791. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, L., H. Schaefer, F. Joux, C. Courties, G. Muyzer, and P. Lebaron. 2000. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat. Microb. Ecol. 23:1-11. [Google Scholar]
- 4.Bernhard, A. E., D. Colbert, J. McManus, and K. G. Field. 2005. Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol. Ecol. 52:115-128. [DOI] [PubMed] [Google Scholar]
- 5.Biegala, I. C., G. Kennaway, E. Alverca, J.-F. Lennon, D. Vaulot, and N. Simon. 2002. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescent in situ hybridization and confocal microscopy. J. Phycol. 38:404-411. [Google Scholar]
- 6.Brown, M. V., M. S. Schwalbach, I. Hewson, and J. A. Fuhrman. 2005. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 7:1466-1479. [DOI] [PubMed] [Google Scholar]
- 7.Brussaard, C. P. D. 2004. Viral control of phytoplankton populations—a review. J. Eukarot. Microbiol. 51:125-138. [DOI] [PubMed] [Google Scholar]
- 8.Brussaard, C. P. D., X. Marie, J. D. van Bleijswijk, and M. J. Veldhuis. 2005. A mesocosm study of Phaeocystis globosa (Prymnesiophyceae) population dynamics. II. Significance for the microbial food web. Harmful Algae 4:875-893. [Google Scholar]
- 9.Buchan, A., J. M. Gonzàlez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 12.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fandino, L. B., L. Riemann, G. F. Steward, R. A. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23:119-130. [Google Scholar]
- 14.Franklin, M. P., I. R. McDonald, D. G. Bourne, N. J. Owens, R. C. Upstill-Goddard, and J. C. Murrell. 2005. Bacterial diversity in the bacterioneuston (sea surface microlayer): the bacterioneuston through the looking glass. Environ. Microbiol. 7:723-736. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman, J. A., and C. C. Overney. 1998. Marine microbial diversity studied via 16S rRNA sequences: cloning results from coastal waters and counting of native archaea with fluorescent single probes. Aquat. Microb. Ecol. 32:3-15. [Google Scholar]
- 17.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-203. In E. Stackebrandt and M. Goodfellow (ed.), Modern microbiological methods: nucleic acids techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 18.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, J. M., R. Simo, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedros-Alio, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossart, H. P., F. Levold, M. Allgaier, M. Simon, and T. Brinkhoff. 2005. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7:860-873. [DOI] [PubMed] [Google Scholar]
- 21.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY.
- 22.Hamasaki, K., A. Taniguchi, Y. Tada, R. A. Long, and F. Azam. 2007. Actively growing bacteria in the Inland Sea of Japan identified by combined bromodeoxyuridine immunocapture and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 73:2787-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriques, I. S., A. Almeida, A. Cunha, and A. Correia. 2004. Molecular sequence analysis of prokaryotic diversity in the middle and outer section of the Portuguese estuary Ria de Aveiro. FEMS Microbiol. Ecol. 49:269-279. [DOI] [PubMed] [Google Scholar]
- 24.Huber, J. A., H. P. Johnson, D. A. Butterfield, and J. A. Baross. 2006. Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8:88-99. [DOI] [PubMed] [Google Scholar]
- 25.Imai, I., M.-C. Kim, K. Nagasaki, S. Itakura, and Y. Ishida. 1998. Relationship between dynamics of red tide-causing raphidophycean flagellates and algicidal micro-organisms in the coastal sea of Japan. Phycol. Res. 46:139-146. [Google Scholar]
- 26.Imai, I., T. Sunahara, T. Nishikawa, Y. Hori, R. Kondo, and S. Hiroishi. 2001. Fluctuation of the red tide flagellates Chattonella spp. (Raphidophyceae) and the algicidal bacterium Cytophaga sp. in the Seto Inland Sea, Japan. Mar. Biol. 138:1043-1049. [Google Scholar]
- 27.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 28.Koizumi, Y., H. Kojima, K. Oguri, H. Kitazato, and M. Fukui. 2004. Vertical and temporal shifts in microbial communities in the water column and sediment of saline meromictic Lake Kaiike (Japan), as determined by a 16S rDNA-based analysis, and related to physicochemical gradients. Environ. Microbiol. 6:622-637. [DOI] [PubMed] [Google Scholar]
- 29.Lau, W. W., and E. V. Armbrust. 2006. Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ. Microbiol. 8:1688-1702. [DOI] [PubMed] [Google Scholar]
- 30.Lebaron, P., N. Parthuisot, and P. Catala. 1998. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl. Environ. Microbiol. 64:1725-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maki, T., and I. Imai. 2001. Relationships between intracellular bacteria and the bivalve killer dinoflagellate Heterocapsa circularisquama (Dinophyceae). Fish. Sci. 67:794-803. [Google Scholar]
- 33.Mayali, X., and F. Azam. 2004. Algicidal bacteria in the sea and their impact on algal blooms. J. Eukarot. Microbiol. 51:139-144. [DOI] [PubMed] [Google Scholar]
- 34.Miller, T. R., and R. Belas. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648-1659. [DOI] [PubMed] [Google Scholar]
- 35.Morris, R. M., K. Longnecker, and S. J. Giovannoni. 2006. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8:1361-1370. [DOI] [PubMed] [Google Scholar]
- 36.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nealson, K. H., and J. C. Venter. 2007. Metagenomics and the global ocean survey: what's in it for us, and why should we care? ISME J. 1:185-187. [DOI] [PubMed] [Google Scholar]
- 38.Oppenheimer, C. H., and C. E. ZoBell. 1952. The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J. Mar. Res. 11:10-18. [Google Scholar]
- 39.Paerl, H. W., and K. K. Gallucci. 1985. Role of chemotaxis in establishing a specific nitrogen-fixing cyanobacterial-bacterial association. Science 227:647-649. [DOI] [PubMed] [Google Scholar]
- 40.Park, M. G., W. Yih, and D. W. Coats. 2004. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J. Eukarot. Microbiol. 51:145-155. [DOI] [PubMed] [Google Scholar]
- 41.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, O. Guadayol, A. Malits, and C. Marrase. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pommier, T., B. Canback, L. Riemann, K. H. Bostrom, K. Simu, P. Lundberg, A. Tunlid, and A. Hagstrom. 2007. Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16:867-880. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 45.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 46.Rappe, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811-826. [Google Scholar]
- 47.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riemann, L., J. Titelman, and U. Bamstedt. 2006. Links between jellyfish and microbes in a jellyfish dominated fjord. Mar. Ecol. Prog. Ser. 325:29-42. [Google Scholar]
- 49.Rooney-Varga, J., M. W. Giewat, M. C. Savin, S. Sood, M. LeGresley, and J. L. Martin. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49:163-175. [DOI] [PubMed] [Google Scholar]
- 50.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y.-H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sala, M. M., V. Balague, C. Pedros-Alio, R. Massana, J. Felipe, L. Arin, H. Illoul, and M. Estrada. 2005. Phylogenetic and functional diversity of bacterioplankton during Alexandrium spp. blooms. FEMS Microbiol. Ecol. 54:257-267. [DOI] [PubMed] [Google Scholar]
- 52.Sapp, M., A. Wichels, K. H. Wiltshire, and G. Gerdts. 2007. Bacterial community dynamics during the winter-spring transition in the North Sea. FEMS Microbiol. Ecol. 59:622-637. [DOI] [PubMed] [Google Scholar]
- 53.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 55.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 56.Simmons, S. L., S. M. Sievert, R. B. Frankel, D. A. Bazylinski, and K. J. Edwards. 2004. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 70:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simu, K., and A. Hagstrom. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simu, K., K. Holmfeldt, U. L. Zweifel, and Å. Hagström. 2005. Culturability and coexistence of colony-forming and single-cell marine bacterioplankton. Appl. Environ. Microbiol. 71:4793-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staley, J. T., and A. Konopka. 1985. Measurements of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 61.Sukhanova, I. N., M. V. Flint, G. Hibaum, V. Karamfilov, A. I. Kopylov, E. Matveeva, T. N. Rat'kova, and A. F. Sazhin. 1988. Exuviaella cordata red tide in Bulgarian coastal waters (May to June 1986). Mar. Biol. 99:1-8. [Google Scholar]
- 62.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods), 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 63.Vanholme, B., J. De Meutter, T. Tytgat, M. Van Montagu, A. Coomans, and G. Gheysen. 2004. Secretions of plant-parasitic nematodes: a molecular update. Gene 332:13-27. [DOI] [PubMed] [Google Scholar]
- 64.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the black sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winter, C., M. M. Moeseneder, and G. J. Herndl. 2001. Impact of UV radiation on bacterioplankton community composition. Appl. Environ. Microbiol. 67:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]