Abstract
Understanding the survival, multiplication, and transmission to seeds of plant pathogenic bacteria is central to study their pathogenesis. We hypothesized that the type III secretion system (T3SS), encoded by hrp genes, could have a role in host colonization by plant pathogenic bacteria. The seed-borne pathogen Xanthomonas fuscans subsp. fuscans causes common bacterial blight of bean (Phaseolus vulgaris). Directed mutagenesis in strain CFBP4834-R of X. fuscans subsp. fuscans and bacterial population density monitoring on bean leaves showed that strains with mutations in the hrp regulatory genes, hrpG and hrpX, were impaired in their phyllospheric growth, as in the null interaction with Escherichia coli C600 and bean. In the compatible interaction, CFBP4834-R reached high phyllospheric population densities and was transmitted to seeds at high frequencies with high densities. Strains with mutations in structural hrp genes maintained the same constant epiphytic population densities (1 × 105 CFU g−1 of fresh weight) as in the incompatible interaction with Xanthomonas campestris pv. campestris ATCC 33913 and the bean. Low frequencies of transmission to seeds and low bacterial concentrations were recorded for CFBP4834-R hrp mutants and for ATCC 33913, whereas E. coli C600 was not transmitted. Moreover, unlike the wild-type strain, strains with mutations in hrp genes were not transmitted to seeds by vascular pathway. Transmission to seeds by floral structures remained possible for both. This study revealed the involvement of the X. fuscans subsp. fuscans T3SS in phyllospheric multiplication and systemic colonization of bean, leading to transmission to seeds. Our findings suggest a major contribution of hrp regulatory genes in host colonization processes.
The phyllosphere (i.e., the environment of leaf surfaces) (21) provides important niches for microbes such as fungi and bacteria with diverse lifestyles, including epiphytes, saprophytes, and pathogens. While colonizing leaves, microorganisms are exposed to a harsh environment subjected to multiple stresses, including desiccation, UV irradiation, and nutrient limitation (4, 28). Xanthomonas fuscans subsp. fuscans (40) and Xanthomonas axonopodis pv. phaseoli (44, 45) are causal agents of common bacterial blight of bean (Phaseolus vulgaris L) (46). During colonization of the bean phyllosphere, X. fuscans subsp. fuscans forms aggregates (biofilms) that protect it from stresses and maintain population sizes (22). Under conditions of low relative humidity (RH) that limit bacterial multiplication, X. fuscans subsp. fuscans can achieve a complete biological cycle on its susceptible host, from the sown seed to the harvested seed, without causing any macroscopic symptoms (10). Consequently, such an asymptomatic pathogenic life cycle during which bacteria not only survive but also multiply to colonize their host is of great pathogenic and ecological importance. It could provide a discrete inoculum for future outbreaks of common bacterial blight of bean and a means of survival for the bacteria under conditions that are not favorable for its multiplication.
Pathogen transmission is one of the most important parameters for fitness (14, 30). It combines the ability to survive outside the host prior to infection, multiplication on the host, dispersion, and transmission to new ecological niches, including host seeds. Three seed infection pathways have bee described for seed-borne pathogens (29). Seeds can be internally contaminated via the host xylem, as occurs for viruses, some fungi, and a few bacteria. This can result in the contamination of the seeds, often through the hilum (1). Seeds may also become infested via the stigma, where bacteria move through the stylar tissues to the embryo; this was recently demonstrated for bacterial fruit blotch of watermelon (47). An external infection occurs via flowers and fruits as a consequence of contact of the seed with bacterial populations on symptomatic tissue or during threshing with residues carrying large bacterial populations (50). The molecular determinants involved in active mechanisms of bacterial transmission to seeds in the absence of symptoms remain unknown.
The hrp genes are one of the major pathogenicity determinants of most plant pathogenic bacteria. These genes form a cluster, conserved in plant and animal pathogenic bacteria, that encodes proteins which form a molecular syringe, the type III secretion apparatus (18, 37). This type III secretion system (T3SS) allows the secretion and the injection of bacterial virulence proteins, called effectors, directly in the host cell cytoplasm. Surprisingly, it has been shown that this T3SS is also necessary for leaf-associated colonization of bean by Pseudomonas syringae pv. syringae (20). No similar studies have been undertaken for other pathogens with an epiphytic growth phase or for other steps of host colonization, such as transmission to seeds.
The objectives of the work presented here were to characterize the hrp cluster of X. fuscans subsp. fuscans and to determine the role of hrp genes in the survival and the multiplication of this bacterium in the bean phyllosphere and in the transmission to bean seeds.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Xanthomonad strains were grown at 28°C in 10% TSA medium (1.7 g liter−1 tryptone, 0.3 g liter−1 soybean peptone, 0.25 g liter−1 glucose, 0.5 g liter−1 NaCl, 0.5 g liter−1 K2HPO4, and 15 g liter−1 agar) supplemented with the appropriate antibiotics. Escherichia coli cells were cultivated at 37°C in Luria-Bertani medium (10 g liter−1 tryptone, 5 g liter−1 yeast extract, 5 g liter−1 NaCl, 15 g liter−1 agar) and, for in planta studies, on 100% TSA medium (17 g liter−1 tryptone, 3 g liter−1 soybean peptone, 2.5 g liter−1 glucose, 5 g liter−1 NaCl, 5 g liter−1 K2HPO4, and 15 g liter−1 agar) supplemented with rifamycin. Antibiotics were used at the following final concentrations: rifamycin, 50 mg liter−1; kanamycin (Km), 25 mg liter−1; rifampin (Rif), 50 mg liter−1; and chloramphenicol (Cm), 12.5 mg liter−1. For in planta studies, media were supplemented with 50 mg liter−1 cycloheximide and 10 mg liter−1 propiconazole to inhibit fungal growth. To prepare inocula, strains were grown for 48 h in appropriate media supplemented with appropriate antibiotics. Bacterial cells were scraped from plates and suspended in sterile distilled water. Suspensions were calibrated to 1 × 108 CFU ml−1 and adjusted to the desired final concentrations with sterile distilled water.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pVO155 | Derivative plasmid of pUC19 containing the promoterless gus (uidA) gene; Kmr Ampr | 36 |
| pRK600 | Cmr, traRK2, oriColE1 | 16 |
| pIJ3225 | hrp cluster of X. campestris pv. campestris cloned in pLAFR1 | 3 |
| Wild-type strains | ||
| CFBP4834-R | X. fuscans subsp. fuscans CFBP4834-R wild-type strain; yellow strain producing the typical fuscous pigment; Rifr | 22 |
| ATCC 33913 | X. campestris pv. campestris ATCC 33913 wild-type strain; Rifr | 11 |
| 85.10 | X. axonopodis pv. vesicatoria wild-type strain; Rifr | 6 |
| 306 | X. citri subsp. citri wild-type strain | 11 |
| K-12 | Escherichia coli wild-type strain | 12 |
| DH5α | E. coli F φ980dlacZ:M15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR (lacZYA-argF)U169 | 19 |
| C600 | E. coli C600 wild-type strain; Rifr | 34 |
| Mutants | ||
| 4834HRCJ | CFBP4834-R hrcJ(420)a::pVO155; Rifr Kmr | This study |
| 4834HRCR | CFBP4834-R hrcR(320)::pVO155; Rifr Kmr | This study |
| 4834HRCT | CFBP4834-R hrcT(799)::pVO155; Rifr Kmr | This study |
| 4834HRCV | CFBP4834-R hrcV(1501)::pVO155; Rifr Kmr | This study |
| 4834HRPB2 | CFBP4834-R hrpB2(298)::pVO155; Rifr Kmr | This study |
| 4834HRPG | CFBP4834-R hrpG(625)::pVO155; Rifr Kmr | This study |
| 4834HRPX | CFBP4834-R hrpX(459)::pVO155; Rifr Kmr | This study |
| 33913HRCU | ATCC 33913 hrcU::pVO155; Rifr Kmr | Gift from M. Arlat |
| 33913HRPX | ATCC 33913 hrpX::pVO155; Rifr Kmr | Gift from M. Arlat |
Numbers in parentheses indicate the position from the start codon of pVO155 insertion in the target gene in X. citri subsp. citri 306.
Molecular biology techniques.
Chromosomal DNA was extracted with the Nucleospin tissue kit (Macherey-Nagel Hoerdt, France). Plasmid preparations were performed with the Wizard Plus Minipreps DNA purification systems (Promega). Restriction enzymes, DNA ligase, and GoTaq DNA polymerase (Promega) were used according to the manufacturer's recommendations. PCRs were done in 20-μl volumes containing 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.5 μM of each primer, and 0.4 U μl−1 GoTaq polymerase (final concentrations) and 4 μl of a boiled bacterial suspension (1 × 107 CFU ml−1). PCR conditions were 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C; and 7 min at 72°C. Southern hybridizations were performed using gene- and vector-specific PCR fragments as probes. Probes were labeled using the PCR digoxigenin labeling mix (Roche Applied Science, France). Genomic DNA was digested with BamHI and transferred to Hybond N+ nylon membranes (Amersham) according to supplier's instructions. Blots were hybridized for 16 h at 42°C in 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 20% sodium dodecyl sulfate (SDS), 2% blocking reagent, and 0.1% N-lauryl-sarcosine, followed by washes in 2× SSC-0.1% SDS and 0.1× SSC-0.1% SDS at 68°C. The probes were detected using the Fab fragments of an antidigoxigenin antibody conjugated with alkaline phosphatase and nitroblue tetrazolium chloride-5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt solution according to the supplier's instructions (Roche Applied Science, France).
Construction of hrp gene disruptions in X. fuscans subsp. fuscans.
For specific inactivation of CFBP4834-R genes, a plasmid integration mutagenesis strategy was used as previously described (36). For each target gene, primers were designed based on the consensus sequence from sequenced genomes of xanthomonads. Sequences of oligonucleotide primers are listed in Table 2. The PCR fragment was ligated into the suicide vector pVO155 (36), and this construction was introduced into E. coli DH5α. PCR fragments cloned into pVO155 were sequenced to verify their identity. The plasmid was transferred into strain CFBP4834-R by triparental mating (17) using the mobilizing E. coli K-12(pRK600) (16).
TABLE 2.
PCR primers used in this study to construct and validate mutations in hrp genes of X. fuscans subsp. fuscans CFBP4834-R
| Target gene | PCR primer
|
Relevant characteristic | PCR fragment size (bp) | |
|---|---|---|---|---|
| Namea | Nucleotide sequenceb | |||
| hrcJ | HrcJ-F | GGACTAGTCCTGCTGCATGCGGGCGTGGA | SpeI site | 335 |
| HrcJ-R | CCGCTCGAGCGGGAAAGCGGGTCGTTGTTGG | XhoI site | ||
| HrcJ-ext-F | CATGACGCTCATTCCTCCTGT | To be used with ProR | 891 | |
| hrcR | HrcR-F | GGACTAGTCCGCTGGTGGTCATCATGCTGG | SpeI site | 293 |
| HrcR-R2 | CCGCTCGAGCGGTGTTTGAGGAGGAATTGC | XhoI site | ||
| HrcR-ext-R | TCACCGATAGCTCAGAACCAGG | To be used with ProF | 756 | |
| hrcT | HrcT-F | GGACTAGTCCCCTCGCAGGGCGTGTCGCTG | SpeI site | 329 |
| HrcT-R2 | CCGCTCGAGCGGGTCATCTGCACATTGTTGTA | XhoI site | ||
| HrcT-ext-R | CGTTGGCGGCATCGTGCAAT | To be used with ProF | 850 | |
| hrcV | HrcV-F | CGGGATCCCGATTCGCAAAAACGCCCCTGATT | BamHI site | 454 |
| HrcV-R | GGTCTAGAGCCAGCAGACGCCGCAACACAT | XbaI site | ||
| HrcV-ext-F | TGGCCAACGAGAACGAGACAAG | To be used with ProR | 789 | |
| hrpB2 | HrpB2-F | GGACTAGTCCATGACGCTCATTCCTCCTGT | SpeI site | 357 |
| HrpB2-R | CCGCTCGAGCGGGCATCAACTTGATCTGCT | XhoI site | ||
| HrcB2-ext-R | GGAAAGCGGGTCGTTGTTGG | To be used with ProF | 896 | |
| hrpG | HrpG-F | GGACTAGTCCTGCGAGCTGCTGATCTTCGAT | SpeI site | 467 |
| HrpG-R | CCGCTCGAGCGGTTGTAGATGTGCTGCTCCATGGT | XhoI site | ||
| HrpG-ext-F | CACCAACCAGCATCCTGCTC | To be used with ProR | 1,680 | |
| hrpX | HrpX-F | GGACTAGTCCGCCTACAGCTACATGATCACCAA | SpeI site | 206 |
| HrpX-R2 | CCGCTCGAGCGGCTTCGGCCAGCAGTTCGT | XhoI site | ||
| HrpX-ext-R | TTACCGCTGCAAGGTCTCCATCGG | To be used with ProF | 446 | |
| ProR | TTCACGGGTTGGGGTTTCTACA | pVO155 primer | ||
| ProF | GAGATCCCCAGCCCGCCTAATG | pVO155 primer | ||
For each target gene, the two first primers indicated were designed to construct the mutation. The third primer, which needs to be used with ProF or ProR, was designed for the validation of the mutation and for the specific identification of mutated strains.
Underlined sequences indicate the relevant restriction site.
Disruption of each target gene was verified by PCR using one primer in the plasmid (ProR or ProF) and a second primer in the genome outside the recombinant region (Table 2) and Southern blotting using the recombinant fragment as a specific probe. Single insertion was confirmed by Southern blotting using a vector-specific probe (Table 2).
For the phenotypic characterization of the mutants, the in vitro growth rate of each hrp mutant was compared to that of the wild-type strain CFBP4834-R. Growth curves were established by growing strains at an initial concentration of 1 × 107 CFU ml−1 in 100-well honeycomb microtiter plates (Thermo Electron, France) in 10% TSB (1.7 g liter−1 tryptone, 0.3 g liter−1 soybean peptone, 0.25 g liter−1 glucose, 0.5 g liter−1 NaCl, 0.5 g liter−1 K2HPO4). Plates were incubated at 28°C with continuous shaking (120 rpm) over a period of 2 to 3 days. Growth measurements were done automatically every 2 h by optical density measurements at 600 nm using the Bioscreen C instrument (Labsystems, Helsinki, Finland). Noninoculated wells were used as aseptic controls. The experiment was repeated three times for each strain.
The stability of the constructions was verified in vitro by testing the Km resistance of the bacteria. Liquid cultures at initial concentration of 1 × 106 CFU ml−1 of each strain were cultured in 10% TSB without antibiotic selection pressure at 28°C under constant agitation (120 rpm), and 24 h later they were diluted 10-fold and grown up again; this step was repeated for four days. Finally, bacterial populations were plated on selective (10% TSA supplemented with rifamycin and Km) and nonselective (10% TSA) media and population densities were compared. Three independent cultures of every strain were analyzed, and the experiment was repeated three times. The stability of the mutation was also verified in planta by comparison of the bacterial population densities on selective and nonselective media the day of inoculation and at the last sampling date. To confirm stability of the construction versus antibiotic resistance acquisition and to check cross-contamination among treatments, the identity of 10 colonies per plant that developed on the nonselective medium was verified by PCR using specific primers (Table 2).
A functional complementation experiment with our 4834HRCT mutant was conducted by providing in trans the pIJ3225 plasmid, which is pLAFR1 carrying the hrp cluster of Xanthomonas campestris pv. campestris strain 8004 on a 29,200-bp EcoRI-EcoRI DNA fragment (3).
Plant material.
In planta experiments were conducted with a variety of dry bean (P. vulgaris cv. Flavert) susceptible to common bacterial blight and pepper (Capsicum annuum cv. ECW10R). Seeds were sown in 10- by 10- by 18-cm pots (one seed per pot) containing soil substrate (Neuhaus humin substrat S NF 11-44-551; Proveg, La Rochelle, France). Peppers and beans were grown in growth chambers with 16 h of light (OSRAM; 2/3 metal halide arc discharge lamp type HQi-BT 400W and 1/3 high-pressure sodium lamp type NAV-T 400W) at 25°C (28°C for pathogenicity tests) and 8 h of darkness at 20°C (22°C for pathogenicity tests) and under high (95%) RH. For experiments on phyllosphere colonization and transmission to seeds, the RH was decreased to 50% from 2 days after inoculation. For all experiments, plants were watered three times per week, and once a week water was supplemented with 0.3 g liter−1 nitrogen-phosphorus-potassium fertilizer (18:14:18). Plant inoculations were carried out under quarantine at UMR PaVé, Centre INRA, Beaucouzé, France.
Pathogenicity and hypersensitivity tests.
Pathogenicity tests on bean were performed by grazing the surface of a young trifoliate leaf with cotton gauze soaked in a suspension calibrated at 1 × 107 CFU ml−1. One leaf per plant and three plants per strain were inoculated. Symptoms were recorded daily for 11 days following inoculation. These tests were repeated at least three times for every bacterial strain.
Hypersensitivity tests were performed on pepper by infiltrating bacterial suspensions adjusted to 1 × 108 CFU ml−1 into leaves of 3-week-old plants. The presence or absence of a necrosis localized at the point of inoculation was scored 2 days after inoculation. Every strain was infiltrated three times on a plant, and three plants were inoculated per strain.
Dynamics of bacterial population densities on bean leaves.
Plants at the first trifoliate stage (32) were spray inoculated until runoff with bacterial suspensions at 1 × 106 CFU ml−1 and with sterile distilled water as a control. The environmental conditions used for these experiments and the absence of wounding did not favor disease expression. Spray inoculation of plants is, however, satisfactory for studying bacterial colonization and dispersal. For every strain, the first trifoliate leaf of five plants was collected 3 h and at 1, 4, and 11 days after inoculation. Each leaf was weighed and ground individually (Stomacher 80; Seward, London, United Kingdom) for 2 min at maximum power in 5 ml of distilled water. Every sample and appropriate dilutions were spiral plated (Spiral Biotech, Bethesda, MD) on selective medium to enumerate the inoculated strain. Samples from control plants were plated on 100% TSA to quantify bacterial indigenous population densities. Primary leaves were imprinted on appropriate media with appropriate antibiotics at every sampling date. To avoid cross-contamination, plants receiving a similar treatment were grouped in the growth chamber and were separated by polypropylene walls from other treatments. In each experiment, treatments were randomly distributed, and experiments were repeated at least three times.
Inoculations of beans at flower bud stage and analyses of bacterial transmission to seeds.
Bean plants at the flower bud stage (32) were spray inoculated until runoff with bacterial suspensions at 1 × 105 CFU ml−1. Bacterial population densities in flower buds from 5 plants were quantified at 3 h after spray inoculation, and those in seeds from 10 plants per experiment were quantified at 6 weeks after inoculation. The same experimental design as for phyllosphere colonization experiments was used. Samples (flower buds or seeds) were bulked for each plant. Bulks of flower buds were weighed and ground (Stomacher 80; Seward, London, United Kingdom) for 2 min at maximum power in 5 ml of distilled water. Asymptomatic pods were aseptically dissected under a laminar flow hood such that the seeds did not come in contact with the external portion of the pod or with any instruments that had contacted the external pod surface. Seeds were weighed and soaked overnight at 4°C in 2 ml of sterile distilled water per g of seeds (5.56 seeds g−1). Samples were then vigorously shaken. To quantify population densities of strains with mutations in hrp genes and to look for reversion events, aliquots of 500 μl of samples were spread plated and appropriate dilutions were spiral plated on 10% TSA-rifamycin or 10% TSA-rifamycin-Km medium. For every sample, the identity of 10 colonies grown on 10% TSA-rifamycin was confirmed by PCR with appropriate primers (Table 2). For other strains, aliquots of 500 μl of samples and appropriate dilutions were plated on appropriate medium to quantify bacterial population densities.
To compare the occurrences of the vascular and the floral pathways in bacterial transmission to seeds, two different inoculation methods were used. On a first set of plants, direct flower bud inoculation was performed by depositing 20 μl of an inoculum of 1 × 106 CFU ml−1 per flower bud on three groups of flower buds per plant, taking every possible precaution to avoid dispersion of the inoculum on leaves. After inoculum drying, inoculated flower buds were enclosed in transparent cellophane bags to avoid any subsequent contamination of leaves by contact with inoculated flowers. On a second set of plants, three groups of flower buds per plant were protected with transparent cellophane bags before plants were inoculated by spraying the phyllosphere with an inoculum of 1 × 105 CFU ml−1. Bags remained in situ until sampling at harvest time. Three hours after inoculations, bacterial population densities were quantified on leaves bulked for each plant when flower buds were inoculated by depositing drops of inoculum and on the third trifoliate leaf for spray-inoculated plants. Bacterial population densities were also quantified in inoculated and protected flower buds. At harvest (6 weeks after inoculation) bacterial population densities in pods and seeds were quantified as described above for leaf populations. Ten plants per strain and per treatment were analyzed on the day of inoculation and 30 plants per strain and per treatment at harvest. The same experimental design as for phyllosphere colonization experiments was used.
The dynamics of population densities of CFBP4834-R and 4834HRCV were determined in reproductive organs of plants inoculated by depositing 20 μl of an inoculum of 1 × 106 CFU ml−1 per flower bud. Flower buds and pods were sampled at 3 h and at 1, 2, 3, 4, 15, 28, and 35 days after inoculation on five plants per strain and per sampling date. Sample analyses were performed as described above.
Statistical analyses.
Statistical analyses were performed using Statbox Pro software (Grimmer Logiciels, Optima France). Log-transformed data were analyzed with Kruskal-Wallis and Mann-Whitney tests. Comparisons of transmission frequencies were based on Pearson's χ2 test. To compare paired population densities quantified on selective and nonselective media, Wilcoxon's signed-ranks test for two groups was used (41).
Nucleotide sequence accession numbers.
The hrp genes in CFBP4834-R have been assigned NCBI accession numbers EU215387, EU215388, and EU215389.
RESULTS
Bacterial colonization of the bean phyllosphere and transmission to bean seeds are functions of the type of interaction.
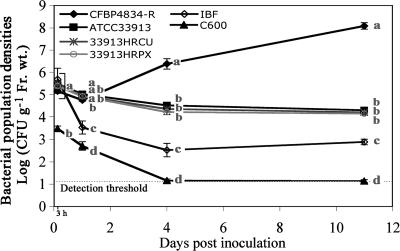
In order to characterize the role of hrp genes in the survival and the multiplication of X. fuscans subsp. fuscans in the bean phyllosphere, we first determined standard bacterial behaviors by the dynamics of bacterial population densities in null (E. coli C600), compatible (X. fuscans subsp. fuscans CFBP4834-R), and incompatible (X. campestris pv. campestris ATCC 33913) interactions. Despite the same initial inoculum concentration (1 × 106 CFU ml−1), E. coli C600 population densities quantified in leaves at 3 hours after inoculation were about 0.01-fold those of CFBP4834-R and ATCC 33913 (Fig. 1), showing that E. coli C600 was impaired in its adhesion to bean leaves or was affected early by the environmental conditions of the phyllosphere. Moreover, E. coli C600 was not able to multiply in the phyllosphere and was not recovered after a few days. Only X. fuscans subsp. fuscans CFBP4834-R was able to multiply on bean leaves, reaching high population densities of around 1 × 108 CFU g−1 of fresh weight at 11 days after the bacteria were inoculated. Throughout the experiment, population densities of X. campestris pv. campestris ATCC 33913 remained stable at 1 × 105 CFU g−1 of fresh weight and were significantly (P < 0.05) higher than those of the indigenous bacterial flora. No symptoms were observed on any aerial parts of beans during experiments.
FIG. 1.
Colonization of bean by wild-type strains X. fuscans subsp. fuscans (CFBP4834-R), X. campestris pv. campestris (ATCC 33913), and E. coli C600; by strains with mutations in hrp genes (33913HRCU and 33913HRPX); and by indigenous bacterial flora (IBF). Bacterial population densities were determined on bean leaves sampled at 3 h and 1, 4, and 11 days after spray inoculation (1 × 106 CFU ml−1). Means and SEMs were calculated for five leaves per sampling date. Mean population densities followed by different letters are significantly (P < 0.05) different on the basis of the Mann-Whitney test.
X. fuscans subsp. fuscans CFBP4834-R, X. campestris pv. campestris ATCC 33913, and E. coli C600 were also separately inoculated on beans at the flower bud stage to determine their rate of transmission to the new generation of seeds. No seeds of the 30 inoculated plants were contaminated with E. coli C600 (Table 3). X. fuscans subsp. fuscans CFBP4834-R was transmitted to the seeds at high frequency, whereas the frequency of X. campestris pv. campestris ATCC 33913 transmission to seeds was significantly (P < 0.05) lower, with much lower population densities on contaminated seeds than for X. fuscans subsp. fuscans CFBP4834-R. Thus, three different behaviors were recorded: (i) in the null interaction, there was no bacterial survival and no bacterial transmission to seeds; (ii) in the incompatible interaction, bacteria survived in the phyllosphere and were transmitted to seeds at low frequencies and with low population sizes; and (iii) in the compatible interaction, bacteria multiplied on leaves and were transmitted to seeds at high frequencies and with high bacterial population sizes.
TABLE 3.
Frequencies of bacterial transmission to bean seeds and mean bacterial population densities after spray inoculation (1 × 105 CFU ml−1) of bean at the flower bud stage
| Strain | Frequencya | Mean bacterial population density (SEM) on contaminated harvest, log CFU g−1 (fresh wt) |
|---|---|---|
| CFBP4834-R | 0.87 | 6.63 (0.33) |
| ATCC 33913 | 0.23* | 1.91 (0.47) |
| C600 | 0* | NDb |
| 4834HRCJ | 0.17* | 2.18 (0.72) |
| 4834HRCR | 0.40* | 1.71 (0.24) |
| 4834HRCT | 0.17* | 0.96 (0.14) |
| 4834HRCV | 0.37* | 2.29 (0.39) |
| 4834HRPB2 | 0.27* | 1.81 (0.28) |
| 4834HRPG | 0.07* | 1.85 (0.38) |
| 4834HRPX | 0.07* | 1.31 (0.17) |
| 33913HRCU | 0.13* | 1.15 (0.27) |
| 33913HRPX | 0.23* | 1.70 (0.53) |
A total of 30 plants were analyzed. *, significantly different from value for the wild-type strain CFBP4834-R by χ2 test.
ND, not determined.
Characterization of the T3SS of X. fuscans subsp. fuscans strain CFBP4834-R.
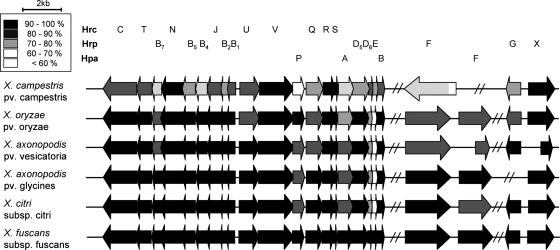
As the X. fuscans subsp. fuscans genome is not yet sequenced, specific primers of hrp genes were designed based on consensus sequences obtained by comparison of hrp cluster sequences of Xanthomonas citri subsp. citri strain 306 (11), X. axonopodis pv. glycines strain 8ra (24), X. axonopodis pv. vesicatoria strain 85-10 (42), Xanthomonas oryzae pv. oryzae strain KACC10331 (25), and X. campestris pv. campestris strain ATCC 33913 (38). This allowed the identification and the sequencing of a complete set of hrp genes in CFBP4834-R. Sequence analysis of the Hrp proteins of CFBP4834-R showed that they were more similar to the Hrp proteins of X. citri subsp. citri (strain 306) and X. axonopodis pv. glycines (strain 8ra) than to those of X. axonopodis pv. vesicatoria (strain 85-10) or to those of X. campestris pv. campestris (strain ATCC 33913) (Fig. 2). In contrast, the sequence of the surface-exposed domain of the pilin HrpE was more closely related to that of X. campestris pv. campestris (strain ATCC 33913) than to that of X. citri subsp. citri (strain 306).
FIG. 2.
Schematic overview of the hrp genes from different sequenced xanthomonads in comparison with X. fuscans subsp. fuscans hrp genes. Genes of the following strains are shown: X. campestris pv. campestris strain ATCC 33913 (11), X. axonopodis pv. vesicatoria strain 85-10 (42), X. citri subsp. citri strain 306 (11), X. axonopodis pv. glycines strain 8ra (24), X. oryzae pv. oryzae strain KACC10331 (25), and X. fuscans subsp. fuscans strain CFBP4834-R. Arrows indicate the sizes, positions, and orientations of the hrp, hrc, and hpa genes. The identity of each protein sequence with its homolog in X. fuscans subsp. fuscans is presented by use of a black/gray color scale. X. fuscans subsp. fuscans amino acid sequences were compared using the NCBI BLAST website http://www.ncbi.nlm.nih.gov/BLAST/with default parameters.
In order to evaluate the role of the T3SS both in colonization processes in the absence of symptoms and in transmission of X. fuscans subsp. fuscans to bean seeds, we constructed strains with mutations in the hrpB2, hrcJ, hrcR, hrcT, hrcV, hrpG, and hrpX genes (Table 1). PCR amplifications using specific primers (Table 2) confirmed that pVO155 unique insertions were at the correct positions. Southern blot hybridizations confirmed the single plasmid insertion event. Population densities of each strain with mutations enumerated on nonselective medium were not (P > 0.05) higher that those enumerated on selective medium, showing that all constructions were stable both in vitro and in planta. Growth rates of the wild-type CFBP4834-R and of every strain with a mutation in hrp genes were similar in 10% TSB, indicating that the mutation of a given hrp gene did not impair the in vitro growth of the corresponding strain. Furthermore, no reversion was observed in vitro after 54 generations without selection pressure (data not shown).
When inoculated onto bean plants, none of the strains with mutations in hrp genes was able to cause any water-soaking symptoms compared to the wild-type strain (data not shown). Moreover, following infiltrations into resistant pepper leaves carrying the BsT and Bs1 resistance genes, none of the strains with mutations in hrp genes induced a hypersensitive response (HR), the typical necrotic lesion associated with plant resistance. In contrast, the wild-type X. campestris pv. campestris strain ATCC 33913 carrying the avrBs1 gene (11) induced the expected classical HR, and the wild-type strain CFBP4834-R of X. fuscans subsp. fuscans showed the same weak spotty HR as described by Escolar and colleagues (13). Pathogenicity of the 4834HRCT strain with a mutation in the hrcT gene was restored by complementation (data not shown). Together, these results showed that X. fuscans subsp. fuscans strain CFBP4834-R had a functional T3SS.
Impact of mutations in hrp genes of CFBP4834-R on bacterial multiplication and bacterial survival in the phyllosphere.
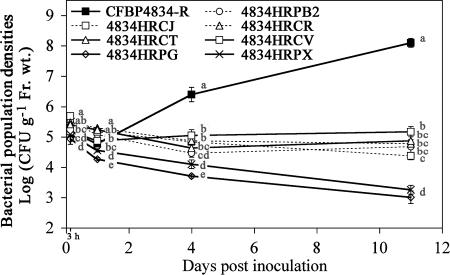
Individual inoculation of strains with mutations in hrp genes and the wild-type strain resulted in three different behaviors during the asymptomatic colonization of the bean phyllosphere (Fig. 3). First, in the compatible interaction, CFBP4834-R population densities increased from the first day after inoculation and reached 1 × 108 CFU g−1 of fresh weight at 11 days after inoculation. Mean CFBP4834-R population densities at 11 days after inoculation were significantly higher (P < 0.05) than those determined on the first day after inoculation. Second, strains (4834HRPB2, 4834HRCJ, 4834HRCR, 4834HRCT, and 4834HRCV) with mutations in structural hrp genes (hrpB2, hrcJ, hrcR, hrcT, and hrcV) were not able to multiply efficiently throughout the experiment, and their population densities stabilized at 1 × 105 CFU g−1 of fresh weight, as observed previously in the incompatible interaction for ATCC 33913 (Fig. 1). For these strains, their mean population densities on the 11th day after inoculation were not significantly (P > 0.05) higher than those determined on the first day after inoculation. To gain insight into the milieu colonized by the strains, we imprinted the surface of leaves on agar medium. Imprinting of leaves inoculated by strain 4834HRCV with a mutation in the hrcV gene (Fig. 4) did not show any substantial differences in surface populations compared to leaves inoculated with the wild-type strain CFBP4834-R. Surface populations illustrated by number of colonies sometimes seemed even higher than what was observed for the wild-type strain. Similar results were obtained for other strains with mutations in structural hrp genes. Third, the behavior of strains (4834HRPX and 4834HRPG) with mutations in the regulatory hrp genes (hrpX and hrpG) was the most affected, with a decrease of their population densities throughout the experiment. Their mean population densities determined on the first day after inoculation were significantly (P > 0.05) higher than those determined 11 days after inoculation. Imprinting of leaves inoculated by strain 4834HRPG, with a mutation in the hrpG gene (Fig. 4), showed a lower number of colonies on leaves than for the wild-type strain CFBP4834-R and strains with mutations in structural hrp genes (Fig. 4).
FIG. 3.
Colonization of bean by X. fuscans subsp. fuscans strain CFBP4834-R and strains with mutations in hrp genes. Bacterial population densities were determined on bean leaves sampled at 3 h and 1, 4, and 11 days after spray inoculation (1 × 106 CFU ml−1). Means and SEMs were calculated for five leaves per sampling date. Mean population densities followed by different letters are significantly (P < 0.05) different on the basis of the Mann-Whitney test.
FIG. 4.
Imprinting of bean leaves (upper and lower surfaces) during phyllosphere colonization by X. fuscans subsp. fuscans strain CFBP4834-R and strains mutated in the hrcV (4834HRCV) or hrpG (4834HRPG) gene after spray inoculation (1 × 106 CFU ml−1). Three leaves per strain and per sampling date are presented. Leaves were sampled at 3 h after inoculation (day 0) and 1 and 11 days later.
The dynamics of the bacterial population densities of two strains of X. campestris pv. campestris, one with a mutation in a structural hrp gene (hrcU) and a second with a mutation in a regulatory hrp gene (hrpX), were quantified in the bean phyllosphere in comparison with the wild-type strain ATCC 33913. Strains of X. campestris pv. campestris with mutations in hrp genes behaved similarly to their parental strain in the bean phyllosphere throughout the 11 days of the experiment (Fig. 1). Their population densities were not significantly different (P < 0.05) from each other and from those of ATCC 33913 at any sampling date. This absence of differences among population densities indicated that a functional T3SS did not play any role in X. campestris pv. campestris survival in the nonhost phyllosphere.
Analysis of transmission to bean seeds for strains with mutations in hrp genes.
The rates of transmission of each strain of X. fuscans subsp. fuscans CFBP4834-R with mutations in hrp genes to the bean seeds were significantly (P < 0.05) altered compared to those of transmission of the wild-type strain (Table 3). The strains with mutations in hrp genes also showed significantly (P < 0.05) lower population densities on contaminated seeds (around 1 × 102 CFU g−1 of fresh weight) than the wild-type strain CFBP4834-R (4.3 × 107 CFU g−1 of fresh weight). Strains (4834HRPG and 4834HRPX) with mutations in regulatory genes (hrpG and hrpX) appeared to be the most altered in their ability to be transmitted to the bean seeds, with very low frequencies (0.07 for each) and low final population densities on seeds. CFBP4834-R strains with mutations in hrp genes were transmitted to bean seeds at frequencies similar to those for X. campestris pv. campestris ATCC 33913 in the incompatible interaction with bean. ATCC 33913 strains with mutations in hrp genes were transmitted to bean seeds at frequencies (around 0.20) similar to those for their parental strain. This result indicates that in this incompatible interaction, a functional T3SS is not required for transmission to seeds.
Analysis of pathways used by CFBP4834-R and strains with mutations in hrp genes for transmission to seeds.
To monitor the bacterial transmission to seeds by the vascular pathway, reproductive organs were protected before spray inoculation of leaves. Using this inoculation method, only wild-type strain CFBP4834-R of X. fuscans subsp. fuscans was able to be transmitted to pods and seeds with high frequencies (0.85 and 0.63, respectively), whereas none of 4834HRPG, 4834HRCT, and 4834HRCV strains were able to be transmitted to pods or seeds. The mean CFBP4834-R population density on contaminated pods was 4.47 × 103 CFU g−1 (standard error of the mean [SEM], 3.08 CFU g−1) of fresh weight, and that on contaminated seeds was 158 CFU g−1 (SEM, 2.07 CFU g−1) of fresh weight.
To monitor bacterial transmission to seeds by the floral pathway, an inoculum was deposited directly in flower buds. These experiments showed that CFBP4834-R was also able to be transmitted to both pods and seeds with high frequencies (0.9). CFBP4834-R strains with mutations in regulatory hrp gene (4834HRPG) and in structural hrp genes (4834HRCT and 4834HRCV) showed high transmission rates to pods, with frequencies of 0.67, 0.52, and 0.89, respectively. These strains could be transmitted to seeds with lower frequencies (0.1, 0.2, and 0.04, respectively) than CFBP4834-R. Under our asymptomatic conditions, transmission to seeds by contact with pod symptoms was prevented, and great care was taken to avoid any seed contamination with pod tissue while collecting seeds.
The lower bacterial transmission to pods is not a consequence of a lower initial colonization of the strains with mutations in hrp genes in flowers compared to CFBP4834-R. The dynamics of flower contamination following flower bud inoculation were similar (P > 0.05) for CFBP4834-R and 4834HRCV until the fourth day after inoculation. Precisely, the mean population densities of CFBP4834-R were 2.33 × 105, 4.05 × 105, 3.58 × 106, and 1.43 × 106 CFU g−1 of fresh weight, and those of 4834HRCV were 2.16 × 105, 1.63 × 106, 3.48 × 106, and 3.16 × 105 CFU g−1 of fresh weight, at days 1 to 4 after inoculation, respectively. On the 15th, 28th, and 35th days after inoculation, population densities were significantly (P < 0.05) lower on pods colonized by 4834HRCV (2.26 × 104, 2.29 × 104, and 1.28 × 105 CFU g−1 of fresh weight, respectively) than on pods colonized by CFBP4834-R (1.94 × 107, 2.77 × 107, and 3.51 × 108 CFU g−1 of fresh weight, respectively). Populations were 3 orders of magnitude lower for the strain with a mutation in the hrcV gene than for the wild type at each sampling date.
DISCUSSION
In this study, we showed that a classical pathogenicity determinant, the T3SS encoded by the hrp gene cluster, is involved in X. fuscans subsp. fuscans early and late colonization processes but not in its survival on its host. The T3SS is also required for a high frequency of pathogen transmission to the seeds. However, low rates of bacterial transmission to seeds are independent of a functional T3SS, as is bacterial survival on nonhost plants. Previous studies have demonstrated that alteration of the T3SS in other bacterial pathogens abolishes pathogenicity (8, 24, 27, 52) and in planta multiplication (7, 27). Our experimental approach focused on the measurement of phyllosphere colonization processes and transmission to seeds by wild-type X. fuscans subsp. fuscans and strains with mutations in hrp genes in low-RH conditions with low initial bacterial population densities (below 1 × 106 CFU g−1 of fresh weight). These conditions do not favor disease expression for X. fuscans subsp. fuscans (22) and were used to reproduce the most commonly observed situation in a temperate climate for a bacterial plant pathogen, i.e., asymptomatic colonization of the host. Indeed, the interactions between a susceptible host and a virulent plant pathogenic bacterium may only rarely result in disease symptoms (21). We show here that the absence of disease manifestation does not compromise active host colonization by X. fuscans subsp. fuscans, including the phyllosphere and the reproductive organs. This behavior of X. fuscans subsp. fuscans is not restricted to survival; it is therefore different from that of X. campestris pv. campestris on bean in these environmental conditions.
During bean phyllosphere colonization, X. fuscans subsp. fuscans colonizes both external and internal compartments of the plant (22, 51). It is probable that efficient asymptomatic bean colonization by X. fuscans subsp. fuscans was mainly due to an endophytic colonization. Indeed, we observed with leaf imprinting that the number of colonies of X. fuscans subsp. fuscans that could be removed by printing the leaf surface on agar medium decreased in the days following spray inoculation whereas, the total population densities of X. fuscans subsp. fuscans in the phyllosphere significantly increased, suggesting an important endophytic colonization or the aggregation of X. fuscans subsp. fuscans in biofilms that were tightly adherent to the leaf surface. Jacques and collaborators (22) already demonstrated that X. fuscans subsp. fuscans aggregates in biofilms on the bean leaf surface. Endophytic colonization of the leaf parenchyma may be particularly well supported by the presence of more nutrients in the internal compartment of leaves than on the leaf surface (31).
Strains with alterations in the T3SS were impaired in leaf colonization. Leaf imprinting showed that similar numbers of colonies were recovered from the surface of the leaf for strains with mutations in structural hrp genes and the wild-type strain, whereas strains with mutations in hrp genes established total phyllosphere population densities that were 1,000-fold lower than those of the wild-type strain. This means that the decrease probably concerned mainly the endophytic compartment. Interestingly, similar conclusions were obtained for other phytopathogens such as P. syringae and Erwinia amylovora. P. syringae pv. tomato hrp mutants are impaired in the endophytic colonization of their host (7). Furthermore, it has been shown that structural hrp genes are induced inside leaf tissue and not on the leaf surface (9) and that the elicitor of hrp genes via PrhA in Ralstonia solanacearum is a nondiffusible plant cell wall component (2).
We demonstrated that X. fuscans subsp. fuscans strains deficient in the regulatory genes hrpG and hrpX were more affected than strains deficient in the T3SS structural genes in their colonization capacities, suggesting that HrpG and HrpX regulate additional genes beyond the T3SS in X. fuscans subsp. fuscans. Moreover, the population size of 4834HRPG, a strain mutated in the hrpG gene, was slightly but significantly lower than that of the wild type at 3 h after leaf inoculations, suggesting that the strain was impaired in leaf adhesion. This could have affected its potential for later leaf colonization. Interestingly, we also showed that this strain (4834HRPG) was also hypermotile (data not shown). It has been shown for Ralstonia solanacearum that HrpG positively regulates genes involved in attachment and protection response functions (43). Those authors proposed that HrpG serves as a molecular switch between saprophytic and pathogenic lifestyles (43). Indeed, there is an opposite regulation in the phytopathogenic bacterium Erwinia amylovora between the virulence-associated T3SS and the flagellar system (9). Moreover, it was also demonstrated that nutrient acquisition in X. campestris pv. campestris could involve plant carbohydrate scavenging by TonB-dependent receptors and that some TonB-dependent receptors could be under the regulation of the hrpG gene but also are independent of a functional T3SS (5). It is therefore tempting to hypothesize that nutrient acquisition during saprophytic development of these plant pathogenic bacteria is dependent on HrpG regulation but also independent of a functional T3SS. Alternatively, as a consequence of the positive regulation of bacterial adhesion by HrpG (43), alterations in the leaf colonization process for strains with mutations in hrp genes could also result from altered capacities to aggregate in adherent biofilms on the leaf surface. Surprisingly, X. fuscans subsp. fuscans strains deficient in the regulatory genes hrpG and hrpX were still able to be transmitted to seeds. This perhaps is linked to the very different chemical and physical natures of these two environments (phyllosphere and flower buds). This is also coherent with other reports dealing with the pleiotropic phenotype of strains with mutations in hrp regulatory genes (35, 43).
We confirmed that the translated sequences of the X. fuscans subsp. fuscans hrp genes shared strong homology with Hrp proteins of other X. axonopodis pathovars sensu Vauterin et al. (22, 45). The percentage of identity corroborated the predicted phylogeny of xanthomonads, namely, that X. fuscans subsp. fuscans was more closely related to X. citri subsp. citri and X. axonopodis pv. glycines than to X. axonopodis pv. vesicatoria and was more closely related to X. oryzae pv. oryzae than to X. campestris pv. campestris (39, 44). However, the Hrp pilus subunit, HrpE, seemed to have evolved differently than other Hrp proteins. We found that the surface-exposed domain of the X. fuscans subsp. fuscans HrpE was more closely related to that of X. campestris pv. campestris than to that of X. citri subsp. citri. A structure in three domains is proposed for HrpE: (i) a domain containing the T3S signal in the N terminus, (ii) a surface-exposed domain, and (iii) a polymerization domain in the C terminus (48). Weber and Koebnik (49) showed that the C terminus is subjected to purifying selection and the surface-exposed domain to positive selection, corresponding to an evolutionary adaptation of this surface structure to avoid recognition by the plant defense system (49). Curiously, we found poor homology (30%) with that of X. axonopodis pv. phaseoli, which is also a bean pathogen and was until recently considered to belong to same species and pathovar (22, 45).
From both ecological and agricultural perspectives, transmission of a pathogen to the next generations of its host is a major critical step. We are not aware of any other study designed to look explicitly at the effect of loss of a major class of pathogenicity determinants on the transmission of a pathogen to the seeds of its host. One pathway for bacterial transmission to seeds (i.e., seed pollution) was suppressed in our experimental approach by avoiding contact of seeds with symptoms (the environmental conditions did not allow symptom development) or contaminated pod tissue (by delicate extraction of seeds from pods). The two remaining pathways for bacterial transmission to seeds are the vascular pathway, in which bacteria colonize reproductive organs through the vascular system, and the floral pathway, in which bacteria colonize the pistil and the ovary (29) to finally reach and contaminate the seeds. We found that alteration of the T3SS drastically decreased transmission to seeds by X. fuscans subsp. fuscans. This could be the result of low bacterial population densities of strains with mutations in hrp genes but also of transmission pathways not available for such strains limiting contamination of seeds. Our experiments involving leaf inoculation associated with protection of flower buds showed that strains with mutations in hrp genes were completely unable to use the vascular pathway for transmission to seeds. For the wild-type strain, vascular transmission to seeds was responsible for the contamination of seeds for more than 50% of the plants. By direct flower bud inoculations, X. fuscans subsp. fuscans hrp strains with mutations in hrp genes could be transmitted to seeds through the stylar tissues. Floral transmission to seeds via the stylar tissue was demonstrated for Acidovorax avenae subsp. citrulli in watermelon blossoms (26) and for a bacterial biocontrol agent (15). Floral transmission to seeds is, however, less efficient for strains with mutations in hrp genes than for the wild-type strain. This was not a consequence of a low bacterial installation on flower buds, as population densities remained similar for the wild-type strain and 4834HRCV for at least 4 days following inoculation of flower buds.
On the basis of our results, we propose different stages to describe colonization of aerial plant parts by bacteria. First, bacteria adhere on the plant surface after arrival. This step was not possible for E. coli on bean leaves, based on the 100-fold-lower population densities for E. coli immediately following inoculation. Second, bacteria survive in the phyllosphere. This survival is certainly limited to the surface of the leaf. Indeed, based on the comparison of the population densities quantified in the phyllosphere and the number of colonies found on the leaf surface by leaf imprinting for X. campestris pv. campestris, strains mutated in structural hrp genes, and the wild-type strain of X. fuscans subsp. fuscans, we conclude that the survival of X. campestris pv. campestris and of strains mutated in structural hrp genes was restricted mainly to the leaf surface. This basic epiphytic competence depends partially on the master hrp regulators (HrpG and HrpX) but not on a functional T3SS. On the basis of the model proposed by Jones and Dangl (23), it may be hypothesized that bean defense reactions induced after recognition of some plant-associated molecular patterns of these strains could act to limit internal colonization by X. campestris pv. campestris and strains of X. fuscans subsp. fuscans with mutations in hrp structural genes. Meanwhile, the X. fuscans subsp. fuscans wild-type strain may inject effectors through its T3SS to suppress the plant defense reactions or prevent its recognition. Strains of X. fuscans subsp. fuscans with mutations in hrp structural genes and X. campestris pv. campestris are able to colonize new organs (data not shown) and contaminate reproductive organs such as flowers and seeds. The frequencies of such events and the associated population densities, however, are low. It could be hypothesized that X. campestris pv. campestris transmission to seeds operates by a kind of saprophytism via the floral structures because of the abundance of nutrients in these organs (33). Furthermore, Ngugi and Scherm (33) suggest that there are no inducible defense responses in flowers. Third, in the case of a compatible interaction, an increase in the density of the bacterial population colonizing the host is dependent on a functional T3SS even in the absence of symptoms. This colonization is mainly endophytic and leads to an efficient transmission of the pathogen to the seeds.
Together, the results reported in this study indicate that hrp genes are implicated in early and late stages of host phyllosphere colonization by X. fuscans subsp. fuscans and in transmission to host seeds. This new finding opens questions about the genes regulated by the master regulators HrpG and HrpX and about the physical role of the T3SS in these processes.
Acknowledgments
This work was supported by a grant from Conseil Régional des Pays de la Loire.
We are grateful to M. Arlat and E. Lauber for providing strains and the mutagenesis protocol. We thank J. Benard and P. Horeau for plant production; S. Domecyn, J. Menat, and N. Sommerlatt for assistance; and G. Beattie and T. Boureau for critical review of the manuscript.
Footnotes
Published ahead of print on 1 March 2008.
REFERENCES
- 1.Agarwal, V. K., and J. B. Sinclair. 1987. Principles of seed pathology, vol. 1. CRC Press Inc., Boca Raton, FL.
- 2.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlat, M., C. L. Gough, C. E. Barber, C. Boucher, and M. J. Daniels. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:593-601. [DOI] [PubMed] [Google Scholar]
- 4.Beattie, G. A., and S. E. Lindow. 1995. The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 33:145-172. [DOI] [PubMed] [Google Scholar]
- 5.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonas, U., R. E. Stall, and B. Staskawicz. 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218:127-136. [DOI] [PubMed] [Google Scholar]
- 7.Boureau, T., J. Routtu, E. Roine, S. Taira, and M. Romantschuk. 2002. Localization of hrpA-induced Pseudomonas syringae pv. tomato DC3000 in infected tomato leaves. Mol. Plant Pathol. 3:451-460. [DOI] [PubMed] [Google Scholar]
- 8.Buttner, D., and U. Bonas. 2002. Getting across bacterial type III effector proteins on their way to the plant cell. EMBO J. 21:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesbron, S., J. P. Paulin, M. Tharaud, M. A. Barny, and M. N. Brisset. 2006. The alternative sigma factor HrpL negatively modulates the flagellar system in the phytopathogenic bacterium Erwinia amylovora under hrp-inducing conditions. FEMS Microbiol. Lett. 257:221-227. [DOI] [PubMed] [Google Scholar]
- 10.Darrasse, A., C. Bureau, R. Samson, C. Morris, and M.-A. Jacques. 2007. Contamination of bean seeds by Xanthomonas axonopodis pv. phaseoli associated with low bacterial densities in the phyllosphere under field and greenhouse conditions. Eur. J. Plant Pathol. 119:203-215. [Google Scholar]
- 11.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 12.Devoret, R., and M. Blanco. 1970. Mutants of Escherichia coli K12 (λ)+ non-inducible by thymine deprivation. Mol. Gen. Genet. 107:272-280. [Google Scholar]
- 13.Escolar, L., G. Van Den Ackerveken, S. Pieplow, O. Rossier, and U. Bonas. 2001. Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol. Plant Pathol. 2:287-296. [DOI] [PubMed] [Google Scholar]
- 14.Fenton, A., J. P. Fairbairn, R. Norman, and P. J. Hudson. 2002. Parasite transmission: reconciling theory and reality. J. Anim. Ecol. 71:893-905. [Google Scholar]
- 15.Fessehaie, A., and R. R. Walcott. 2005. Biological control to protect watermelon blossoms and seed from infection by Acidovorax avenae subsp. citrulli. Phytopathology 95:413-419. [DOI] [PubMed] [Google Scholar]
- 16.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 18.Gurlebeck, D., F. Thieme, and U. Bonas. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233-255. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Hirano, S. S., A. O. Charkowski, A. Collmer, D. K. Willis, and C. D. Upper. 1999. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc. Natl. Acad. Sci. USA 96:9851-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques, M. A., K. Josi, A. Darrasse, and R. Samson. 2005. Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field-grown beans. Appl. Environ. Microbiol. 71:2008-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, J. D. G., and J. L. Dangl. 2006. The plant immune system. Proc. Natl. Acad. Sci. USA 444:323-329. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. G., B. K. Park, C. H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessl, J. T., A. Fessehaie, and R. R. Walcott. 2007. Colonization of female watermelon blossoms by Acidovorax avenae ssp. citrulli and the relationship between blossom inoculum dosage and seed infestation. J. Phytopathol. 155:114-121. [Google Scholar]
- 27.Lindgren, P. B., R. C. Peet, and N. J. Panopoulos. 1986. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J. Bacteriol. 168:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maude, R. B. 1996. Seedborne diseases and their control: principles & practice. CAB International, Oxon, United Kingdom.
- 30.McCallum, H., N. Barlow, and J. Hone. 2001. How should pathogen transmission be modelled? Trends Ecol. Evol. 16:295-300. [DOI] [PubMed] [Google Scholar]
- 31.Mercier, J., and S. E. Lindow. 2000. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael, T. E. 1994. The bean plant, p. 1-4. In R. Hall (ed.), Compendium of bean diseases. APS Press, St. Paul, MN.
- 33.Ngugi, H. K., and H. Scherm. 2006. Biology of flower-infecting fungi. Annu. Rev. Phytopathol. 44:261-282. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 35.Occhialini, A., S. Cunnac, N. Reymond, S. Genin, and C. Boucher. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18:938-949. [DOI] [PubMed] [Google Scholar]
- 36.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 37.Puhler, A., M. Arlat, A. Becker, M. Gottfert, J. P. Morrissey, and F. O'Gara. 2004. What can bacterial genome research teach us about bacteria-plant interactions? Curr. Opin. Plant Biol. 7:137-147. [DOI] [PubMed] [Google Scholar]
- 38.Qian, W., Y. Jia, S. X. Ren, Y. Q. He, J. X. Feng, L. F. Lu, Q. Sun, G. Ying, D. J. Tang, H. Tang, W. Wu, P. Hao, L. Wang, B. L. Jiang, S. Zeng, W. Y. Gu, G. Lu, L. Rong, Y. Tian, Z. Yao, G. Fu, B. Chen, R. Fang, B. Qiang, Z. Chen, G. P. Zhao, J. L. Tang, and C. He. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rademaker, J. L. W., F. J. Louws, M. H. Schultz, U. Rossbach, L. Vauterin, J. Swings, and F. J. de Bruijn. 2005. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology 95:1098-1111. [DOI] [PubMed] [Google Scholar]
- 40.Schaad, N. W., E. Postnikova, G. H. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2005. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov. X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 28:494-518. [DOI] [PubMed] [Google Scholar]
- 41.Sokal, R. R., and F. J. Rohlf. 1969. Biometry. The principles and practice of statistics in biological research. W. H. Freeman & Co., San Francisco, CA.
- 42.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Buttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klosgen, T. Patschkowski, C. Ruckert, O. Rupp, S. Schneiker, S. C. Schuster, F. J. Vorholter, E. Weber, A. Puhler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valls, M., S. Genin, and C. Boucher. 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 45.Vauterin, L., J. Rademaker, and J. Swings. 2000. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677-682. [DOI] [PubMed] [Google Scholar]
- 46.Vidaver, A. K. 1993. Xanthomonas campestris pv. phaseoli: cause of common bacterial blight of bean, p. 40-44. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman & Hall, London, United Kingdom.
- 47.Walcott, R. R., R. D. Gitaitis, and A. C. Castro. 2003. Role of blossoms in watermelon seed infestation by Acidovorax avenae subsp. citrulli. Phytopathology 93:528-534. [DOI] [PubMed] [Google Scholar]
- 48.Weber, E., and R. Koebnik. 2005. Domain structure of HrpE, the Hrp pilus subunit of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 187:6175-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, E., and R. Koebnik. 2006. Positive selection of the Hrp pilin HrpE of the plant pathogen Xanthomonas. J. Bacteriol. 188:1405-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weller, D. M., and A. W. Saettler. 1980. Evaluation of seedborne Xanthomonas phaseoli and Xanthomonas phaseoli var. fuscans as primary inocula in bean blights. Phytopathology 70:148-152. [Google Scholar]
- 51.Zaumeyer, W. J. 1930. The bacterial blight of beans caused by Bacterium phaseoli. Technical bulletin 186. U.S. Department of Agriculture, Washington, DC.
- 52.Zou, L. F., X. P. Wang, Y. Xiang, B. Zhang, Y. R. Li, Y. L. Xiao, J. S. Wang, A. R. Walmsley, and G. Y. Chen. 2006. Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72:6212-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]