Abstract
The alphaproteobacterium Nitrobacter hamburgensis X14 is a gram-negative facultative chemolithoautotroph that conserves energy from the oxidation of nitrite to nitrate. Sequencing and analysis of the Nitrobacter hamburgensis X14 genome revealed four replicons comprised of one chromosome (4.4 Mbp) and three plasmids (294, 188, and 121 kbp). Over 20% of the genome is composed of pseudogenes and paralogs. Whole-genome comparisons were conducted between N. hamburgensis and the finished and draft genome sequences of Nitrobacter winogradskyi and Nitrobacter sp. strain Nb-311A, respectively. Most of the plasmid-borne genes were unique to N. hamburgensis and encode a variety of functions (central metabolism, energy conservation, conjugation, and heavy metal resistance), yet ∼21 kb of a ∼28-kb “autotrophic” island on the largest plasmid was conserved in the chromosomes of Nitrobacter winogradskyi Nb-255 and Nitrobacter sp. strain Nb-311A. The N. hamburgensis chromosome also harbors many unique genes, including those for heme-copper oxidases, cytochrome b561, and putative pathways for the catabolism of aromatic, organic, and one-carbon compounds, which help verify and extend its mixotrophic potential. A Nitrobacter “subcore” genome was also constructed by removing homologs found in strains of the closest evolutionary relatives, Bradyrhizobium japonicum and Rhodopseudomonas palustris. Among the Nitrobacter subcore inventory (116 genes), copies of genes or gene clusters for nitrite oxidoreductase (NXR), cytochromes associated with a dissimilatory nitrite reductase (NirK), PII-like regulators, and polysaccharide formation were identified. Many of the subcore genes have diverged significantly from, or have origins outside, the alphaproteobacterial lineage and may indicate some of the unique genetic requirements for nitrite oxidation in Nitrobacter.
Nitrification is a two-step process by which ammonia is converted to nitrate via nitrite. Nitrification plays a key role in transformation of fertilizer nitrogen in agricultural systems and is a key component of nitrogen removal in wastewater treatment. Production of soluble inorganic nitrogen by nitrification can lead to the contamination and eutrophication of terrestrial and aquatic ecosystems, while the gaseous products of nitrifier denitrification, nitric oxide (NO) and nitrous oxide (N2O), destroy stratospheric ozone and greatly contribute to global warming (26, 54, 61). Nitrite-oxidizing bacteria (NOB) participate in the process of nitrification by converting nitrite (NO2−), the end product of ammonia oxidation, into nitrate (NO3−) according to the following reaction; NO2− + H2O → NO3− + 2H+ + 2e−. Nitrite also functions as an electron donor for the reduction of NAD+ via reverse electron flow as well as for the generation of ATP by oxidative phosphorylation (17).
Although several phylogenetically distinct genera (Nitrospira, Nitrobacter, Nitrococcus, and Nitrospina) carry out NO2− oxidation, most of what is known about the physiology and biochemistry of NOB has been derived from studies of the genus Nitrobacter. As facultative chemolithoautotrophs, Nitrobacter spp. oxidize nitrite and fix carbon dioxide (CO2) via the Calvin-Benson-Basham pathway. However, they also have the ability to assimilate a narrow range of simple organic carbon (C) compounds (pyruvate, acetate, α-ketoglutarate, and glycerol) in the absence of nitrite (3, 52). The effectiveness of organotrophy varies among members of the genus Nitrobacter. For example, in Nitrobacter winogradskyi, the highest growth rates are achieved using nitrite as an energy source, yet organic carbon additions to nitrite-containing cultures can increase the final growth yield and protein content after nitrite is depleted (52). In contrast, the highest growth rates reported for Nitrobacter hamburgensis X14 are achieved in media containing both nitrite and organic carbon (6, 7, 56). The genetic and full enzymatic bases for these differing phenotypes have not been explored.
Nitrobacter is a member of the family Bradyrhizobiacaea and is closely related (97 to 98% identity of 16S rRNA) to the Bradyrhizobium and Rhodopseudomonas genera. Rhodopseudomonas palustris can grow either chemotrophically or phototropically and is arguably one of the most metabolically versatile bacteria known (38), and the plant symbiont Bradyrhizobium japonicum has one of the largest prokaryote genomes. However, neither of these close Nitrobacter relatives has the ability to use nitrite as an energy source. The availability of genome sequences from these three genera has provided an opportunity to gain insight into the basis of what distinguishes the “metabolically limited” Nitrobacter from its close “physiologically versatile” relatives and has led to a better understanding of the physiological requirements for nitrite oxidation.
Here we present an analysis of the genome sequence from N. hamburgensis X14 and a comparative analysis of the genomes within the genus Nitrobacter. Included in our Nitrobacter comparative analysis are the genomes of Nitrobacter winogradskyi Nb-255 and Nitrobacter sp. strain Nb-311A (NB311A). Curiously, NB311A was isolated near the west coast of central Africa and grows in seawater (B. Ward, unpublished results), yet its 16S rRNA gene sequence is 100% identical to that of the soil isolate N. winogradskyi. N. winogradskyi was previously sequenced and analyzed (53), and a draft sequence of NB311A was recently made available, for which no published reports currently exist. In addition, we included in our analysis five complete genomes from Rhodopseudomonas palustris and three from Bradyrhizobium japonicum, which are also available in public databases. Placed in the context of these Bradyrhizobiaceae members, the genetic framework for determining the metabolic variations and similarities that exist within Nitrobacter is analyzed and the genomic basis of lithoautotrophy versus trophic flexibility among closely related bacteria is explored.
MATERIALS AND METHODS
Construction, sequencing, and assembly.
Genomic DNA from N. hamburgensis X14 was isolated; purified; sheared into approximately 3-kb, 8-kb, and 40-kb fragments; and ligated into the pUC18, pMCL200, and pCC1Fos cloning vectors, respectively. After amplification, double-ended plasmid sequencing reactions were performed at the DOE Joint Genome Institute using ABI 3730xl DNA analyzers and MegaBACE 4500 genetic analyzers as previously described (9, 16; http://www.jgi.doe.gov/).
Processing of sequence traces, base calling, and assessment of data quality were performed with PHRED and PHRAP (14, 15) at an estimated error rate of <2.5 bp per Mbp sequenced. After quality control of the 66,403 total initial reads of the draft sequence, 58,661 sequences were assembled, producing an average of 11-fold genome coverage. The reads were assembled into 52 high-quality draft sequence contigs, visualized with CONSED (23), and linked into 18 larger scaffolds using paired-end sequence information. Gaps between linked contigs were closed either by walking on gap-spanning clones or with PCR products generated from genomic DNA, while physical (or uncaptured) gaps were closed by combinatorial PCR. Sequence finishing and polishing added 2,360 reads, and final quality assessment of the completed genome was completed as previously described (9).
Genome analysis and annotation.
Automated gene modeling for N. hamburgensis was completed by combining results from the Critica, Generation, and Glimmer modeling packages. Automated annotation was produced by searching all predicted peptides against TIGRFAMs (HMMPfam trusted cutoffs), PRIAM (rpsblast 1e−30 cutoff), PFAM (HMMPfam trusted cutoffs), InterPro (interproscan default cutoffs), and COGs (rpsblast 1e−10 cutoff). Proteins that failed to return a definitive result with the aforementioned profile searches were annotated on the basis of BLASTP searches against the KEGG and SwissprotTREMBL (1e−5 cutoff) peptide databases. In the manual curation phase, the protein set was also searched against GenBank's nonredundant database and the PROSITE, EcoCYC, and MetaCYC databases to further assess function. Gross truncations of peptides (n = 171), base insertions or deletions that produced frameshifts (n = 153), or internal stop codons (n = 23) that resulted in coding sequences (CDSs) that were ≤80% of the best BLASTP match were considered pseudogenes.
NB311A was isolated by Stan Watson in 1968 from surface waters in the tropical Eastern Atlantic Ocean off the coast of West Africa (J. Waterbury, personal communication). The draft genome sequence of NB311A was downloaded from a public database at the Venter Institute (https://research.venterinstitute.org/moore/) and reannotated using the same criteria as for N. hamburgensis and N. winogradskyi to facilitate an accurate comparative analysis.
Cross genomic analysis.
Orthologous and paralogous groups were determined using OrthoMCL version 1.4. A peptide database of 11,704 sequences of the CDS translations from Nitrobacter hamburgensis X14 (accession no. CP000319), Nitrobacter winogradskyi Nb-255 (CP000114), and Nitrobacter sp. strain NB311A (AAMY01000000) was assembled. To determine which CDS translations were conserved in all three species, an all-versus-all analysis was performed using BLASTP (BLASTALL 2.2.13) with seg, with an E value threshold of 1 × 10−5. The results were processed by OrthoMCL (mcl-06-21) using an inflation factor of 1.5. The output of OrthoMCL was parsed to separate orthologous and paralogous groups. Duplicated genes/paralogs were counted as a single “gene type.” The clustered orthologous/paralogous data set displayed an average E value of 1 × 10−113 and an average identity of 86%. NCBI Taxplot software (http://www.ncbi.nlm.nih.gov/sutils/taxik2.cgi?isbact = 1) was used to compare the similarity between the conserved proteins in the three Nitrobacter genomes using N. winogradskyi as the query genome with the cutoff set at 10.
Nitrobacter core and subcore construction.
The OrthoMCL output was filtered to produce a list of ortholog/paralog groups which contained genes from all three Nitrobacter species. The resulting list is the core set of genes shared by the three Nitrobacter species. The Nitrobacter core genes or CDSs were extracted into a separate data set and were subjected to BLASTP searches (1e−10 and 1e−20 cutoffs) against a database consisting of Bradyrhizobium japonicum (GenBank/EMBL/DDBJ accession numbers BA000040, CP000494, and CU234118) and Rhodopseudomonas palustris (BX571963, CP000250, CP000283, CP000301, and CP000463) sequences (46111 peptides). Core sequences without a match (≥1e−10) were designated the Nitrobacter-specific subcore. The subcore was curated manually to remove the remaining peptides (n = 7) that were still found to be conserved in B. japonicum or R. palustris that were not filtered out by the e−10 cutoff. Additional manual searches for conserved gene clusters added seven peptides to the subcore using the following criteria: (i) the peptides were present in the 1e−20 subcore list, (ii) the peptides were adjacent to a peptide(s) in the 1e−10 database, and (iii) the peptide sequence identity to similar peptides in the R. palustris/B. japonicum database was <50%.
Nucleotide sequence accession numbers.
The sequence and annotation of the complete N. hamburgensis X14 chromosome are available at GenBank/EMBL/DDBJ using accession number CP000319. The sequences of the N. hamburgensis plasmids pPB13, pPB12, and pPB11 are available as plasmid 1, plasmid 2, and plasmid 3, using accession numbers CP000320, CP000321, and CP000322, respectively.
RESULTS AND DISCUSSION
Genome overview.
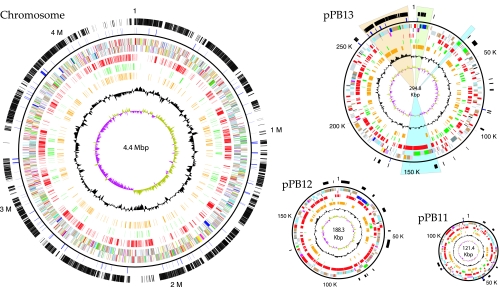
The N. hamburgensis X14 genome consists of a 4,406,967-bp chromosome (61.7% G+C) and three plasmids: pPB11 (121,408 bp, 61.7% G+C), pPB12 (188,318 bp, 61.2% G+C), and pPB13 (294,829 bp, 60.4% G+C). Based on BLAST analysis, N. hamburgensis shares more total CDSs with NB311A (1,434) than with N. winogradskyi (974). In contrast, when conserved proteins were analyzed for similarity, 76% of the proteins in N. winogradskyi were more similar to those of NB311A than to those of N. hamburgensis (S. R. Starkenburg, unpublished results). This analysis is consistent with the fact that N. winogradskyi and NB311A have identical 16S rRNA gene sequences that are approximately 98% identical to the N. hamburgensis 16S rRNA gene.
The genome of N. hamburgensis is much larger than those of the other two Nitrobacter species, containing ∼1.6 and 0.9 Mbp more genetic material than N. winogradskyi and NB311A, respectively (Table 1). Several features of the N. hamburgensis genome account for its relatively large size. First, a disproportionately large number of pseudogenes and paralogs were identified, which account for 20% of the genome. 347 CDSs (∼8% of the sequence space) were identified as pseudogenes in N. hamburgensis, compared to 21 pseudogenes in N. winogradskyi (pseudogene annotation was not completed for the draft genome of Nitrobacter sp. strain NB311A). N. hamburgensis also has the largest number of paralogs (634 genes in 251 groups), approximately 30% more than NB311A and over twice the number found in N. winogradskyi. Second, the majority of the genes carried on the N. hamburgensis plasmids do not have orthologs in the N. winogradskyi or NB311A genome (Fig. 1). As presented below, these relatively large plasmids harbor a number of functions that are uniquely beneficial to N. hamburgensis and clearly distinguish it from N. winogradskyi and NB311A.
TABLE 1.
General genome characteristics
| Strain | Origin | % 16S rRNA gene identitya | No. of chromosome bases | % GC | No. of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total genes | Genes without predicted function | Pseudogenes | Paralogs | Paralog groups | Plasmids | |||||
| N. hamburgensis X14 | Soil | 98 | 4,406,967 | 61.61 | 4,716 | 1,848 | 347 | 634 | 251 | 3b |
| N. winogradskyi Nb-255 | Soil | 100 | 3,402,093 | 62.05 | 3,118 | 993 | 21 | 283 | 74 | 0 |
| Nitrobacter sp. strain Nb-311Ac | Marine | 100 | ∼4,105,362 | 62 | 4,256 | 1,461 | NDd | 478 | 143 | ND |
16S rRNA identity relative to N. winogradskyi Nb-255.
pPB13, 294,829 bp; pPB12, 188,318 bp; pPB11, 121,408 bp.
Draft sequence.
ND, not determined (an accurate count of the pseudogenes or the presence of plasmids was not possible from the unfinished, draft sequence of NB311A).
FIG. 1.
Genome of Nitrobacter hamburgensis X14 (ATCC 25391). The outer circle depicts the locations of genes conserved in all three sequenced Nitrobacter genomes. Genes that are conserved in Nitrobacter but not R. palustris or B. japonicum (the subcore) are indicated in the second circle. The third and fourth circles depict predicted protein-encoding and structural RNA genes in N. hamburgensis on the plus and minus strands, respectively (green, energy metabolism; red, DNA replication; magenta, transcription; yellow, translation; orange, amino acid metabolism; dark blue, carbohydrate metabolism; pale red, nucleotide metabolism; black, coenzyme metabolism; cyan, lipid metabolism; light blue, cellular processes; brown, general function; gray, hypothetical and conserved hypothetical genes; pale green, structural RNAs). Genes “unique” to N. hamburgensis (not present in N. winogradskyi or NB311A) are depicted in circle five (red). The sixth and seventh circles indicate the locations of all annotated pseudogenes (green) and paralogs (orange), respectively. The eighth circle indicates GC bias, and the ninth circle indicates GC skew. The highlighted region of pPB13 depicts the locations of key gene clusters: autotrophic island (peach), RuBisCO and pentose phosphate pathway genes (green), and the cytochrome oxidases (aa3 and bd-ubiquinol types) (blue).
Several “unique” (i.e., not found in the other Nitrobacter genome sequences) genomic islands, many of which appear to have been acquired by conjugation or transduction, also help account for the genomic and physiological separation of N. hamburgensis and N. winogradskyi. For example, a 240-kb island (Nham_3756 to Nham_4008; Fig. 1; chromosome locus 4.13 to 4.37 Mbp), which contains a high concentration of pseudogenes and paralogs, appears to have originated from plasmid or phage sources, since it harbors putative plasmid replication initiator proteins (Nham_3835 and Nham_3863), a partitioning protein (Nham_3861), and phage integrase and phage-related proteins (Nham_3842 and Nham_4008). Similarly, a portion of a 4.2-kb phage element (rep28; see Table S1 in the supplemental material) is found replicated on the N. hamburgensis chromosome and contains a phage-related methylase, an uncharacterized phage protein, and two hypothetical proteins found only in N. hamburgensis. These and other similar elements lie within other large chromosomal regions (Nham_0784 to Nham_0835, Nham_0838 to Nham_0943, and Nham_1145 to Nham_1186) that are not present in the N. winogradskyi genome and appear to have integrated into N. hamburgensis (or to have been lost from the other Nitrobacter genomes) since delineation from a common ancestor. Based on genome sequence data in REBASE (47), N. hamburgensis and N. winogradskyi also encode an above-average quantity of restriction-modification (RM) systems (11 in N. hamburgensis [2.39 RM genes per Mbp] and 9 in N. winogradskyi [2.64 RM genes per Mbp]), the majority of which are type II RM systems. These RM systems should provide defense against genome contamination by phage or other foreign DNA. On the other hand, N. hamburgensis appears to have been a successful target of phages at some point in its evolutionary history. Recent evidence indicates that type II RM complexes may function as “selfish” mobile genetic elements and can promote homologous recombination in the host bacteria (as a defense mechanism), resulting in more genomic rearrangements and diversity (32, 33, 44). Clearly, unraveling the complex history and role of phages, RM systems, and other mobile genetic elements (transposons) in the diversification and evolution of these closely related Nitrobacter species will require further investigation.
Interspecies comparisons.
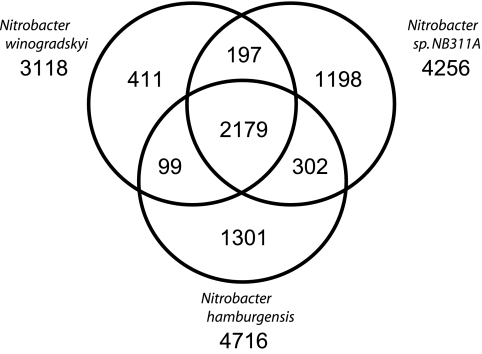
A global comparative analysis of all genes was completed to assess both the common and unique elements of each of the sequenced Nitrobacter species (Fig. 2). A total of 2,179 “gene types” (excluding paralogs/gene duplications) were conserved in each Nitrobacter genome. The majority (86%) of the genes in the genome of N. winogradskyi are conserved in either N. hamburgensis or NB311A; therefore, most of the genome-inferred metabolic potential of N. winogradskyi described previously can be extrapolated to these other species (53). In contrast, N. hamburgensis and NB311A collectively carry approximately 2,801 gene types not found in N. winogradskyi (approximately one-third of each genome; 1,301 and 1,198 gene types, respectively). Surprisingly, although the N. hamburgensis genome is >900 kb larger than the NB311A draft sequence, our analysis indicated that NB311A harbors roughly the same number of gene types (3,881 versus 3,876) as N. hamburgensis.
FIG. 2.
Global gene conservation in Nitrobacter. Each circle represents the total number of gene types in each genome. Overlapping regions depict the number of genes types shared between the respective genomes. The numbers outside the circles indicate the total number of genes identified in each genome, including paralogs/gene duplications.
The organization of unique genes by COG groups revealed similar gene distribution patterns for all three species, suggesting that many of these genes (based solely on COG groupings) appear to be functional analogs (S. R. Starkenburg, unpublished results). Nevertheless, manual analysis of genome-specific sequences revealed that each genome did contain unique genetic material (Table 2), which potentially confers specific functions relevant to the ecological niche of each bacterium. Many of the unique N. hamburgensis genes/functions are discussed below. With regard to N. winogradskyi, of the 411 genes not found in either of the other two Nitrobacter species, only 124 could be assigned a putative function, including an alkane-sulfonate monoxygenase, two nitrate/sulfonate/bicarbonate ABC transporters, and synthesis genes for the pyrroloquinoline quinone cofactor.
TABLE 2.
Unique genes and putative functional biases in the genus Nitrobacter
| Organism | Category (functions) |
|---|---|
| N. winogradskyi Nb-255 | Transport (NO3−/sulfonate/CO32− [2], iron uptake systems, nickel/cobalt, PO43− porin, uncharacterized ABC transporter components), miscellaneous (histidine biosynthesis, multiple FecIR genes, pyrroloquinoline quinone biosynthesis) |
| N. hamburgensis X14 | Transport (ammonia permease, K+ transport, uncharacterized ABC transport components), carbon metabolism (formate dehydrogenase, carbon monoxide dehydrogenase-like, l-lactate dehydrogenase, malate dehydrogenase, pyruvate-formate lyase, homogentisate/phenylacetate degradation), energetics (cytochrome c oxidase, cytochrome bd ubiquinol oxidase, cytochome b561 [4], cytochrome P460, flavoredoxin reductase, sulfur oxidation genes [soxXYZAB], nitric oxide reductase [sNOR]), miscellaneous (type II secretion, serine proteases, conjugal transfer, heavy metal resistance, arsenite oxidase) |
| Nitrobacter sp. strain Nb-311A | Transport (TonB systems, Ca2+/Na2+ antiporter, Cl− channel, chromate, uncharacterized ABC transporter components, Mg/cobalt, sulfate permeases, Co/Zn/Cd efflux), replication (DNA replication/repair, DNA polymerases IV and III, minCDE septum formation), miscellaneous (polysaccharide synthesis and export, UspA stress genes, ectoine synthase, cation ATPases, pyoverdine synthesis, hydroxymate siderophore syn. [IucC family]) |
Several unique gene clusters were identified in NB311A (Table 2), including some that may be indicative of adaptation to a marine lifestyle. NB311A uniquely possesses genes that encode a chloride channel (NB311A_05795), an Na+/Ca2+ antiporter (NB311A_09276), and several cation-dependent ATPases. NB311A also harbors a four-gene cluster (NB311A_1874 to NB311A_1879), which may code for synthesis of ectoine-like osmoprotectants. The putative NB311A ectoine synthesis peptides have 47, 57, 51, and 50% identity to the ectoine synthesis proteins (EctABCD) in the moderate halophile Chromohalobacter salexigens DSM 3043, which has been shown to produce ectoines as osmo- and thermoprotectants (21, 22, 60). All three genomes were found to encode a putative Na+/H+ antiporter (nhaA), which is required for Escherichia coli to survive high-salt stress (2, 51). Consequently, we carried out a preliminary investigation of the salt tolerance of N. hamburgensis and N. winogradskyi by monitoring growth and nitrite consumption in cultures amended with 0, 100, 200, 400, and 600 mM NaCl (culture medium amended with 600 mM NaCl is equivalent to the salinity of seawater). N. winogradskyi grew normally in standard growth medium amended with 0 to 200 mM NaCl. In cultures amended with 400 mM NaCl, the growth rate decreased by 50%, yet the growth yield of N. winogradskyi was unaffected. Growth was not observed when cultures where amended with 600 mM NaCl, even though the cells continued to oxidize nitrite. N. hamburgensis was less salt resistant, as growth was inhibited at >200 mM NaCl, although the cells continued to oxidize nitrite in cultures containing up to 400 mM NaCl. Nitrite oxidation of N. hamburgensis was completely inhibited by 600 mM NaCl (S. R. Starkenburg, unpublished results). In light of these results, all members of the genus Nitrobacter appear to be quite halotolerant, although many of the aforementioned genes in NB311A presumably provide additional means to manage osmotic stress and may enable NB311A to thrive in marine coastal environments.
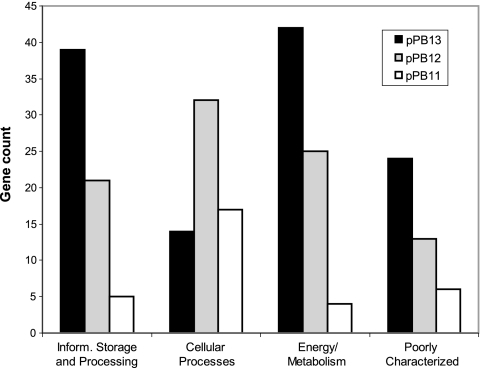
Plasmid analysis.
Previous reports indicated that N. hamburgensis contains three plasmids, designated pPB11, pPB12, and pPB13 (24, 35), yet little was known about how these plasmids support the lifestyle of N. hamburgensis. Each of the plasmid genes was analyzed for functional content through clustering of orthologous groups of proteins (COG) (Fig. 3). The largest plasmid, pPB13, was found to be biased toward carbon/energy metabolism (28 genes; see below) and information storage/processing, although most of the genes in the latter category encode transposases (30 genes). Conversely, the small plasmid pPB11 is dominated by conjugation/pilus formation genes; part of this region (∼2.5 kb) appears to have been duplicated within pPB11, and a larger (∼9.8-kb) portion has been duplicated in pPB13. pPB12 appears to be a functional hybrid of the other two plasmids, containing gene clusters for conjugation and energy and carbon metabolism, plus a suite of genes for heavy metal resistance, including those for heavy metal efflux (Nham_4358 and Nham_4359; Nham_4404 and Nham_4405; Nham_4529 to Nham_4535), mercury resistance (Nham_4416 to Nham_4421), and copper resistance (Nham_4364 and Nham_4365; Nham_4380; Nham_4382 to Nham_4385; Nham_4397 to Nham_4399; Nham_4422 to Nham_4424; Nham_4492). A putative arsenite oxidase gene and accessory genes were also located on pPB12 (Nham_4425 to Nham_4432), although it is unknown whether these genes are involved in arsenic detoxification or are linked to a respiratory chain. A few additional transport functions encoded on pPB12 include a CDS for a P-type ATPase Mg2+ importer (Nham_4377), a predicted TrkA K+ transporter (Nham_4433), and a putative arabinose transporter (Nham_4434). Notably, the only copy in the genome of an ATP-dependent glucokinase (Nham_4371) is located on pPB12.
FIG. 3.
Functional distribution of N. hamburgensis plasmid genes based on COG assignments. Genes in COG groups J, K, and L are included in “information storage and processing.” Genes in COG groups D, V, T, M, N, U, and O are included in “cellular processes.” Genes in COG groups, G, E, F, H, I, P, and Q are included in “metabolism.” Genes in COG groups R and S are included in “poorly characterized.” Of the 494 plasmid genes, 242 could be assigned to a COG group. The remaining plasmid genes were excluded from the analysis.
A significant feature of pPB13 is the presence of a large, ∼28-kb gene cluster which may be indispensable for autotrophic growth of this organism (Fig. 1). This “autotrophic island” encodes the large and small subunits of a type I ribulose-bisphosphate carboxylase (RuBisCO) (Nham_4332 to Nham_4333), the sole complement of genes necessary for carboxysome formation, and includes a tandem repeat of a four-gene cluster which contains a chain L-like subunit of complex I and a PII protein. Although most of the plasmid-borne genes are unique to N. hamburgensis, ∼21 kb of this ∼28-kb autotrophic island is conserved in the chromosomes of N. winogradskyi and NB311A (Fig. 1). This gene cluster was most likely acquired by the ancestor to all three Nitrobacter genomes via a lateral gene transfer event, because the GC content of this island is 4.6% and 3.4% higher than the GC averages of the plasmids and genome, respectively, and the genes in the autotrophic island are most similar and syntenous to the RuBisCO and carboxysome gene clusters in the Betaproteobacteria, Nitrosomonas eutropha, and Thiobacillus spp.
Many examples of DNA exchange and duplication (beyond insertion sequence element transposition) between the plasmids and the chromosome have been identified (see Table S1 in the supplemental material). In addition to the autotrophic island described above, several other Calvin cycle enzymes are also located on pPB13. A 6.1-kb region (rep18) encodes a second, nonparalogous copy of a type I RuBisCO (see “C1 carbon metabolism” below) and single copies of fructose-1,6-bisphosphatase, phosphoribulokinase, and ketose-bisphosphate aldolase. This second autotrophic island is >99% identical to a region on the chromosome (Nham_3749 to Nham_3754). Interestingly, this similarity does not extend to an upstream and divergently transcribed LysR-type regulator (Nham_4044 in pPB13, Nham_3755) in the chromosome, which may be responsible for the differential regulation of these paralogous gene clusters. In addition to the autotrophic islands, pPB13 carries two more regions nearly identical to chromosomal loci, including a 6.7-kb gene cluster (Nham_4077 to Nham_4081, rep12) which includes genes for an aconitase and a DNA-binding ferritin-like protein. This 6.7-kb gene cluster is >99% identical to the chromosomally located gene cluster (Nham_0912 to Nham_0916). pPB12 also carries two additional loci (Nham_4530 to Nham_4359; Nham_4404 and Nham_4405) that are similar to chromosomal locus Nham_1848 to Nham_1854 and encode products that are also involved in heavy metal resistance.
Regulation and signaling.
To assess the genomic repertoire of signaling and regulatory capacity, the pertinent genes in Nitrobacter were compared with respective genes from the alphaproteobacterial relatives, B. japonicum and R. palustris, for which these data were available. The N. hamburgensis genome encodes a moderate abundance of signaling proteins, more than the other Nitrobacter strains but about one-half and one-third of the signaling capacities in R. palustris and B. japonicum, respectively. In contrast, N. hamburgensis and NB311A contain about half the number of fecI-like extracytoplasmic transcription factors (ECFs) (a subfamily of σ70) as in N. winogradskyi. Many of the ECF genes in N. winogradskyi and NB311A are proximal to fecR and/or siderophore receptor genes, suggesting that most of these proteins function to positively regulate iron uptake. Surprisingly, the N. hamburgensis genome is completely devoid of fecR homologs and contains fewer siderophore receptor genes, and none of the ECF proteins are adjacent to iron-related proteins. Thus, N. hamburgensis appears to have evolved a different iron management strategy than either N. winogradskyi or NB311A and presumably relies solely on the global iron regulator FUR to control intracellular iron levels instead of responding to extracellular iron concentrations via FecIR.
In comparison to those of B. japonicum and R. palustris, the Nitrobacter genomes contain fewer genes encoding proteins with EAL and GGDEF domains, which likely function in the synthesis and hydrolysis of the intracellular signaling compound cyclic diguanylate (49). The Nitrobacter genomes contained a similar number of genes encoding PAS/PAC-domain proteins as B. japonicum, which often function as redox sensors (62). Because Nitrobacter can grow both aerobically and anaerobically (1, 17, 18), these sensors may be important for the functioning of nitrification aggregates at the oxic/anoxic interface by sensing a low redox potential in the environment to induce gene expression needed to switch to an anaerobic metabolism such as nitrate respiration.
Histidine protein kinases (HPKs) and response regulator proteins (RRs) constitute two-component regulatory systems that are often dedicated to the sensing and mitigation of environmental stress conditions. With a total of 46 HPKs and 45 RRs, N. hamburgensis has a respectable complement of potentially functional two-component proteins that are expressed from genes arranged in tandem (20 paired HPK and RR genes) as well as singletons (26 unpaired HPK genes and 24 unpaired RR genes). Some of the genes encoding chemotaxis components were grouped with HPKs and RRs with EAL, GGDEF, or GAF domains, suggesting an interconnection between chemotaxis and other signal transduction systems.
Sulfur Metabolism.
All sequenced Nitrobacter species have the capacity to assimilate sulfur by reducing sulfate to sulfide, which is then incorporated into cysteine. The use of reduced sulfur as an energy source has not been reported for Nitrobacter, but R. palustris can grow photolithoautotrophically using thiosulfate as an electron donor (48, 58). Intriguingly, N. hamburgensis has a small operon containing several genes associated with dissimilatory sulfur oxidation, soxXYZA_B (Nham_3671 to Nham_3676). The genes in this cluster encode three of the four main protein complexes which catalyze the oxidation of reduced sulfur, i.e., SoxYZ, SoxAX, and SoxB (19, 20, 46, 57). The fourth protein complex, SoxCD, which completes the oxidation of thiosulfate by oxidizing sulfur to sulfate (20), is not present in N. hamburgensis. With respect to experimentally validated gene products, the arrangement and protein identity of the N. hamburgensis soxXYZA_B gene cluster are most similar to those for genes found in Chlorobium. Similarly, Chlorobium spp. do not contain a classical SoxCD complex but still have the ability to oxidize thiosulfate or sulfide anaerobically using an alternate sulfur oxidase/dehydrogense (12). Indeed, two genes annotated as encoding subunits of a sulfite oxidase (Nham_1093 and Nham_1094), are found elsewhere in the N. hamburgensis genome. Unlike the other sox-like genes, these two genes are conserved in N. winogradskyi (but not NB311A) and have some sequence similarity with soxCD in sulfur oxidizers such as the obligate sulfur oxidizer Thiomicrospira crunogena XCL-2 (50). Whether any of these putative N. hamburgensis sox genes function in respiratory sulfur oxidation, detoxification, or assimilation remains unknown and awaits further experimentation.
Dissimilatory nitrogen metabolism.
Nitrite-dependent lithotrophic growth in Nitrobacter is catalyzed by a reversible nitrite oxidoreductase (NXR). NXR is a heterodimer containing alpha (NxrA) and beta (NxrB) subunits and is evolutionarily related to the Nar-type dissimilatory nitrate reductase (30). Similar to N. winogradskyi, multiple copies of nxrA (n = 3) and nxrB (n = 2) are carried in the N. hamburgensis genome, but only one central gene cluster (Nham_3443 to Nham_3451) encodes the putative accessory proteins of NXR. In addition to nxrA and nxrB, several genes in this cluster are conserved in all three Nitrobacter genomes, including homologs to nitrate reductase accessory proteins NarJI (Nham_3446 and Nham_3447), a peptidyl prolyl cis-trans-isomersase (nxrX, Nham_3448), a cytochrome c (Nham_3450), and putative proteins involved in the transport of nitrite and/or nitrate (narK, Nham_3444; TDT family of transporters, Nham_3443).
Some Nitrobacter species have been shown to grow anaerobically using nitrate as a terminal electron acceptor when coupled to the oxidation of simple organic compounds (1, 18), and the terminal product of denitrification in Nitrobacter is reported to be nitrous oxide (17, 18). As was the case for N. winogradskyi, additional homologs of nitrate reductase genes other than NXR were not identified in N. hamburgensis or NB311A, and likewise, a gene cluster encoding a nirK-type nitrite reductase is conserved in all three Nitrobacter genomes. Together these enzymes presumably function to reduce nitrate to nitric oxide under anaerobic conditions. Each genome also contains a putative flavin mononucleotide-dependent nitroreductase which, if functional, could be involved in detoxification or respiration. In contrast, a gene cluster (Nham_2710 to Nham_2712) encoding a nitric oxide reductase homologous to sNOR in the heme-copper oxidase superfamily was found only in N. hamburgensis and not the other two Nitrobacter genomes (8). Similarly, a cytochrome P460 (cytL, Nham_2497) whose translated product contains an infrequently used heme-coordination motif, CGxxCH (13), was found only in N. hamburgensis. Although the function of cytochrome P460 has not been resolved, related proteins within the larger cytochrome P460 family have been proposed to mediate the transfer of NO during denitrification and/or to protect bacteria from NO toxicity (13, 45). An NO detoxification mechanism via oxidation would be advantageous for Nitrobacter, because instead of forming nitrous oxide (via sNOR) or ammonia [via NAD(P)H-siroheme nitrite reductase, Nham_2963 to Nham_2965], cytochrome P460 may recycle NO back to nitrite. Understanding of the physiological functions of nirK and cytochrome P460 in nitrogen oxide metabolism of Nitrobacter awaits experimentation.
Assimilatory nitrogen metabolism.
In terms of nitrogen assimilation, each NOB genome contains an assimilatory nitrite reductase (nirBD), which permits the production of ammonia from nitrite. Assimilatory nitrate reductases were not found in any Nitrobacter genome. In contrast to the case for N. winogradskyi, which lacks an ammonia permease or urea catabolic genes (53), the N. hamburgensis and NB311A genomes both contain ammonia permeases (Nham_0084) and two genes annotated as urea carboxylase (Nham_2041; COG1984) and allophanate hydrolase (Nham_2040; COG2049). The latter two enzymes putatively contribute to ATP-dependent urea amidolyase activity. All NOB genomes lack genes encoding a classical urease (ATP-independent urea hydrolase) or a urea transporter identified previously in several ammonia-oxidizing bacteria (34); however, N. hamburgensis contains five clusters of genes that encode branched-chain amino acid (urea/short-chain amide) ABC transport systems. Although it would be a costly solution given the ATP dependence of all the enzymes, N. hamburgensis may obtain urea from the environment or from salvaging protein-nitrogen via the urea cycle, thereby providing the means to regulate its internal pH or provide associated ammonia-oxidizing bacteria with ammonia and CO2.
C1 carbon metabolism.
Previous DNA hybridization studies concluded that N. hamburgensis contained two copies of the large subunit of RuBisCO (cbbL), one encoded on pPB13 and the other on the chromosome (24). Three copies of RuBisCO were identified in the N. hamburgensis genome. Two sequence-divergent copies of a type I RuBisCO are located on pPB13 (Nham_4049 and Nham_4050; Nham_4332 and Nham_4333). The third set of RuBisCO-encoding genes (Nham_3750 and Nham_3751) found on the chromosome is identical to the RuBisCO genes Nham_4049 and Nham_4050 on pPB13. Intriguingly, both sequence-divergent RuBisCO copies are preceded by PII-like regulatory proteins, and this arrangement is conserved in all three Nitrobacter genomes. PII proteins are ubiquitous in bacteria and are most commonly known for their role in controlling nitrogen assimilation (39). These particular PII homologs may be serving a regulatory role in carbon fixation, or alternatively, a coupled regulatory link between carbon and nitrogen can be envisioned. Although coordination of nitrogen and carbon metabolism is not uncommon, finely tuned control of nitrogen and carbon assimilation via PII-type regulators may be crucial for Nitrobacter to thrive on an energy-limited substrate such as nitrite.
Most strikingly, the N. hamburgensis genome contains four gene clusters, plus a lone CDS, encoding multiple homologs of molybdopterin-containing carbon monoxide dehydrogenase (Mo-CODH). In contrast, only one Mo-CODH homolog is encoded in N. winogradskyi. The largest of these clusters (Nham_2601 to Nham_2608) has high similarity and gene synteny to those identified in the N. winogradskyi (53), Nitrobacter sp. strain 311A, Bradyrhizobium japonicum USDA 110, and Rhodopseudomonas palustris CGA009 genomes (Table 3). This unusual Mo-CODH gene cluster lacks a CDS for coxL/cutL, the large subunit of Mo-CODH (53). However, these lone CDSs found in the same genomes are highly conserved and contain the AYRGAGR active site and other motifs of form II CoxL proteins (29). Although growth on, or utilization of, CO by R. palustris has not been reported, B. japonicum USDA 110 is capable of aerobic growth on CO as a sole carbon and energy source (40), albeit at a very low rate. B. japonicum USDA 110 can also oxidize CO at the expense of nitrate reduction, but without growth, under anaerobic conditions (28). The N-terminal sequence of CoxL purified from B. japonicum USDA 110 apparently matched the translated sequence of blr0336 (M. Lorite, unpublished results), indicating that this gene cluster is expressed for carboxydotrophy. Whether any Nitrobacter sp. is capable of CO metabolism remains to be determined; however, N. hamburgensis has the largest and most diverse Mo-CODH-like gene inventory thus far among the Nitrobacter sp. genomes. In contrast to N. winogradskyi, N. hamburgensis contains several genes encoding cytochrome b561, an important electron transfer component in aerobic carboxidotrophic bacteria (42, 43). In fact, the N. hamburgensis genome contains more complete copies of these Mo-CODH-like genes than it does of NXR, which further suggests that these proteins may play a vital, yet unknown role in the lifestyle of Nitrobacter.
TABLE 3.
CODH gene clusters in Nitrobacter, B. japonicum, and R. palustris
| Subunit | N. hamburgensis gene cluster |
N. winogradskyi Nb-255
|
Nitrobacter sp. strain Nb-311A
|
B. japonicum USDA 110
|
R. palustris CGA009
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Nearest neighbor | % Protein sequence identity | Nearest neighbor | % Protein sequence identity | Nearest neighbor | % Protein sequence identity | Nearest neighbor | % Protein sequence identity | ||
| CoxG | Nham_2608 | Nwin_2206 | 77 | NB311A_18136 | 77 | bll5666 | 84 | RPA3804 | 81 |
| CoxS/CutS | Nham_2607 | Nwin_2205 | 93 | NB311A_18141 | 91 | bll5665 | 91 | RPA3803 | 90 |
| CoxM/CutM | Nham_2606 | Nwin_2204 | 83 | NB311A_18146 | 85 | bll5664 | 84 | RPA3802 | 84 |
| ATPase | Nham_2605 | Nwin_2203 | 87 | NB311A_18151 | 89 | bll5663 | 87 | RPA3801 | 86 |
| CoxE | Nham_2604 | Nwin_2202 | 80 | NB311A_18156 | 79 | bll5662 | 76 | RPA3800 | 79 |
| CoxF | Nham_2603 | Nwin_2201 | 85 | NB311A_18161 | 86 | bll5661 | 89 | RPA3799 | 88 |
| CoxF | Nham_2602 | Nwin_2200 | 86 | NB311A_18166 | 86 | bll5660 | 79 | RPA3798 | 80 |
| MobA-like | Nham_2601 | Nwin_2199 | 89 | NB311A_18171 | 90 | bll5659 | 77 | RPA3797 | 76 |
| CoxL/CutL | Nham_1307 | Nwin_1079 | 86 | NB311A_11547 | 87 | bll5914 | 78 | RPA3974 | 79 |
| CoxS/CutS | Nham_1039 | NPa | NB311A_01949 | 91 | blr5209 | 79 | NP | ||
| CoxM/CutM | Nham_1040 | NP | NB311A_01944 | 89 | blr5210 | 71 | NP | ||
| CoxL/CutL | Nham_1041 | NP | NB311A_01939 | 88 | blr5211 | 67 | NP | ||
| CoxS/CutS | Nham_1453 | NP | NP | blr0335 | 81 | RPA4666 | 82 | ||
| CoxL/CutL | Nham_1454 | NP | NP | blr0336 blr0337 | 84 | RPA4667 | 85 | ||
| CoxM/CutM | Nham_1455 | NP | NP | 72 | RPA4668 | 74 | |||
| CoxL/CutL | Nham_1181b | NP | NP | NP | NP | ||||
| CoxS/CutS | Nham_1182c | NP | NP | NP | NP | ||||
| CoxL/CutL | Nham_1183d | NP | NP | NP | NP | ||||
NP, not present.
Nearest neighbor for Nham_1181, CutL from Magnetospirillum magnetotacticum MS-1 (Magn03009301) (45%).
Nearest neighbor for Nham_1182, ferredoxin from Burkholderia ambifaria (BambDRAFT_3191) (68%).
Nearest neighbor for Nham_1183, cytochrome c from Mesorhizobium sp. strain BNC1 (MesoDRAFT_4421) (46%).
Unlike the genomes of N. winogradskyi and NB311A, the genome of N. hamburgensis contains four contiguous gene clusters, two in the forward (Nham_3124 to Nham_3128; Nham_3132 and Nham_3133) and two in the reverse (Nham_3129 to Nham_3131; Nham_3134 and Nham_3135) orientation, that resemble genes for formate dehydrogenase (FDH), the TorD chaperone, and enzymes for biosynthesis of the molybdopterin cofactor. Strains of N. winogradskyi have been shown to oxidize formate (41, 59), although the activity was thought to be catalyzed by NXR (11). Similar three-subunit metalloenzyme FDHs have been shown to catalyze the anaerobic oxidation of formate, which is produced from pyruvate, to CO2 (27). Several Nitrobacter spp., including N. hamburgensis, are capable of both aerobic and anaerobic growth on pyruvate with O2 or nitrate as the electron acceptor, respectively (5). Thus, it remains to be determined whether this putative FDH functions in aerobic or anaerobic environments.
Heterotrophy.
Growth of N. winogradskyi and N. hamburgensis is enhanced in the presence of C2, C3, and C5 carbon molecules (3, 4, 52), although growth on hexose sugars or aromatic compounds has not been reported for any Nitrobacter strain. The genome of N. winogradskyi appears to be devoid of active transporters for sugars, and a classical glycolysis pathway could not be constructed because a phosphofructokinase gene could not be identified (53). In contrast, the inability of N. hamburgensis or NB311A to metabolize hexose sugars is less obvious, given that a complete glycolysis pathway was identified in both N. hamburgensis and NB311A. Additional sleuthing for genes involved in C6 metabolism revealed two plasmid-encoded copies of glucose-methanol-choline oxidoreductase genes (Nham_4194 and Nham_4244) and a unique gene cluster with homology to an ABC-type general sugar transporter (Nham_1206 to Nham_1209), which together provide the in silico potential to transport and initiate oxidation of hexoses.
Many other genes which may further extend the organic carbon substrate range of N. hamburgensis were identified. A gene cluster encoding the subunits of a putative glycolate oxidase and a cytochrome c550 (glcDEF; Nham_3202 to Nham_3205) were identified in all Nitrobacter genomes. Genetic and functional evidence indicates that a glyoxylate bypass of the tricarboxylic acid cycle operates in Nitrobacter, and thus, glyoxylate (the product of glcDEF) could serve as an additional carbon source. Two NAD-independent (flavin adenine dinucleotide/flavin mononucleotide-dependent) oxidoreductases (Nham_4010 and Nham_1112) were also located, which potentially encode putative d- or l-isomer-specific lactate dehydrogenases. If functional, these enzymes could potentially oxidize d- and/or l-lactate to pyruvate, providing both energy and carbon to the cell. Indeed, both d- and l-lactate dehydrogenase activities were observed in R. palustris (25). Limited metabolism of aromatic carbon compounds may also be possible given that a cluster of 19 genes that encode the pathways for homogentisate and phenylacetate metabolism (Nham_0920 to Nham_0938) were identified. As is the situation for most potential organic substrates, candidate genes for the transport and uptake of the aforementioned compounds were not readily identified in these bacteria.
Gene clusters encoding respiratory terminal oxidases and cytochromes are more abundant in N. hamburgensis than in the other two Nitrobacter species. Previous investigations have indicated that N. hamburgensis has a higher specific affinity for oxygen under suboxic conditions than N. winogradskyi (36, 37), and distinct respiratory chains and terminal oxidases have been suggested to function in N. hamburgensis, as different b- and c-type cytochromes are present during autotrophic growth and heterotrophic growth, respectively (6, 31). In addition to the aa3-type cytochrome c oxidase discussed below, four b-type cytochrome genes plus a plasmid-carried cytochrome bd-ubiquinol oxidase gene were found exclusively in the N. hamburgensis genome. The product of these quinol oxidase genes is a likely source of the major b-type cytochrome previously isolated from heterotrophically grown N. hamburgensis (31). N. hamburgensis contains three gene clusters encoding three copies of an aa3-type cytochrome c oxidase. The two chromosomally encoded cytochrome c oxidase gene clusters (Nham_0255 to Nham_0261; Nham_3457 to Nham_3463) are 100% identical at the protein sequence level and are homologous to two gene clusters encoding the only cytochrome c oxidases in N. winogradskyi (53). The plasmid-encoded cytochrome c oxidase gene cluster (pPB13; Nham_4177 to Nham_4180) contained genes for subunits I to IV, and the CDSs are 42, 32, 27, and 0% identical at the protein sequence level to chromosomal homologs of subunits I, II, III, and IV, respectively. Curiously, a molybdopterin oxidoreductase (Nham_4186 to Nham_4188) with sequence similarity to polysulfide reductases and nrf-type nitrite reductase is also encoded in the plasmid-borne cytochrome c oxidase gene cluster. Another separate gene cluster containing subunits I and II of cytochrome c oxidase plus an adjacent senC gene was identified (Nham_2710 to Nham_2712) and, as mentioned above, is homologous to sNOR-type nitric oxide reductases. The sNOR gene cluster is absent from the N. winogradskyi and NB311A genomes but is present in all of the ammonia-oxidizing and some sulfur-oxidizing bacterial genomes (55). This gene cluster was up-regulated in a nitrite reductase-deficient mutant of Nitrosomonas europaea, indicating involvement of sNOR in the nitrosative stress response (10).
Nitrobacter subcore inventory.
To gain further insight into NOB physiology, a final comparative analysis of the Nitrobacter genomes and all of the sequenced strains of R. palustris and B. japonicum was conducted. The facts that R. palustris and B. japonicum are metabolically versatile, do not use nitrite as an energy source, and are close evolutionary relatives to Nitrobacter provide a unique opportunity to explore the genetic foundation of being a nitrite oxidizer. Using the Nitrobacter core genome as the query database, all core genes with high sequence identity to a gene(s) in any strain of R. palustris or B. japonicum were removed. About 94% of the Nitrobacter core was conserved in either R. palustris or B. japonicum, leaving 116 gene types uniquely conserved in each Nitrobacter genome. Within the 116-gene Nitrobacter “subcore,” 75 genes were identified as encoding hypothetical proteins or contained domains of unknown function (see Table S2 in the supplemental material). Forty-six peptides (all hypothetical) had no match in the KEGG genome database (E value cutoff of ≥1e−5), and of the remaining 70 subcore peptides that had homologs in other prokaryotic genomes, the majority (n = 61) were most similar to peptides found within other Proteobacteria.
Among the functionally annotated subcore gene set (n = 41) (Table 4), two gene clusters appear to encode polysaccharide synthesis proteins (Nham_1195 to Nham_1201; Nham_1054 and Nham_1055), specifically glycosyl transferases, hydrolases, and sialic-acid based homopolysaccharide formation genes, some of which have little sequence similarity to any alphaproteobacterial peptides. Approximately half of the subcore genes with functional annotations appear to be associated with nitrite metabolism, transport, and regulation, including the gene cluster encoding the subunits of the NXR and the c-type cytochromes and a putative NsrR-like regulatory protein adjacent to nirK. In addition to the main NXR gene cluster which encodes the critical enzyme system for nitrite oxidation in the genus Nitrobacter, the scattered, lone copies of nxrA and nxrB were also included in the subcore inventory. How (or if) these additional nxrA and nxrB gene duplications function remains to be experimentally determined.
TABLE 4.
Nitrobacter subcore genesa
| Category and putative function | Gene(s) in:
|
||
|---|---|---|---|
| N. hamburgensis | N. winogradskyi | NB311A | |
| Polysaccharide biosynthesis | |||
| Lipopolysaccharide biosynthesis protein | Nham_1054 | Nwi_0538 | NB311A03749 |
| Glycosyl transferase, family 2 | Nham_1055 | Nwi_0539 | NB311A03744 |
| Glycosyl transferase | Nham_3303 | Nwi_0540 | NB311A03739 |
| Capsule polysaccharide biosynthesis protein | Nham_1195 | Nwi_0641 | NB311A16262 |
| Acylneuraminate cytidylyltransferase | Nham_1201 | Nwi_0645 | NB311A16252 |
| Hypothetical protein | Nham_1200 | Nwi_0646 | NB311A16247 |
| NUDIX hydrolase | Nham_1197 | Nwi_0648 | NB311A16237 |
| Haloacid dehalogenase-like hydrolase | Nham_1198 | Nwi_0649 | NB311A16232 |
| NUDIX hydrolase | Nham_1199 | Nwi_0650 | NB311A16227 |
| Nitrite metabolism, transport | |||
| Hypothetical protein | Nham_3451 | Nwi_0772 | NB311A09094 |
| Cytochrome c, class I | Nham_3450 | Nwi_0773 | NB311A09084 |
| nxrA1; nitrite oxidoreductase, alpha subunit | Nham_3449 | Nwi_0774 | NB311A09079 |
| nxrX; nitrite oxidoreductase, subunit X | Nham_3448 | Nwi_0775 | NB311A09069 |
| nxrB1; nitrite oxidoreductase, beta subunit | Nham_3447 | Nwi_0776 | NB311A09069 |
| nxrD; nitrite oxidoreductase, delta subunit | Nham_3446 | Nwi_0777 | NB311A09054 |
| nxrG; nitrite oxidoreductase, gamma subunit | Nham_3445 | Nwi_0778 | NB311A09049 |
| narK; nitrite/nitrate major facilitator superfamily transporter | Nham_3444 | Nwi_0779 | NB311A09044 |
| TDT family transport protein | Nham_3443 | Nwi_0780 | NB311A09039 |
| Cytochrome c, class IC (adjacent to nirK) | Nham_3282 | Nwi_2649 | NB311A15177 |
| Cytochrome c, class I | Nham_3283 | Nwi_2650 | NB311A15182 |
| Cytochrome c biogenesis factor | Nham_3285 | Nwi_2652 | NB311A15192 |
| nsrR; transcriptional regulator, BadM/Rrf2 family | Nham_3286 | Nwi_2653 | NB311A15197 |
| Nitrite reductase [NAD(P)H] large subunit, NirD | Nham_2964 | Nwi_0720 | NB311A01969 |
| nxrB homolog | Nham_3289 | Nwi_0965 | NB311A10815 |
| nxrA homolog(s) | Nham_0951, Nham_2961 | Nwi_2068 | NB311A17691 |
| Regulatory genes | |||
| PII-like regulatory protein | Nham_4043, Nham_4324, Nham_4330 | Nwi_1989, Nwi_2931 | NB311A00865 |
| Transcriptional regulator, LuxR family | Nham_2943 | Nwi_0957 | NB311A10770 |
| Transcription regulatory protein GAL11 domain | Nham_2946 | Nwi_0960 | NB311A10785 |
| Transcription factor jumonji/aspartyl beta-hydroxylase | Nham_1520 | Nwi_1272 | NB311A10046 |
| Miscellaneous | |||
| Sulfite reductase (NADPH) hemoprotein, beta subunit | Nham_0683 | Nwi_0591 | NB311A03464 |
| Major facilitator superfamily MFS_1 | Nham_1829 | Nwi_1437 | NB311A17399 |
| Polyphosphate glucokinase | Nham_2098 | Nwi_1575 | NB311A17319 |
| Phosphoesterase, PA-phosphatase related | Nham_2161 | Nwi_1638 | NB311A18878 |
| Isoprenylcysteine carboxyl methyltransferase | Nham_1746 | Nwi_1821 | NB311A06266 |
| Luciferase-like protein | Nham_0958 | Nwi_2071 | NB311A16699 |
| TonB-dependent receptor | Nham_0957 | Nwi_2072 | NB311A16694 |
| TadE-like protein | Nham_3156 | Nwi_2536 | NB311A14567 |
| Cupin 2, conserved barrel | Nham_3657 | Nwi_0906 | NB311A18960 |
| Na+/H+ antiporter NhaA | Nham_4598 | Nwi_2853 | NB311A16719 |
| Putative serine/threonine protein phosphatase | Nham_3383 | Nwi_3015 | NB311A06978 |
| Nucleoside phosphorylase | Nham_3696 | Nwi_3067 | NB311A02747 |
| Additional hypothetical proteins (75) | |||
| Phage genes (6) | |||
All of the genes with a functional annotation had at least one homolog (E value of ≤1e−5) in a genome other than R. palustris or B. japonicum. Of the 75 hypothetical proteins, 41 had no match to any peptide in the KEGG genome database (see Table S2 in the supplemental material).
Strikingly, the R. palustris and/or B. japonicum genomes contain homologs of several genes in the Nitrobacter subcore inventory, yet these genes are orthologous to genes outside the Alphaproteobacteria lineage. For example, R. palustris and B. japonicum both contain nirK, yet the Nitrobacter ncgABC-nirK gene cluster is syntenous to and has higher sequence similarity with the ncgABC-nirK gene cluster in ammonia-oxidizing Nitrosomonas spp., suggesting that it was horizontally transferred between the niche-sharing Nitrobacter and Nitrosomonas bacteria (8). Similarly, R. palustris and B. japonicum contain PII protein-encoding genes, yet the Nitrobacter PIIs adjacent to RuBisCO are more similar to those in the Betaproteobacteria, Nitrosomonas eutropha, and the denitrifying Thiobacillus denitrificans. Furthermore, the R. palustris and B. japonicum genomes contain periplasmic nitrate reductase (NapABC) but do not encode NarGH-type nitrate reductase (evolutionarily related to NXR), and the closest protein homologs of the Nitrobacter NXR are the putative nitrite oxidoreductase in Nitrococcus mobilis (∼68% protein sequence identity), a nitrite-oxidizing gammaproteobacterium, and NarGH in the deltaproteobacterium Geobacter metallireducens (∼59% protein sequence identity). In sum, the subcore appears to carry a collection of genes which are not indicative of its evolutionary origins but instead reflect the ecological niche (nitrification and denitrification) of Nitrobacter achieved through assimilation, modification, and expression of genes acquired from more distant bacterial lineages.
In conclusion, the genome sequence of N. hamburgensis, compared with those of N. winogradskyi and NB311A, has helped verify and narrow the genetic basis of nitrite oxidation in this famous Alphaproteobacteria lineage. Simultaneously, many putative gene candidates were identified which may expand the metabolic capabilities within the genus Nitrobacter and account for the phenotypic variations that are known to exist between N. winogradskyi and N. hamburgensis. Although the genome of N. hamburgensis is the largest of the Nitrobacter genomes, it is less organized and more fragmented, given its seemingly high content of pseudogenes, paralogs, mobile genetic elements, and phage remnants. Nevertheless, N. hamburgensis appears to have maintained a greater level of metabolic flexibility, especially in terms of organic and inorganic carbon use. Extensive duplications of several gene clusters (e.g., NXR, terminal oxidases, CODH, RuBisCO, etc.) in N. hamburgensis imply an increase in metabolic capacity and/or the ability to differentially express paralogous gene clusters based on environmental stimuli. In contrast, the N. winogradskyi genome is smaller, contains fewer paralogs, and may be restricting the organism to chemolithoautotrophic metabolism. Future genetic comparisons between additional Nitrobacter strains and other lineages of nitrite oxidizers, such as Nitrospira and Nitrococcus, will further advance our understanding of lithotrophic metabolism and the role of NOB in nitrification.
Supplementary Material
Acknowledgments
Sequencing was funded by the U.S. Department of Energy's Office of Biological and Environmental Research and carried out at the Joint Genome Institute. Additional funding was provided to S. R. Starkenburg by Subsurface Biosphere Integrative Graduate Education and Research Traineeship (IGERT) grant 0114427-DGE at Oregon State University from the National Science Foundation's Division of Graduate Education. M. G. Klotz, M. E. Gentry and A. T. Poret-Peterson were supported, in part, by incentive funds provided by the University of Louisville-EVPR office and the National Science Foundation (EF-0412129). Genome finishing was completed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory, University of California, under contract W-7405-Eng-48. Computational annotation was carried out at Oak Ridge National Laboratory.
Special thanks go to John Waterbury, the Moore Foundation, and The Venter Institute for submitting, funding, and sequencing the draft genome of NB311A.
Footnotes
Published ahead of print on 7 March 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahlers, B., W. Konig, and E. Bock. 1990. Nitrite reductase activity in Nitrobacter vulgaris. FEMS Microbiol. Lett. 67:121-126. [Google Scholar]
- 2.Arkin, I. T., H. Xu, M. O. Jensen, E. Arbely, E. R. Bennett, K. J. Bowers, E. Chow, R. O. Dror, M. P. Eastwood, R. Flitman-Tene, B. A. Gregersen, J. L. Klepeis, I. Kolossvary, Y. Shan, and D. E. Shaw. 2007. Mechanism of Na+/H+ antiporting. Science 317:799-803. [DOI] [PubMed] [Google Scholar]
- 3.Bock, E. 1976. Growth of Nitrobacter in the presence of organic matter. II. Chemoorganotrophic growth of Nitrobacter agilis. Arch. Microbiol. 108:305-312. [DOI] [PubMed] [Google Scholar]
- 4.Bock, E., H. P. Koops, H. Harms, and B. Ahlers. 1991. The biochemistry of nitrifying organisms, p. 171-200. In J. M. Shively and L. L. Barton (ed.), Variations in autotrophic life. Academic Press, San Diego, CA.
- 5.Bock, E., H.-P. Koops, U. C. Möller, and M. Rudert. 1990. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch. Microbiol. 153:105-110. [Google Scholar]
- 6.Bock, E., H. P. Koops, and H. Harms. 1986. Cell biology of nitrifiers, p. 17-38. In J. I. Prosser (ed.), Nitrification, vol. 20. IRL, Oxford, United Kingdom. [Google Scholar]
- 7.Bock, E., H. Sundermeyer-Klinger, and E. Stackebrandt. 1983. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch. Microbiol. 136:281-284. [Google Scholar]
- 8.Cantera, J. J., and L. Y. Stein. 2007. Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9:765-776. [DOI] [PubMed] [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, C. M.-H., T. Yan, X. Liu, L. Wu, J. Zhou, and L. Y. Stein. 2006. Transcriptome of Nitrosomonas europaea with a disrupted nitrite reductase (nirK) gene. Appl. Environ. Microbiol. 72:4450-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobley, J. G. 1984. Oxidation of nitrite and formate in Nitrobacter membrane preparations: evidence that both reactions are catalyzed by the same enzyme, p. 169-183. In W. R. Strohl and O. H. Tuovinen (ed.), Microbial chemoautotrophy. Ohio State University Press, Columbus.
- 12.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmore, B. O., D. J. Bergmann, M. G. Klotz, and A. B. Hooper. 2007. Cytochromes P460 and c′-beta; a new family of high-spin cytochromes c. FEBS Lett. 581:911-916. [DOI] [PubMed] [Google Scholar]
- 14.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 15.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 17.Freitag, A., and E. Bock. 1990. Energy conservation in Nitrobacter. FEMS Microbiol. Lett. 66:157-162. [Google Scholar]
- 18.Freitag, A., M. Rudert, and E. Bock. 1987. Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol. Lett. 48:105-109. [Google Scholar]
- 19.Friedrich, C. G., F. Bardischewsky, D. Rother, A. Quentmeier, and J. Fischer. 2005. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8:253-259. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Estepa, R., M. Argandona, M. Reina-Bueno, N. Capote, F. Iglesias-Guerra, J. J. Nieto, and C. Vargas. 2006. The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J. Bacteriol. 188:3774-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Estepa, R., D. Canovas, F. Iglesias-Guerra, A. Ventosa, L. N. Csonka, J. J. Nieto, and C. Vargas. 2006. Osmoprotection of Salmonella enterica serovar Typhimurium by Nγ-acetyldiaminobutyrate, the precursor of the compatible solute ectoine. Syst. Appl. Microbiol. 29:626-633. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 24.Harris, S., A. Ebert, E. Schutze, M. Diercks, E. Bock, and J. M. Shively. 1988. Two different genes and gene products for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCOase) in Nitrobacter hamburgensis. FEMS Microbiol. Lett. 49:267-271. [Google Scholar]
- 25.Horikiri, S., Y. Aizawa, T. Kai, S. Amachi, H. Shinoyama, and T. Fujii. 2004. Electron acquisition system constructed from an NAD-independent d-lactate dehydrogenase and cytochrome c2 in Rhodopseudomonas palustris no. 7. Biosci. Biotechnol. Biochem. 68:516-522. [DOI] [PubMed] [Google Scholar]
- 26.Houghton, J., Y. Ding, D. Griggs, M. Noguer, and P. J. van der Linden. 2001. Climate change 2001: the scientific basis. Cambridge University Press, Cambridge, United Kingdom.
- 27.Jormakka, M., B. Byrne, and S. Iwate. 2003. Formate dehydrogenase—a versatile enzyme in changing environments. Curr. Opin. Struct. Biol. 13:418-423. [DOI] [PubMed] [Google Scholar]
- 28.King, G. M. 2006. Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol. 56:1-7. [DOI] [PubMed] [Google Scholar]
- 29.King, G. M., and C. F. Weber. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5:107-118. [DOI] [PubMed] [Google Scholar]
- 30.Kirstein, K., and E. Bock. 1993. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch. Microbiol. 160:447-453. [DOI] [PubMed] [Google Scholar]
- 31.Kirstein, K. O., E. Bock, D. J. Miller, and D. J. D. Nicholas. 1986. Membrane-bound b-type cytochromes in Nitrobacter. FEMS Microbiol. Lett. 36:63-67. [Google Scholar]
- 32.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi, I. 1999. Homologous recombination and sex as a strategy against selfish genes attacking the genome. Ann. N. Y. Acad. Sci. 870:354-356. [DOI] [PubMed] [Google Scholar]
- 34.Koper, T. E., A. F. El-Sheikh, J. M. Norton, and M. G. Klotz. 2004. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 70:2342-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft, I., and E. Bock. 1984. Plasmids in Nitrobacter. Arch. Microbiol. 140:79-82. [Google Scholar]
- 36.Laanbroek, H. J., P. Bodelier, and S. Gerards. 1994. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch. Microbiol. 161:156-162. [Google Scholar]
- 37.Laanbroek, H. J., and S. Gerards. 1993. Competition for limiting amounts of oxygen between Nitrosomonas europaea and Nitrobacter winogradskyi grown in mixed continuous cultures. Arch. Microbiol. 159:453-459. [Google Scholar]
- 38.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 39.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349-377. [DOI] [PubMed] [Google Scholar]
- 40.Lorite, M. J., J. Tachil, J. Sanjuán, O. Meyer, and E. J. Bedmar. 2000. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 66:1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malavolta, E., C. C. Delwiche, and W. D. Burge. 1962. Formate oxidation by cell-free preparations from Nitrobacter agilis. Biochim. Biophys. Acta 57:347-351. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, O., L. Gremer, R. Ferner, M. Ferner, H. Dobbek, M. Gnida, W. Meyer-Klaucke, and R. Huber. 2000. The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase. Biol. Chem. 381:865-876. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, O., K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut. 1990. Utilization of carbon-monoxide by aerobes—recent advances. FEMS Microbiol. Rev. 87:253-260. [Google Scholar]
- 44.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 45.Pearson, A. R., B. O. Elmore, C. Yang, J. D. Ferrara, A. B. Hooper, and C. M. Wilmot. 2007. The crystal structure of cytochrome P460 of Nitrosomonas europaea reveals a novel cytochrome fold and heme-protein cross-link. Biochemistry 46:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reijerse, E. J., M. Sommerhalter, P. Hellwig, A. Quentmeier, D. Rother, C. Laurich, E. Bothe, W. Lubitz, and C. G. Friedrich. 2007. The unusual redox centers of SoxXA, a novel c-type heme-enzyme essential for chemotrophic sulfur-oxidation of Paracoccus pantotrophus. Biochemistry 46:7804-7810. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2005. REBASE—restriction enzymes and DNA methyltransferases Nucleic Acids Res. 33:D230-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rolls, J. P., and E. S. Lindstrom. 1967. Induction of a thiosulfate-oxidizing enzyme in Rhodopseudomonas palustris. J. Bacteriol. 94:784-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 50.Scott, K. M., S. M. Sievert, F. N. Abril, L. A. Ball, C. J. Barrett, R. A. Blake, A. J. Boller, P. S. Chain, J. A. Clark, C. R. Davis, C. Detter, K. F. Do, K. P. Dobrinski, B. I. Faza, K. A. Fitzpatrick, S. K. Freyermuth, T. L. Harmer, L. J. Hauser, M. Hugler, C. A. Kerfeld, M. G. Klotz, W. W. Kong, M. Land, A. Lapidus, F. W. Larimer, D. L. Longo, S. Lucas, S. A. Malfatti, S. E. Massey, D. D. Martin, Z. McCuddin, F. Meyer, J. L. Moore, L. H. Ocampo, Jr., J. H. Paul, I. T. Paulsen, D. K. Reep, Q. Ren, R. L. Ross, P. Y. Sato, P. Thomas, L. E. Tinkham, and G. T. Zeruth. 2006. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4:e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano, R. 1996. Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int. Rev. Cytol. 165:1-52. [DOI] [PubMed] [Google Scholar]
- 52.Smith, A. J., and D. S. Hoare. 1968. Acetate assimilation by Nitrobacter agilis in relation to its “obligate autotrophy.” J. Bacteriol. 95:844-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Starkenburg, S. R., P. S. Chain, L. A. Sayavedra-Soto, L. Hauser, M. L. Land, F. W. Larimer, S. A. Malfatti, M. G. Klotz, P. J. Bottomley, D. J. Arp, and W. J. Hickey. 2006. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72:2050-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein, L. Y., and Y. L. Yung. 2003. Production, isotopic composition, and atmospheric fate of biologically produced nitrous oxide. Annu. Rev. Earth Planet. Sci. 31:329-356. [Google Scholar]
- 55.Stein, L. Y., D. J. Arp, P. M. Berube, P. S. G. Chain, L. Hauser, M. S. M. Jetten, M. G. Klotz, F. W. Larimer, J. M. Norton, H. J. M. Op den Camp, M. Shin, and X. Wei. 2007. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ. Microbiol. 9:2993-3007. [DOI] [PubMed]
- 56.Steinmuller, W., and E. Bock. 1976. Growth of Nitrobacter in the presence of organic matter. I. Mixotrophic growth. Arch. Microbiol. 108:299-304. [DOI] [PubMed] [Google Scholar]
- 57.Stout, J., G. Van Driessche, S. N. Savvides, and J. Van Beeumen. 2007. X-ray crystallographic analysis of the sulfur carrier protein SoxY from Chlorobium limicola f. thiosulfatophilum reveals a tetrameric structure. Protein Sci. 16:589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Truper, H. G., and J. F. Imhoff. 1989. Genus Rhodopseudomonas Kluyver and van Niel in Czurda and Maresch 1937, 119AL, p. 1672-1677. In J. T. Staley, M. P. Bryant, N. Pfennnig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 59.Van Gool, A., and H. Laudelout. 1966. Formate utilization by Nitrobacter winogradskyi. Biochim. Biophys. Acta 127:295-301. [DOI] [PubMed] [Google Scholar]
- 60.Vargas, C., M. Jebbar, R. Carrasco, C. Blanco, M. I. Calderon, F. Iglesias-Guerra, and J. J. Nieto. 2006. Ectoines as compatible solutes and carbon and energy sources for the halophilic bacterium Chromohalobacter salexigens. J. Appl. Microbiol. 100:98-107. [DOI] [PubMed] [Google Scholar]
- 61.Wrage, N., G. L. Velthof, M. L. van Beusichem, and O. Oenema. 2001. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 36:229-236. [Google Scholar]
- 62.Zhulin, I., B. Taylor, and R. Dixon. 1997. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.