Abstract
“Dehalococcoides” bacteria can reductively dehalogenate a wide range of halogenated organic pollutants. In this study, DNA microarrays were used to monitor dynamic changes in the transcriptome as “Dehalococcoides ethenogenes” strain 195 transitioned from exponential growth into stationary phase. In total, 415 nonredundant genes were identified as differentially expressed. As expected, genes involved with translation and energy metabolism were down-regulated while genes involved with general stress response, transcription, and signal transduction were up-regulated. Unexpected, however, was the 8- to 10-fold up-regulation of four putative reductive dehalogenases (RDases) (DET0173, DET0180, DET1535, and DET1545). Another unexpected finding was the up-regulation of a large number of genes located within integrated elements, including a putative prophage and a multicopy transposon. Finally, genes encoding the dominant hydrogenase-RDase respiratory chain of this strain (Hup and TceA) were expressed at stable levels throughout the experiment, providing molecular evidence that strain 195 can uncouple dechlorination from net growth.
The remediation of groundwater resources contaminated with chlorinated organic pollutants is an ongoing environmental challenge. To address this challenge, one promising strategy is to utilize anaerobic bacteria that dechlorinate these pollutants via reductive pathways (34). “Dehalococcoides” bacteria are of particular interest for such applications for a number of reasons. First, they are the only known microorganisms that can completely dechlorinate tetrachloroethene (PCE) and trichloroethene (TCE) to nontoxic end products (34). Second, Dehalococcoides bacteria can dehalogenate a wide range of other halogenated organic pollutants, including both aliphatics and aromatics (1, 2, 6, 9, 16, 22, 37, 45). Finally, Dehalococcoides-based bioremediation systems have been successfully implemented in several field trials, some of which have potential for full-scale applications (15, 32, 36, 43).
Although our physiological understanding of Dehalococcoides bacteria is rapidly progressing, relatively little is known about how these organisms respond to conditions that limit their growth and dechlorination activity. Several studies have investigated conditions where growth is limited by the absence of chlorinated ethene electron acceptors. These studies found that genes encoding reductive dehalogenases (RDases) (27, 28, 31, 42, 51), hydrogenases (40, 42), and a putative formate dehydrogenase (40, 42) are down-regulated during periods of electron acceptor starvation and up-regulated when electron acceptors are available. Other studies have examined the effects of corrinoid deficiency, demonstrating that insufficient corrinoid can limit both growth and dechlorination activity (17, 23). This is consistent with the essential role of corrinoids as cofactors of characterized RDases (35) despite the lack of systems for de novo corrinoid biosynthesis in sequenced isolates (30, 47).
The primary objective of this study was to improve our general understanding of how “Dehalococcoides ethenogenes” strain 195 responds to growth-limiting conditions. Because electron acceptor- and corrinoid-limited growth conditions have been the focus of previous studies (23, 28, 31, 40, 42, 51), a second objective of this study was to identify novel factors that can limit the growth and dechlorination activity of this strain. To accomplish both of these objectives, batch cultures of strain 195 were grown with excess electron acceptor (TCE) and corrinoid, along with excess electron donor, carbon source, and nitrogen source. A DNA microarray was then used to examine temporal changes in the transcriptome as cells transitioned from exponential growth into stationary phase.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
D. ethenogenes strain 195 was grown at 34°C in batch-pure cultures (160-ml bottles with 100 ml liquid volume) with a defined mineral salts medium and an H2/CO2 headspace (80/20, vol/vol) (23, 31). The medium contained 5 mM sodium acetate and 5.5 mM ammonium chloride as the primary sources of carbon and nitrogen, respectively. After autoclaving, 7 μl of liquid TCE was added to each culture bottle using a 10-μl syringe (approximately 78 μmol). Cultures were further amended with a vitamin mixture containing 10 μg of the corrinoid cyanocobalamin (final concentration, 100 μg of cyanocobalamin per liter of medium) (23). This corrinoid concentration is well above the known growth-limiting concentration of 1 μg of cyanocobalamin per liter of medium (23). Cultures were then inoculated with 1 ml of a stock culture (1%, vol/vol) that had completely dechlorinated one 7-μl dose of TCE to vinyl chloride (VC) and ethene.
Growth experiment and cell collection.
A total of 180 parallel bottles containing 100 ml liquid medium were inoculated with 1 ml of stock culture (1%, vol/vol) and amended with 7 μl of TCE as described above. Additional 7-μl doses of TCE were added after complete conversion of TCE to VC and ethene. Two cultures were used to track chlorinated ethenes and Dehalococcoides-specific 16S rRNA genes throughout the time course of the experiment. For 16S rRNA gene analyses, cells were collected from 1-ml culture samples by vacuum filtration using hydrophilic Durapore membrane filters (diameter, 47 mm; pore size, 0.22 μm) (Millipore, Billerica, MA). After filtration, each filter was placed in a 2-ml microcentrifuge tube and stored at −80°C until processing. DNA was extracted from frozen filters, and the quantities of 16S rRNA genes per ml of culture were measured by quantitative PCR (qPCR) as described below.
At various times during the growth experiment, cultures were sacrificed and cells were collected for microarray analyses by vacuum filtration as described above (100 ml of liquid culture per filter). These collection times corresponded to the early-exponential (EE) (90 cultures sacrificed), late-exponential (LE) (30 cultures), transition (TR) (15 cultures), early-stationary (ES) (15 cultures), and late-stationary (LS) (30 cultures) phases. After filtration, the filters were immediately placed in 2-ml microcentrifuge tubes, frozen with liquid nitrogen, and stored at −80°C until processing. The time required to open each culture bottle, filter cells from 100 ml of culture, and freeze the filter with liquid nitrogen was less than 3 min.
RNA and DNA extraction.
RNA was extracted from frozen filters using a modified version of the acid phenol method described previously (27). Briefly, each 2-ml microcentrifuge tube that contained a frozen filter was filled with 250 μl lysis buffer (50 mM sodium acetate, 10 mM EDTA [pH 5.1]), 100 μl 10% sodium dodecyl sulfate, 1.0 ml buffer-equilibrated phenol (pH 4.3) (Sigma-Aldrich, St. Louis, MO), and 1 g 100-μm-diameter zirconia-silica beads (Biospec Products, Bartlesville, OK). Cells were lysed by heating to 65°C for 2 min, bead beating with a Mini Bead Beater (Biospec Products) for 2 min, incubating at 65°C for 8 min, and bead beating again for an additional 2 min. Cellular debris was collected by centrifugation (5 min at 15,000 × g), and the aqueous lysate was transferred to a new nuclease-free microcentrifuge tube. The lysate was extracted twice with 1 volume of phenol-chloroform-isoamyl alcohol (pH 4.3) (125:24:1, vol/vol) and once with 1 volume of chloroform-isoamyl alcohol (24:1, vol/vol) (Sigma-Aldrich). RNA was precipitated by adding 0.5 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol. The precipitate was collected by centrifugation, washed once with 80% ethanol, and resuspended in nuclease-free water. RNA was purified from contaminating DNA by DNase I treatment using the DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. Purified RNA was stored at −80°C prior to further use. The approximate mass of total RNA recovered from 100-ml cultures at each phase was as follows: EE, 0.2 μg; LE, 1.5 μg; TR, 3 μg; ES, 3 μg; LS, 1.5 μg.
DNA was extracted from frozen filters using a phenol-based protocol similar to that for RNA extraction described above. The differences are as follows. First, the phenol was buffer-equilibrated to pH 8.0 (Sigma-Aldrich) rather than pH 4.3. Second, the lysis buffer consisted of 200 mM Tris (pH 8.0), 50 mM EDTA, and 200 mM sodium chloride. Finally, DNA was precipitated by adding 0.1 volume of 3 M ammonium acetate (pH 5.2) and 1 volume of 100% isopropanol.
Affymetrix GeneChip characteristics.
The Affymetrix (Santa Clara, CA) GeneChip microarray applied in this study targets >99% of the 1,642 predicted genes within the genome of strain 195. A complete description of the array has been reported elsewhere, and the description includes an assessment of the quantitative dynamic range (53).
Microarray sample preparation, hybridization, and scanning.
Five micrograms of total RNA was used as starting material for each microarray analysis, where each 5-μg pool of RNA was obtained from an independent set of biological cultures (30 for EE, 10 for LE, 5 for TR, 5 for ES, and 10 for LS). A total of three microarray analyses were performed for every time point. cDNA was synthesized, fragmented, labeled, and hybridized to arrays according to the protocols outlined in section 3 of the Affymetrix GeneChip Expression Analysis technical manual. Hybridized arrays were stained and washed according to standard Affymetrix protocols and were scanned using an Affymetrix Scan3000 scanner.
Microarray data analysis.
All microarray analyses were performed within the R statistical programming environment (www.r-project.org) using packages available from Bioconductor, version 1.9 (www.bioconductor.org) (21). Probe set hybridization signal intensities were calculated using the “mas5” function from the “affy” package (20). This function implemented the MAS 5.0 algorithm (3) with global scaling to a value of 2,500. The average and median coefficients of variation among replicate measurements of individual probe set signal intensities were 8.6% and 5.5%, respectively.
To test the hypothesis that a gene was differentially expressed during the experiment, one-way analysis of variance (ANOVA) was applied using the “ftest” function from the “multtest” package (21). The P values returned from the ANOVA test were then adjusted to correct for multiple hypothesis testing by applying the Benjamini and Hochberg procedure (7) with the “rawp2adjp” function from the “multtest” package (21). This procedure was used to control the false discovery rate below 1%. Differentially expressed genes were further restricted using the following two criteria. First, the absolute hybridization signal intensity had to be greater than 300 for at least one sampling time point. Second, the change in expression between any two sampling time points had to be greater than twofold. Differentially expressed genes meeting all of these criteria were sorted by hierarchical clustering (14) using the “heatplot” function from the “made4” package (11).
qPCR analysis of 16S rRNA genes.
Quantification of Dehalococcoides-specific 16S rRNA genes by qPCR was performed using the protocol, primers, TaqMan probe (Applied Biosystems, Foster City, CA), and absolute standards described by Holmes and coworkers (25).
Chlorinated ethene analyses.
Chlorinated ethenes were resolved and analyzed by gas chromatography using a flame ionization detector and a GC-Gas Pro column (J&W Scientific, Folsom, CA) as described previously (31). The total mass of each chlorinated ethene was calculated using calibration curves generated from standards equilibrated to the culture incubation temperature of 34°C.
Bioinformatics.
Gene annotations were obtained from Seshadri and coworkers (47). Operon predictions were obtained from the Microbes Online Database (www.microbesonline.org) (5, 41).
Microarray data accession numbers.
All hybridization signal intensity measurements have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) (13) under accession numbers GPL6336 (platform) and GSE10185 (series).
RESULTS AND DISCUSSION
Characterization of growth phases.
The primary objective of this study was to improve our understanding of how strain 195 responds to growth-limiting conditions. To accomplish this objective, dechlorination rates, 16S rRNA gene quantities, and transcriptomics were monitored as batch-pure cultures transitioned from exponential growth into the stationary phase. A second objective of this study was to identify novel factors that can limit growth and activity. To address this second objective, growth factors known to be essential for strain 195 were added in excess, including the terminal electron acceptor (TCE), electron donor (hydrogen), carbon source (acetate), nitrogen source (ammonium), and corrinoid (cyanocobalamin).
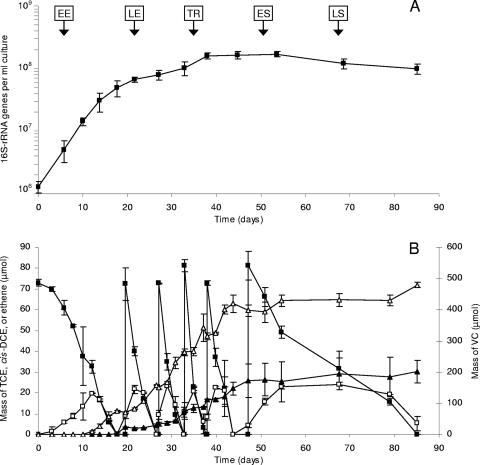
Figure 1 summarizes the temporal dynamics of 16S rRNA gene quantities and dechlorination rates over the duration of the experiment. Three distinct phases were observed. In the first phase (days 0 to 18), cells grew exponentially at a rate of 0.21 per day (generation time, 3.3 days) and with a yield of 3.2 × 107 ± 0.9 × 107 16S rRNA gene copies per μmol of Cl− released. After this phase of exponential growth, the growth rate slowed abruptly to a rate of 0.05 per day (generation time, 14 days) and remained steady thereafter until day 38. After day 38, net growth was no longer evident, while TCE dechlorination continued at decreasing rates (days 38 to 85). The continued dechlorination without a corresponding increase in the 16S rRNA gene copy number indicates that net growth had become uncoupled from dechlorination, a behavior similar to that previously reported for strain 195 (37) and for a Dehalococcoides-containing enrichment culture (28). Over the course of the experiment, approximately 450 μmol of TCE was dechlorinated to VC and ethene without any accumulation of cis-dichloroethene (cis-DCE) (Fig. 1B). Other isomers of DCE (trans-DCE and 1,1-DCE) were never detected in significant quantities (data not shown).
FIG. 1.
Temporal dynamics of Dehalococcoides-specific 16S rRNA genes (A) and chlorinated ethenes (B) in 100-ml batch-pure cultures of strain 195 receiving six consecutive doses of TCE. Vertical arrows in panel A indicate time points when cells were collected for microarray analyses. Symbols: ▪, TCE; □, cis-DCE; ▴, ethene; ▵, VC. All measurements are averages from two biological cultures. Error bars, 1 standard deviation.
Overview of transcriptome dynamics.
To provide insight into the factors controlling the observed shifts in physiology, transcriptomic microarray analyses were performed at different points of the experimental time course. Cells were collected for microarray analyses during the EE, LE, TR, ES, and LS phases, as shown in Fig. 1A.
Figure S1 in the supplemental material summarizes the overall dynamics of the transcriptome as cells transitioned into stationary phase. In this analysis, the total numbers of genes significantly up- or down-regulated relative to the EE phase are reported at each time point. As cells transitioned into the LE and TR phases, relatively few genes were up-regulated (19 and 33 genes, respectively) or down-regulated (45 and 38 genes, respectively). The major shift in gene expression occurred during the onset of the ES phase, with 96 genes up-regulated and 127 genes down-regulated (see Fig. S1 in the supplemental material). Time course studies of other bacteria have also reported that the largest shift in gene expression occurred during the transition into stationary phase (10, 50). As cells progressed further into the LS phase, the numbers of genes up- and down-regulated continued to increase (131 and 217, respectively) with no apparent stabilization of the transcriptome (see Fig. S1 in the supplemental material). Overall, a total of 415 nonredundant genes, corresponding to 27% of the nonredundant genes within the genome, were detected as either up- or down-regulated at some point during the experiment. This proportion of genes exhibiting growth phase-dependent regulation is within the range of 11 to 32% measured in similar investigations of other bacterial species (10, 26, 33, 46, 50).
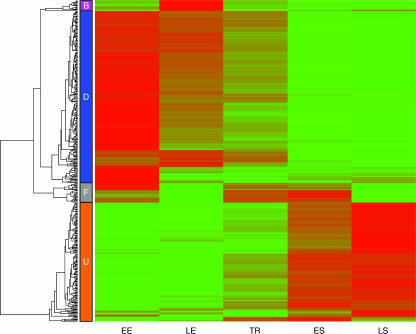
Hierarchical clustering was applied to provide further insight into the temporal dynamics of the transcriptome. This technique sorts genes into groups that exhibit similar patterns of transcription among a set of conditions (14). Four groups of genes that shared similar transcription patterns as cells transitioned from the EE to the LS phases were identified (Fig. 2). These groups are defined as response groups D (down-regulated), U (up-regulated), B (biphasic), and F (fluctuating) and are described in detail below. To facilitate analysis, the genes within each response group were further sorted into COG (clusters of orthologous genes) categories (see Fig. S2 in the supplemental material).
FIG. 2.
Hierarchical clustering and dendrogram of 415 differentially expressed genes (>2-fold; false discovery rate, <0.01). The color gradient from green to red represents increasing microarray hybridization intensity. Four major response groups were detected and are identified by the colored boxes labeled B (biphasic), D (down-regulated), F (fluctuating), and U (up-regulated).
Genes down-regulated as cells transition into stationary phase (response group D).
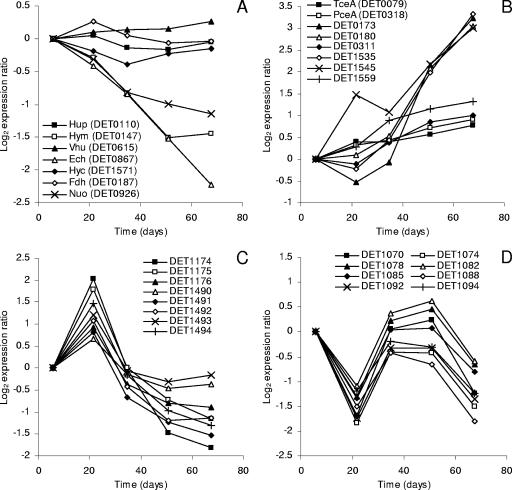
Response group D consists of 222 genes that were down-regulated as cells transitioned from the EE to the LS phase (Fig. 2). The largest category of genes within this response group is related to energy metabolism (see Fig. S2 in the supplemental material), and many of these genes encode oxidoreductases. Specifically, all the genes encoding the membrane-bound Hym (DET0145 to -0148) and Ech (DET0860 to -0868) hydrogenases and most of the genes encoding the NADH-quinone oxidoreductase (Nuo) (DET0925 to -0933) were down-regulated two- to sixfold (Fig. 3A; see also Table S1 in the supplemental material). Although the physiological functions of these oxidoreductases are currently unknown, the Ech hydrogenase has been speculated to carry low-potential electrons for biosynthetic reactions (40), which would be consistent with its observed down-regulation under growth-limiting conditions. Also included within the energy metabolism category were genes encoding an incomplete tricarboxylic acid cycle (DET0448 to -0451 and DET0453 to -0454) and the ATP synthase complex (DET0558 and DET0561 to -0565), all of which were down-regulated two- to fourfold (see Table S1 in the supplemental material). The down-regulation of these systems is expected given the observed reduction in growth rates over the duration of the experiment.
FIG. 3.
Transcription profiles of select genes from each response group. Each expression measurement is the ratio of the hybridization intensity at the specified time to the hybridization intensity observed during the EE phase (day 6). Genes for which results are shown encode oxidoreductases (A), RDases (B), ABC transporters (C), and putative prophage proteins (D).
After energy metabolism, the next largest category of genes within response group D was related to translation and ribosomes (see Fig. S2 in the supplemental material). Included within this category were 11 genes encoding ribosomal components and 6 genes encoding tRNA synthases, all of which were down-regulated two- to fourfold (see Table S1 in the supplemental material). As with genes encoding ATP synthase, the down-regulation of translation-related genes during stationary phase is expected given the reduction in growth rates.
The remaining genes within response group D were broadly distributed among many categories (see Fig. S2 in the supplemental material). Of these, 55 genes were classified within the transport and metabolism categories and encoded a range of different systems, including several ABC-type transporters and pathways involved with lipid and nucleotide metabolism (see Table S1 in the supplemental material). There were also a large number of unclassified genes. These genes were distributed throughout the genome and were strongly biased against integrated elements (IEs). Overall, only 3% of the nonredundant genes from response group D (7 out of 222) were located within IEs (see Table S1 in the supplemental material), while IE-located genes constitute more than 9% of the total nonredundant genome (47).
Genes up-regulated as cells transition into stationary phase (response group U).
Response group U consists of 154 genes that were up-regulated as cells transitioned from the EE to the LS phase (Fig. 2). For this group, the most surprising finding was the increased expression of nine genes in the energy generation category (see Fig. S2 in the supplemental material), six of which encode putative RDases (Fig. 3B). The expression dynamics of these RDases could be sorted into two distinct subgroups. The first subgroup consisted of DET0311 and DET1559, both of which were up-regulated two- to threefold at fairly consistent rates over the duration of the experiment (Fig. 3B). The expression dynamics of these two genes are similar to those for the genes encoding TceA (DET0079) and PceA (DET0318), neither of which met our criteria for differential expression (Fig. 3B). The second subgroup consisted of DET0173, DET0180, DET1535, and DET1545. These RDases distinguished themselves from the first subgroup by having a burst of expression between the TR and LS phases (Fig. 3B). Overall, RDases within subgroup 2 were up-regulated 8- to 10-fold and ranked among the 10 most up-regulated genes within the entire genome (see Table S1 in the supplemental material).
Another striking feature of response group U was that 32% of these genes (49 out of 154) were located within IEs. In fact, at least one gene from each IE was up-regulated during the transition into the ES and LS phases (Table 1). Particularly surprising was the increased expression of all 22 genes from a 22-gene transposon that is present in three identical copies (IEs III, IV, and VI) (Table 1; see also Table S1 in the supplemental material). Also interesting was the up-regulation of 13 genes from IE VII (Table 1; see also Table S1 in the supplemental material), which shows extensive synteny with prophages of Bacillus cereus and other members of the Firmicutes (47). These IEs, discussed above, do not encode RDases or provide any other apparent selective advantage to strain 195 (47). Moreover, they are absent from other sequenced Dehalococcoides isolates, including strains CBDB1 (30) and BAV1 (NCBI accession number NC_009455).
TABLE 1.
Summary of the number, fraction, and response group of genes from each IE exhibiting differential expression
| IEa | Locus namesa | RDasesa | Total no. of ORFsa | No. of ORFs differentially expressed | Fraction of ORFs differentially expressed (%) | Response group(s) |
|---|---|---|---|---|---|---|
| I | DET0063-0091 | Yes | 28 | 2 | 7.1 | U |
| II | DET0155-0169 | Yes | 14 | 2 | 14 | U |
| IIIb | DET0251-0272 | No | 22 | 22 | 100 | U |
| IVb | DET0273-0295 | No | 22 | 22 | 100 | U |
| V | DET0875-0883 | Yes | 9 | 1 | 11 | U |
| VIb | DET0884-0905 | No | 22 | 22 | 100 | U |
| VII | DET1066-1118 | No | 52 | 42 | 81 | D, F, U |
| VIII | DET1472-1478 | No | 7 | 4 | 57 | D, U |
| IX | DET1552-1561 | Yes | 10 | 6 | 60 | U |
See reference 47. ORFs, open reading frames.
IEs III, IV, and VI are identical copies.
A number of genes from response group U have putative functions involved with general stress response systems (see Fig. S2 in the supplemental material, under the posttranslational modification and protein turnover category and the unclassified category). These include genes encoding heat shock-type chaperones (DET0954, DET1286, DET1411) and components of the oxidative stress response and repair systems (DET0198, DET1241, DET1581, DET1623) (see Table S1 in the supplemental material). Also interesting is the concomitant twofold up-regulation of an extracytoplasmic function (ECF)-type sigma factor (DET1348) (see Table S1 in the supplemental material). ECF sigma factors are known to regulate the transcription of various stress responses in other organisms, including responses to heat shock and oxidative stress (24, 38).
In addition to the ECF sigma factor, several other genes from response group U were assigned to the transcription and signal transduction categories. These include a gene encoding a LexA-type regulator (DET0251) that was up-regulated approximately twofold (see Table S1 in the supplemental material). Interestingly, this gene is located within the up-regulated multicopy IE (IEs III, IV, and VI) discussed above. Also included was a TetR-type transcription regulator (DET1580) that was up-regulated twofold only after entering ES phase (see Table S1 in the supplemental material). The timing of this increase in expression is intriguing given that TetR-type regulators have been linked with cell density-sensing regulatory cascades (29, 44). Finally, genes encoding response regulators for four putative RDases were up-regulated (DET0300, DET1524, DET1540, and DET1561) (see Table S1 in the supplemental material). None of these response regulators, however, are predicted to regulate the group of RDases exhibiting the highest levels of up-regulation (8- to 10-fold) discussed above (Fig. 3B) (47).
Genes with biphasic expression dynamics (response group B).
Response group B consists of 14 genes that were characterized by biphasic expression dynamics, with an initial period of up-regulation followed by down-regulation thereafter (Fig. 2). The majority of these 14 genes encode transport systems (see Table S1 in the supplemental material). Six genes (DET1490 to -1495) are located within an operon predicted to encode a peptide ABC transporter, and four genes (DET1173 to -1176) are located within an operon predicted to encode a ferric iron ABC transporter. Most of these genes were up-regulated two- to fourfold during the LE phase and subsequently down-regulated to levels below those initially observed (Fig. 3C). Temporal transcriptomic studies of other bacterial species have reported similar findings. Specifically, genes encoding peptide transporters in Bacillus anthracis (33) and iron transporters in Desulfovibrio vulgaris (10) were similarly up-regulated during the LE and ES phases and down-regulated thereafter. It is noteworthy that the increased expression of genes within response group B coincided with the shift to the sustained lower growth rate observed between days 18 and 38 (Fig. 1A).
Genes with fluctuating expression dynamics (response group F).
Response group F consists of 25 genes that were characterized by fluctuating expression dynamics (Fig. 2). In general, these genes were expressed at relatively high levels during the EE phase, down-regulated during the LE phase, up-regulated during the TR and ES phases, and down-regulated again during the LS phase (Fig. 3D). Strikingly, 24 of these 25 genes are located within IE VII, which shows extensive synteny with prophages of Bacillus cereus (47). These 24 genes are contiguous (DET1070 to -1072, DET1074 to -1094), are transcribed in the same direction, and are predicted to encode proteins involved with phage assembly, including a capsid protein, a tail fiber coating protein, a DNA packaging protein, and an endolysin (see Table S1 in the supplemental material).
Genes expressed at stable levels as cells transition into stationary phase.
A number of genes of interest were expressed at stable levels throughout the time course of the experiment. Most notable were genes encoding the Hup hydrogenase and the TceA RDase (Fig. 3A and B). These oxidoreductases are thought to make up the dominant hydrogenase-RDase respiratory chain of strain 195 when cells are grown with TCE (35, 39, 40). The stable expression of these genes even 30 days after growth had ceased provides molecular-level evidence supporting physiological observations that Dehalococcoides bacteria can uncouple net growth from dechlorination (28, 37).
Another interesting finding was that the genes encoding the Hup, Hyc, and Vhu hydrogenases and the putative Fdh formate dehydrogenase were all expressed at stable levels as cells transitioned into stationary phase (Fig. 3A; see also Table S1 in the supplemental material). The stable expression of Fdh is especially intriguing because previous studies have reported that it is highly expressed relative to other oxidoreductases (39, 40, 42), suggesting that it plays a crucial role in Dehalococcoides physiology.
Other genes that were expressed at stable levels include those encoding a corrinoid ABC transporter (DET0650 to -0652) and a corrinoid salvage pathway (DET0657 to -0660) (see Table S1 in the supplemental material). This is consistent with corrinoid availability not being a factor limiting growth and activity under the conditions investigated. Also notable is the stable expression of a previously studied LexA-encoding gene (DET1640) (see Table S2 in the supplemental material) that is presumed to be involved in DNA repair (18).
Physiological adaptation to growth-limiting conditions.
The primary objective of this study was to characterize the adaptive response of strain 195 to growth-limiting conditions. The general finding from the transcriptomic studies is that genes involved with energy metabolism and translation were down-regulated while genes involved with stress response, transcription, and signal transduction were up-regulated during the transition into stationary phase. These responses are consistent with the stationary-phase responses observed for a wide range of other bacterial species in comparable studies (4, 8, 10, 12, 26, 33, 46, 49, 50, 52).
A second goal of this study was to identify novel factors that can limit the growth and dechlorination activity of strain 195 by adding known growth-limiting factors in excess, including the electron donor, electron acceptor, nitrogen source, carbon source, and corrinoid. Unfortunately, the time course microarray approach used here did not clearly identify a potential factor responsible for the onset of stationary phase. Although predicted iron and peptide ABC-type transporters were transiently up-regulated during the LE phase (Fig. 3C), no transporters were up-regulated during the ES and LS phases after net growth had ceased. One reason for the absence of expression dynamics indicative of a specific nutritional limitation might be that no corresponding adaptive system exists in strain 195.
Factors other than nutrient limitations may also have contributed to the cessation of growth. For instance, the accumulation of VC and ethene (Fig. 1B) might have resulted in product inhibition. Alternatively, a quorum-sensing-type regulatory mechanism might have controlled the maximum cell density. Such a mechanism seems plausible given the up-regulation of a TetR-type response regulator (DET1580) during the ES and LS phases (see Table S1 in the supplemental material). TetR regulators have been implicated in density-sensing regulatory cascades in Pseudomonas species (29, 44), and thus, analysis of a TetR mutant would be a promising avenue for future investigations.
Even though no obvious growth-limiting factor was identified, this investigation yielded important findings about the transition of strain 195 into stationary phase. Particularly surprising was the 8- to 10-fold up-regulation of four RDase-encoding genes (DET0173, DET0180, DET1535, and DET1545) (Fig. 3B). These RDases ranked among the 10 genes with the highest levels of up-regulation, suggesting that they play important roles in the observed stationary-phase response. The absolute hybridization signals of these RDases, however, were substantially lower than that of the tceA gene (DET0079) (see Table S1 in the supplemental material). Even during the LS phase, when these RDases were maximally expressed, only DET1545 had a hybridization signal that was within 1 order of magnitude of the tceA signal (see Table S1 in the supplemental materia). Thus, it appears unlikely that these RDases play crucial roles in TCE dechlorination. Nor do they appear to play a role in DCE or VC dechlorination, since they were not up-regulated until after the concentrations of VC and ethene had stabilized (Fig. 1B and 3B). Thus, it remains unclear why these RDases were up-regulated during stationary phase; characterizations of their substrate ranges are needed in order to elucidate their physiological role.
Another surprising finding was the stable expression of genes encoding the Hup hydrogenase and the TceA RDase, which are thought to comprise the dominant hydrogenase-RDase respiratory chain when strain 195 is grown with TCE (35, 39, 40). One would expect these genes to be down-regulated as rates of growth decline. Instead, these genes were expressed at stable levels even 30 days after net growth had ceased, suggesting a lack of integration between cell growth and their transcriptional control. It should be noted that the substrates of these enzymes, hydrogen and TCE, were still abundant in the culture and may have served to induce their expression. Indeed, previous studies have shown that the presence of TCE is necessary for tceA transcription (19). Studies of gene expression under conditions of TCE and H2 limitation would clarify some of these issues and help determine whether measurements of RDase expression levels can be used to accurately assess the physiological activity of strain 195.
An examination of the expression of other oxidoreductase-encoding genes also revealed interesting transcription dynamics. Previous investigations showed that Hup, Vhu, and Fdh are down-regulated during electron acceptor-limited growth conditions (40, 42). In this study, however, these three oxidoreductases were expressed at stable levels even 30 days after net growth had ceased. Thus, it appears that distinct regulatory systems control the adaptive responses to electron acceptor-limiting conditions and the growth-limiting conditions investigated here. The oxidoreductases that were down-regulated in this study were the membrane-bound Ech and Hym hydrogenases and the Nuo NADH-quinone oxidoreductase. Morris and coworkers also observed lower transcript levels for these three oxidoreductases under electron acceptor-limiting conditions (40). Thus, even though distinct regulatory systems are involved with the adaptation to different growth limitations, significant portions overlap within a general stationary-phase response.
Growth phase-dependent expression of IEs.
This study identified a large number of genes located within IEs that are differentially regulated during the transition into stationary phase (Table 1). Particularly surprising was the up-regulation of 100% of the genes from a 22-gene transposon that is present in three identical copies (IEs III, IV, and VI). However, this transposon does not provide any obvious selective advantage to the cell. It is therefore difficult to interpret these results except to state that, like many transposons, these elements show signs of mobilization under growth-limiting conditions.
Also of interest was the differential regulation of genes from IE VII (DET1066 to -1118) that encode components of a temperate bacteriophage (47). The up-regulated genes are located within two contiguous sections (DET1067 to -1069 and DET1095 to -1104 [see Table S1 in the supplemental material]) and are predicted to encode genes for phage excision and DNA synthesis. These are reminiscent of the classic “early” genes in the bacteriophage lytic cycle (48). The genes showing fluctuating transcript levels are located in a separate contiguous section (DET1070 to -1094 [see Table S1 in the supplemental material]) and are reminiscent of the “late” genes for phage assembly. It is difficult to explain the fluctuating expression patterns of the “late” genes, and factors such as phage excision and replication and the lower energy status of the cells when TCE dechlorination slowed may have played a role. Nevertheless, the fact that these genes within this contiguous section all follow the same pattern shows that their expression is tightly coordinated in a growth phase-dependent manner. Although we did not examine whether this prophage was induced from lysogeny during stationary phase, the induction of this phage might be another factor contributing to the cessation of growth of strain 195.
Supplementary Material
Acknowledgments
We thank Kimberlee West, Patrick Lee, and Todd DeSantis for thoughtful discussions.
This work was supported by the Lawrence Berkeley National Laboratory through the Laboratory Directed Research and Development Program, the National Science Foundation under grant 0504244, and the Superfund Basic Research Program under grant NIEHS ES04705.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adrian, L., S. K. Hansen, J. M. Fung, H. Görisch, and S. H. Zinder. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ. Sci. Technol. 41:2318-2323. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Affymetrix. 2001. Statistical algorithms reference guide. Technical report. Affymetrix, Santa Clara, CA.
- 4.Alsaker, K. V., and E. T. Papoutsakis. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 15:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedard, D. L., K. M. Ritalahti, and F. E. Löffler. 2007. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl. Environ. Microbiol. 73:2513-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 8.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 9.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 10.Clark, M. E., Q. He, Z. He, K. H. Huang, E. J. Alm, X.-F. Wan, T. C. Hazen, A. P. Arkin, J. D. Wall, J.-Z. Zhou, and M. W. Fields. 2006. Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culhane, A. C., J. Thioulouse, G. Perrière, and D. G. Higgins. 2005. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics 21:2789-2790. [DOI] [PubMed] [Google Scholar]
- 12.Denef, V. J., J. Park, T. V. Tsoi, J.-M. Rouillard, H. Zhang, J. A. Wibbenmeyer, W. Verstraete, E. Gulari, S. A. Hashsham, and J. M. Tiedje. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis, D. E., E. J. Lutz, J. M. Odom, R. J. Buchanan, Jr., M. D. Lee, C. L. Bartlett, M. R. Harkness, and K. A. DeWeerd. 2000. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 34:2254-2260. [Google Scholar]
- 16.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 17.Fennell, D. E., J. M. Gossett, and S. H. Zinder. 1997. Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ. Sci. Technol. 31:918-926. [Google Scholar]
- 18.Fernández de Henestrosa, A. R., J. Cuñé, I. Erill, J. K. Magnuson, and J. Barbé. 2002. A green nonsulfur bacterium, Dehalococcoides ethenogenes, with the LexA binding sequence found in gram-positive organisms. J. Bacteriol. 184:6073-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung, J. M., R. M. Morris, L. Adrian, and S. H. Zinder. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73:4439-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier, L., L. Cope, B. M. Bolstad, and R. A. Irizarry. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307-315. [DOI] [PubMed] [Google Scholar]
- 21.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. H. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, J., K. R. Robrock, and L. Alvarez-Cohen. 2006. Microbial reductive debromination of polybrominated diphenyl ethers (PBDEs). Environ. Sci. Technol. 40:4429-4434. [DOI] [PubMed] [Google Scholar]
- 23.He, J., V. Holmes, P. K. H. Lee, and L. Alvarez-Cohen. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, V. F., J. He, P. K. H. Lee, and L. Alvarez-Cohen. 2006. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl. Environ. Microbiol. 72:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J., C.-J. Lih, K.-H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. R., P. K. H. Lee, V. F. Holmes, A. C. Fortin, and L. Alvarez-Cohen. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 31.Lee, P. K. H., D. R. Johnson, V. F. Holmes, J. He, and L. Alvarez-Cohen. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lendvay, J. M., F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, Jr., M. J. Barcelona, E. Petrovskis, R. Hickey, J. M. Tiedje, and P. Adriaens. 2003. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422-1431. [Google Scholar]
- 33.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates III, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Löffler, F. E., and E. A. Edwards. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274-284. [DOI] [PubMed] [Google Scholar]
- 35.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 37.Maymó-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 38.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 39.Morris, R. M., J. M. Fung, B. G. Rahm, S. Zhang, D. L. Freedman, S. H. Zinder, and R. E. Richardson. 2007. Comparative proteomics of Dehalococcoides reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris, R. M., S. Sowell, D. Barofsky, S. Zinder, and R. E. Richardson. 2006. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ. Microbiol. 8:1499-1509. [DOI] [PubMed] [Google Scholar]
- 41.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahm, B. G., R. M. Morris, and R. E. Richardson. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72:5486-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahm, B. G., S. Chauhan, V. F. Holmes, T. W. Macbeth, K. S. Sorenson, and L. Alvarez-Cohen. 2006. Molecular characterization of microbial populations at two sites with differing reductive dechlorination abilities. Biodegradation 17:523-534. [DOI] [PubMed] [Google Scholar]
- 44.Ramos, J. L., M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritalahti, K. M., and F. E. Löffler. 2004. Populations implicated in anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities. Appl. Environ. Microbiol. 70:4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigues, F., M. Sarkar-Tyson, S. V. Harding, S. H. Sim, H. H. Chua, C. H. Lin, X. Han, R. K. M. Karuturi, K. Sung, K. Yu, W. Chen, T. P. Atkins, R. W. Titball, and P. Tan. 2006. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J. Bacteriol. 188:8178-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Dinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 48.Snyder, L., and W. Champness. 2002. Molecular genetics of bacteria, 2nd ed. ASM Press, Washington, DC.
- 49.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waller, A. S., R. Krajmalnik-Brown, F. E. Löffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σs-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West, K. A., D. R. Johnson, P. Hu, T. Z. DeSantis, E. L. Brodie, P. K. H. Lee, H. Feil, G. L. Andersen, S. H. Zinder, and L. Alvarez-Cohen. 21 March 2008. Comparative genomics of Dehalococcoides ethenogenes 195 and an enrichment culture containing unsequenced Dehalococcoides strains. Appl. Environ. Microbiol. doi: 10.1128/AEM.01835-07. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.