Abstract
Incorporation of plant residues strongly enhances the methane production and emission from flooded rice fields. Temperature and residue type are important factors that regulate residue decomposition and CH4 production. However, the response of the methanogenic archaeal community to these factors in rice field soil is not well understood. In the present experiment, the structure of the archaeal community was determined during the decomposition of rice root and straw residues in anoxic rice field soil incubated at three temperatures (15°C, 30°C, and 45°C). More CH4 was produced in the straw treatment than root treatment. Increasing the temperature from 15°C to 45°C enhanced CH4 production. Terminal restriction fragment length polymorphism analyses in combination with cloning and sequencing of 16S rRNA genes showed that Methanosarcinaceae developed early in the incubations, whereas Methanosaetaceae became more abundant in the later stages. Methanosarcinaceae and Methanosaetaceae seemed to be better adapted at 15°C and 30°C, respectively, while the thermophilic Methanobacteriales and rice cluster I methanogens were significantly enhanced at 45°C. Straw residues promoted the growth of Methanosarcinaceae, whereas the root residues favored Methanosaetaceae. In conclusion, our study revealed a highly dynamic structure of the methanogenic archaeal community during plant residue decomposition. The in situ concentration of acetate (and possibly of H2) seems to be the key factor that regulates the shift of methanogenic community.
Plant residues are a by-product of rice production. The incorporation of plant residues into rice field soil is a common practice in Asian agriculture that could help to maintain the soil fertility. However, the incorporation of plant residues strongly enhances the methane emissions from rice fields (8, 13, 38, 39). It has been shown that the decomposition of straw residues in rice field soil is performed by a spatially well-organized consortium: the hydrolysis and primary fermentation reactions are mainly localized on the straw residue while the syntrophic and methanogenic reactions occur in the adjacent soil (14). There was an astonishing richness of archaeal diversity present on rice roots and in the surrounding paddy soil (4, 16, 17, 29). Although it was shown that the structure of methanogenic archaea in the rice field soil remained relatively constant over the growing season of rice plants (24), the incorporation of rice straw selectively enhanced the growth of Methanosarcinaceae and Methanobacteriales and suppressed rice cluster I (RC-I) methanogens and Methanomicrobiales (7). Apparently, the members of the methanogenic community respond differently to the incorporation of organic residues. Rice straw and root residues are the major forms of organic residues incorporated into rice field soils. It was shown that the decomposition of root residues was slower than straw residues (26). However, it is unknown whether such a difference in the rates of decomposition is also reflected in the structure of the residue-decomposing microbiota.
Temperature is another important factor that controls the decomposition of organic materials (6, 22, 25). Glissmann and colleagues (15) found that incubation of lake sediment samples (in situ, 7.5°C) at increasing temperatures from 4°C to 30°C resulted in increasing rates of CH4 and H2 production. Studies on rice field soils gave similar results (12, 25). However, the shift of temperature may change not only the rate of CH4 production but also its pathway. It has been shown that hydrogenotrophic methanogenesis and acetoclastic methanogenesis account for approximately 33% and 67% of total CH4 production at 30°C, respectively (5). However, the contribution of acetoclastic methanogensis increases to approximately 85% at 10°C (6), while the substrate for CH4 production is found to be exclusively H2-CO2 at 50°C (11). The change of methanogenic pathway is reflected in the structure of the methanogenic community. It was revealed that Methanosaetaceae, Methanosarcinaceae, and the thermophilic RC-I methanogens dominated the methanogenic community in Italian rice field soil at 15°C, 30°C, and 50°C, respectively (2, 11, 42).
A number of studies have been conducted to determine the structure of the methanogenic archaeal community in flooded rice soils (24, 29, 34, 40), but few have evaluated the effect of plant residue incorporation (7); and to our knowledge none has determined the effects of residue incorporation at different temperatures. The purpose of the present study was (i) to compare the rates of CH4 production from the decomposition of rice straw relative to root residue, (ii) to determine the structure of the archaeal community during the decomposition processes, and (iii) to evaluate the effect of temperature (15°C, 30°C, and 45°C) and residue type on the dynamics of archaeal community in rice field soil.
MATERIALS AND METHODS
Preparation of soil and residue samples.
Rice field soil and residue samples were collected from an experimental farm at the China National Rice Research Institute in Hangzhou, China (30o04′37″N, 119o54′37″E). The soil sample had the following characteristics as measured by standard methods (32) (per kg of soil): pH 6.7, cation exchange capacity of 14.4 cmol, organic C of 24.2 g, and total N of 2.3 g. Soil samples were air dried and stored at 4°C. Rice root residues and the stems of rice plants were collected at the maturity of plants, dried for 48 h at 60°C, and cut into 0.5-cm-long pieces. Glass bottles (100 ml) were filled with 10 g of dry soil and 20 ml of autoclaved and degassed water. The rice straw and root residues (0.1 g) were added to individual bottles. The bottles were closed with butyl stoppers, evacuated, and flushed with N2 several times. Then, all bottles were shaken vigorously for 30 s and incubated for 90 days at 15°C, 30°C, or 45°C. Each treatment was carried out in triplicate.
Measurement of gases and volatile fatty acids.
Gas samples (0.5 ml) were taken from headspace with a syringe. The concentrations of CH4, CO2, and H2 were analyzed using gas chromatographs (Shanghai Precision and Scientific Instrument, China) equipped with a methanizer and flame ionization detector or with a thermal conductivity detector. Liquid samples (0.5 ml) were taken with sterile syringes and centrifuged for 15 min at 17,949 × g at 4°C. The supernatant was collected, passed through 0.25-μm-pore-size filters, and stored at −20°C. Acetate was analyzed with an HP 6890 gas chromatograph (Agilent Technologies).
DNA extraction from soil samples.
At various time points, triplicate bottles from each treatment were destructively sampled. Soil slurries were mixed thoroughly and then centrifuged at 3,800 × g for 15 min to remove the residue debris. The soil samples were stored frozen at −20°C until DNA extraction.
About 0.5 g of soil (fresh weight) was placed into a 2-ml bead-beating tube containing 0.7 g of 0.1-mm glass beads. After the addition of 660 μl of TNS buffer (0.5 M Tris-HCl, pH 8.0, 0.1 M NaCl, 10% sodium dodecyl sulfate [wt/vol]) and 220 μl of phosphate buffer (112.9 mM Na2HPO4, 7.12 mM NaH2PO4, pH 8.0), the mixture was shaken for 45 s at 6.5 m s−1 in a bead beater, followed by centrifugation at 20,000 × g for 4 min (27). Further purification and precipitation of DNA extracts followed the procedure as described previously (27). The DNA pellet was finally suspended in 0.2 ml of elution buffer (10 mM Tris-HCl, pH 8.5) and stored at −20°C.
Amplification of archaeal 16S rRNA gene fragments and terminal restriction fragment length polymorphism (T-RFLP) analysis.
PCR was carried out using the domain-specific primer set Ar109f [5′-AC(G/T) GCT CAG TAA CAC GT-3′] and Ar915r (5′-GTG CTC CCC CGC CAA TTC CT-3′) (31). The 5′ end of the Ar915r primer was labeled with 6-carboxyfluorescein. The 50-μl reaction mixture contained 1 μl of DNA template (in 1:10 dilution of original extracts), 5 μl of 10× buffer, 3 μl of 25 mM MgCl2, 1 μl of a 10 mM concentration of the deoxynucleoside triphosphates, 0.5 μl of each primer (50 μM), and 2.5 U of Taq DNA polymerase (TaKaRa). The thermal profile for amplification was as follows: 3 min at 94°C; 32 cycles of 60 s at 94°C, 45 s at 52°C, and 90 s at 72°C; and finally 5 min at 72°C. The 6-carboxyfluorescein-labeled PCR product was purified using an agarose gel DNA extraction kit (TaKaRa) and digested at 65°C for 3.5 h by TaqI (Fermentas, Canada). The digestion products were further purified with SigmaSpin Post-Reaction Clean-Up Columns (Sigma), and a portion was mixed with deionized formamide and the internal standard GeneScan-500 LIZ (Applied Biosystems). The mixtures were denatured for 3 min at 95°C, and the DNA fragments were size separated using a 3130xl Genetic Analyzer (Applied Biosystems).
Cloning, sequencing, and phylogenic analysis.
Four clone libraries were constructed: one for the root residue treatment on day 8 at 30°C and three for the straw treatments on day 8 and day 64 at 30°C (one each) and day 90 at 45°C. The PCR amplification used the same primers as indicated above. PCR products were purified and ligated into the pMD19-T vector (TaKaRa) according to the manufacturer's instructions. Plasmids were transformed into Escherichia coli cells, and more than 30 clones were randomly selected from each clone library and sequenced with an ABI 3730xl sequencer using BigDye Terminator cycle sequencing chemistry (Applied Biosystems).
The sequence data were analyzed using the ARB software package (http://www.arb-home.de). The 16S rRNA gene sequences were aligned and integrated into the ARB database consisting of 14,795 complete or partial 16S rRNA sequences. Some sequence data from the GenBank database were imported to include the closest matches that were not yet available in the ARB database. Phylogenetic trees were constructed using the neighbor-joining algorithm according to the protocol of Lueders and Friedrich (29). Highly variable nucleotide positions and those with possible alignment errors were excluded from the phylogenetic analysis by using only positions present in at least 50% of all the sequences compared in this study.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA clones obtained in this study have been deposited in the EMBL nucleotide sequence database under the following accession numbers: AM778233 to AM778272 and AM779889 for clones D8Ar30S2 to D8Ar30S90; AM778273 to AM778301 for clones D8Ar30R1 to D8Ar30R64; AM778302 to AM778355 for clones D64Ar30R1 to D64Ar30R71; and AM778356 to AM778406 for clones D90Ar45R1 to D90Ar45R65.
RESULTS
CH4, H2, and acetate production.
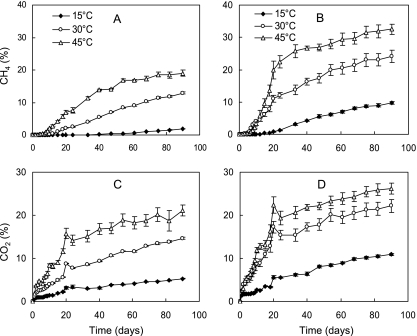
The rates of CH4 production were greater in the straw treatment than root treatment (Fig. 1A and B). After a lag phase, the CH4 production increased linearly between days 7 and 20. The production of CO2 increased without lag phase and was greater in the straw treatment than root treatment (Fig. 1C and D). The lag phase of CH4 production was probably due to the reduction of oxidants such as Fe(III) and sulfate in the soil. Increasing the temperature from 15°C to 45°C significantly enhanced CH4 production. However, during the early period (until day 10), the production of CH4 in the straw treatment exhibited a longer lag phase at 45°C than 30°C. This prolonged lag phase probably resulted from the time required for the thermophilic degraders and methanogens to be activated (12).
FIG. 1.
Effect of temperature on CH4 (A and B) and CO2 (C and D) production in rice field soil after the addition of rice root (A and C) and straw residues (B and D). Data are means ± standard errors (n = 3).
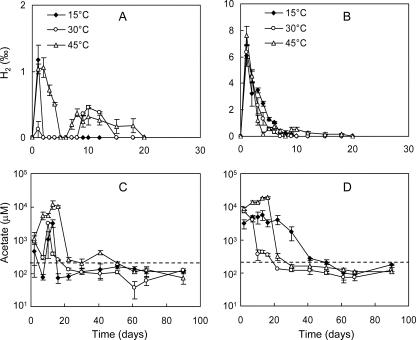
Production of H2 started right from the beginning and reached a maximum at the first day of incubation (Fig. 2A and B). Thereafter, H2 concentrations decreased rapidly. The maximal concentration of H2 was approximately seven times higher in the straw treatment than the root treatment. In the root treatment, H2 in the headspace decreased to the detection limit (106 ppm by volume) on day 2 at either 15°C or 30°C and on day 5 at 45°C (Fig. 2A). Transient accumulation of H2 occurred between days 8 and 15 at 30°C and between days 6 and 20 at 45°C. In the straw treatment, H2 concentration decreased steadily but remained detectable until day 10 at 15°C and 30°C and until day 20 at 45°C (Fig. 2B).
FIG. 2.
Effect of temperature on H2 production after the addition of rice roots (A) or rice straw (B) and on acetate production after the addition of rice roots (C) and rice straw (D). The dashed lines in panels C and D indicate an acetate concentration of 200 μM. Data are means ± standard errors (n = 3).
Acetate accumulated rapidly after incubation. The concentrations reached maxima between days 2 and 16 in the different treatments (Fig. 2C and D). The accumulations were greater in the straw treatments than root treatments and at 45°C than 30°C and 15°C. In the second stage between days 10 and 22, the acetate concentration sharply decreased (to a level of <1 mM). In most cases the concentrations after day 22 were already near to or lower than 200 μM except in the straw treatment at 15°C, where the concentrations remained >1 mM until day 30. In the third stage (after day 22), the acetate remained at low and relatively constant concentrations, indicating a quasi-steady state of acetate dynamics. During this period, the acetate concentrations were relatively lower at 30°C than 15°C.
Structure of the methanogenic archaeal community.
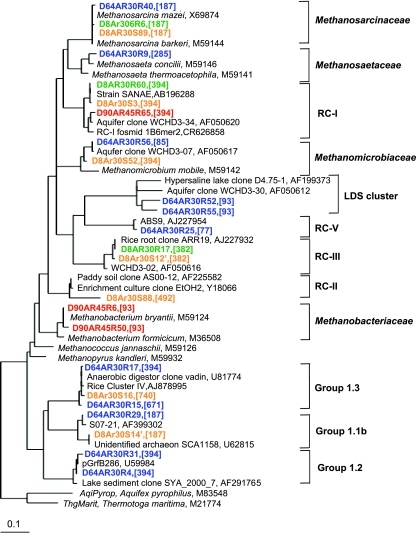
A combination of T-RFLP fingerprinting with cloning and sequencing was used to characterize the dynamics of the archaeal community during the anaerobic decomposition of rice residues. In total, four clone libraries were constructed, and 174 randomly selected clones were sequenced (Table 1). The different clone libraries were created to provide a better resolution of the methanogenic community under different conditions. The phylogenetic analysis of the archaeal sequences showed that the archaeal community consisted of Methanosarcinaceae, Methanosaetaceae, Methanomicrobiales, and Methanobacteriales and of the following yet uncultured archaeal lineages: RC-I, RC-II, RC-III, RC-V, LDS (for Lake Dagow sediment) cluster (15), crenarchaeotal group 1.1b (equivalent to RC-VI) (21, 43), group 1.2 (21), and group 1.3 (equivalent to RC-IV) (Fig. 3). These archaeal lineages except crenarchaeotal group 1.2 have been detected previously in rice field soils (3, 16, 17, 24, 29, 34, 43).
TABLE 1.
Composition of archaeal clone libraries retrieved from anoxic rice field soil during the decomposition of rice root and straw residues at different temperatures
| Clone library | No. of sequences randomly detected
|
|||
|---|---|---|---|---|
| Rice root on day 8 (30°C) | Rice straw
|
|||
| Day 8 (30°C) | Day 64 (30°C) | Day 90 (45°C) | ||
| Methanosarcinaceae | 3 | 20 | 5 | |
| Methanosaetaceae | 9 | 1 | 9 | 1 |
| Methanomicrobiales | 3 | 1 | 15 | |
| Methanobacteriales | 1 | 8 | ||
| RC-I | 2 | 4 | 2 | 37 |
| RC-II | 1 | |||
| RC-III | 1 | 1 | ||
| RC-V | 1 | 4 | ||
| LDS cluster | 4 | |||
| Crenarchaeotal group 1.1b | 7 | 7 | 5 | |
| Crenarchaeotal group 1.2 | 2 | 6 | ||
| Crenarchaeotal group 1.3 | 11 | 1 | 2 | |
| Total | 40 | 29 | 54 | 51 |
FIG. 3.
Phylogenetic relationship of representative archaeal 16S rRNA gene clone sequences generated from soil samples of different incubations. Green, rice straw treatment at 30°C on day 8; orange, rice root treatment at 30°C on day 8; blue, rice straw treatment at 30°C on day 64; red, rice straw treatment at 45°C on day 90. Reference sequences are shown in black. The scale bar represents 10% sequence divergence; GenBank accession numbers of sequences are indicated; the in silico TRF size is given in square brackets.
Seven fragments (77 bp, 85 bp, 93 bp, 187 bp, 285 bp, 382 bp, and 394 bp) were detected as major peaks in the T-RFLP profiles. A combination of in silico sequence analyses and T-RFLP fingerprinting of the representative clones showed that four terminal restriction fragments (TRFs) could be assigned to a single lineage, i.e., 85 bp to Methanomicrobiales, 285 bp to Methanosaetaceae, 77 bp to euryarchaeotal RC-V, and 382 bp to RC-III, whereas three TRFs (93 bp, 187 bp, and 394 bp) were associated with more than one lineage. The 93-bp TRF represented mostly Methanobacteriales but occasionally also the LDS cluster. The 187-bp TRF was characteristic of both the Methanosarcinaceae and crenarchaeotal group 1.1b, and the 394-bp TRF predominantly represented RC-I but often also Methanomicrobiales.
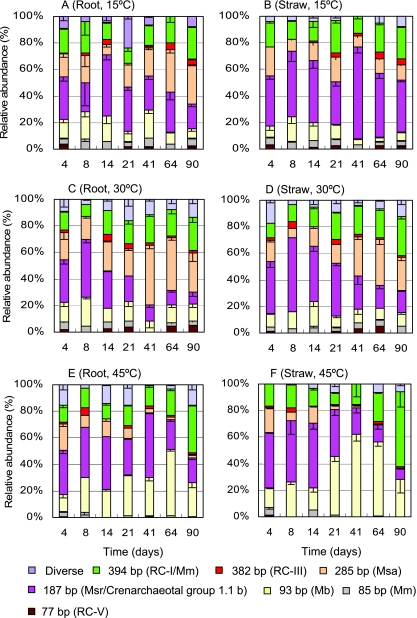
T-RFLP profiles revealed that the residue type and temperature substantially influenced the structure of the archaeal community (Fig. 4). In the root treatment at 15°C (Fig. 4A), the archaeal community consisted of RC-I and Methanomicrobiales (394 bp), Methanosaetaceae (285 bp), Methanosarcinaceae and crenarchaeotal group 1.1b (187 bp), and Methanobacteriales (93 bp). The relative abundance of the 187-bp TRF increased to a maximum in the second week (day 14) and then decreased, while the relative abundance of the 285-bp TRF gradually increased in the later stages. In the straw treatment at 15°C, the relative abundance of the 187-bp TRF remained high throughout the incubation (Fig. 4B). In the straw treatment, the abundance of this TRF was greater and that of the 285-bp TRF was lower than in the root treatment (Fig. 4A and B).
FIG. 4.
The structural dynamics of the archaeal community in anaerobically incubated rice field soil at 15°C, 30°C, and 45°C after the addition of rice root or straw residues. The graphs show the relative abundances of TRFs used as a measure of the composition of the archaeal community. Data are means minus standard error (n = 3). Only major TRFs are shown, and minor archaeal TRFs described previously (4, 35, 41) have been combined as Diverse. Mb, Methanobacteriales; Msa, Methanosaetaceae; Msr, Methanosarcinaceae; Mm, Methanomicrobiales.
In the root treatment at 30°C (Fig. 4C), the 187-bp TRF showed a high abundance only at the early period (up to 45% on day 8) but later was gradually replaced by the 285-bp and 394-bp TRFs. The T-RFLP patterns of the straw treatment at 30°C (Fig. 4D) were similar to those of the root treatment (Fig. 4C), but the gradual replacement of the 187-bp with the 285-bp TRF became apparent only after day 21.
T-RFLP profiles at 45°C (Fig. 4E and F) differed greatly from those at 15°C and 30°C. The Methanosaetaceae-like TRF was detectable in the beginning but hardly in the later stage of incubation. The relative abundance of the 394-bp TRF increased constantly with time and reached 45% and 63% of total fluorescence intensity in the root and straw treatments, respectively. The relative abundance of the Methanobacteriaceae-like TRF remarkably increased at 45°C even though it declined again at the end of incubation. The abundance of the 187-bp TRF at 45°C decreased with time, similar to the result at 30°C. The relative increase in the abundance of the 394-bp and 93-bp TRFs in the straw treatment seemed to be greater than in the root treatment.
The problem that a few lineages shared identical TRFs was partially resolved by comparing the T-RFLP fingerprints with the cloning results. Since the cloning and sequencing showed no presence of crenarchaeotal group 1.1b but a dominance of Methanosarcinaceae in the straw treatment on day 8 at 30°C (Table 1), the increased abundance of the 187-bp TRF in the early stage was very probably due to an increase in the abundance of Methanosarcinaceae. In contrast, the frequency of crenarchaeotal group 1.1b sequences in the day 64 clone library increased (Table 1). Thus, the temporal decrease in the frequency of the 187-bp TRF (Fig. 4C D) was possibly not due to the change in crenarchaeotal group 1.1b but to a decrease in the abundance of Methanosarcinaceae. In the straw treatment at 30°C, the Methanomicrobiales-like sequences increased with incubation (3% in the day 8 library relative to 28% in the day 64 library) while RC-I sequences decreased. Thus, the increase in the abundance of the 394-bp TRF probably indicated the increased presence of Methanomicrobiales rather than RC-I. However, the Methanomicrobiales-like sequences were not detected in the clone library at 45°C (Table 1). Hence, the increase in the abundance of the 394-bp TRF at 45°C most probably represented RC-I.
DISCUSSION
Plant residue is one of the most important carbon sources in soils. The residue particles serve as hotspots for microbial colonization and activity in soil (14, 23). Our study showed that both the activity and structure of the methanogenic archaeal community were affected by the type of plant residue, i.e., root versus straw, and the temperature.
The production rates of H2, acetate, CH4, and CO2 were greater in the straw treatment than root treatment, consistent with a previous study that the degradation of rice straw was much faster than that of root residues (26). The rates of residue decomposition appear to be controlled by the quality of the plant residues. Rice straw consists of cellulose (32 to 37%), hemicelluloses (29 to 37%), lignin (5 to 15%), and inorganic components such as silica (1, 36, 37) while the plant roots generally contain a higher content of recalcitrant components such as lignin and a lower content of decomposable components such as nonstructural carbohydrates (18, 33). Thus, the root residues are more resistant to degradation than the stem residues.
Molecular fingerprinting of the archaeal community indicated that the abundance of Methanosarcinace increased early in the incubations, while Methanosaetaceae became more abundant in the later stages (Fig. 4). The shift in the relative abundance between these two groups was in good agreement with their physiological predictions from pure cultures. Methanosacina spp. have been described as fast-growing and substrate-versatile methanogens, using acetate, methanol and H2-CO2, while Methanosaeta spp. are known as slow-growing organisms utilizing acetate only (19, 20). When growing on acetate, Methanosaeta spp. show a much lower minimum threshold (7 to 70 μM) than Methanosacina spp. (0.2 to 1.2 mM) (19, 20). In our experiment, the fast decomposition of residue material at the beginning resulted in accumulation of both acetate and H2 (Fig. 2), thus stimulating the growth of Methanosarcina spp. The decomposition of straw residue was faster than that of root residue (Fig. 1), resulting in a greater development of Methanosarcinaceae than Methanosaetaceae (Fig. 4). The decrease in the acetate concentration after day 21 to a level of <200 μM coincided with an increasing abundance of Methanosaetaceae in the later stages (Fig. 4C and D). A high acetate concentration in the straw treatment at 15°C (Fig. 2D) was also reflected by a constantly high abundance of Methanosarcinaceae in this treatment. Thus, acetate and probably H2 seem the key factors in regulating the shift between Methanosarcinaceae and Methanosaetaceae during the decomposition of plant residues. The substantial effect of acetate on these two organisms has been also reported previously in rice field soils without residue incorporation (10, 24).
Temperature affected the methanogens both directly and indirectly. Chin et al. (2) showed in a short-term experiment (21 days) that Methanosaetaceae dominated at 15°C while Methanosarcinaceae dominated at 30°C in rice field soil. However, in a long-term experiment (60 to 90 days) where the methanogenic activity already approached quasi-steady-state conditions, Methanosaetaceae became dominant over Methanosarcinaceae at increasingly higher temperatures (10°C to 37°C) (10). It was hypothesized that the direct effect (through membrane fluidity) was possibly responsible for the selection of methanogens in the short-term experiment, while the indirect effect (possibly through acetate concentration) was responsible for the selection under the quasi-steady-state conditions (10). The results of our study are consistent with the observations in the long-term experiment. Apparently, the lower concentration of acetate at 30°C than at 15°C favors the development of Methanosaetaceae over Methanosarcinaceae.
The abundance of Methanobacteriales substantially increased at 45°C. Methanobacteria spp. have been described as hydrogentrophic and sometimes thermophilic methanogens (41, 44). It has also been determined that Methanobacteriales are favored on rice roots when H2 concentrations are high and that the acetoclastic methanogens are partially inhibited (27). In the present experiment, the H2 concentrations remained detectable for a longer period at 45°C than at 15°C and 30°C (Fig. 2A and B), and the acetoclastic Methanosaetaceae appeared to be suppressed at 45°C. Thus, Methanobacteriales were developed. Higher production of H2 in the straw treatment relative to the root treatment was possibly responsible for the greater enhancement of the growth of Methanobacteriales.
Interestingly, the abundance of RC-I methanogens also increased gradually at 45°C. RC-I seems to be of particular importance for CH4 production from root exudates (28). Just recently, a first isolate of this group was obtained from Japanese rice field soil (35). RC-I methanogens have been characterized as thermophilic and hydrogenotrophic methanogens by both culture-dependent and independent techniques (9, 11, 30, 35). However, unlike Methanobacteriales, the RC-I organisms were found to be favored at low H2 concentrations in nature (27). Hence, after day 41 at 45°C, the H2 concentration had possibly already decreased to a level that was limiting for Methanobacteriales but not for RC-I methanogens. However, the H2 concentrations could not be sufficiently resolved in the present experiment to prove this hypothesis.
Although acetoclastic methanogens appeared to be gradually replaced by hydrogenotrophic methanogens at 45°C, acetate did not accumulate. It is thus possible that there exist unknown mechanisms which are responsible for the consumption of acetate under such conditions.
Besides Methanobacteriales and RC-I methanogens, different kinds of the moderately thermophilic methanogens including Methanomicrobiales have been detected in a range of rice field soils (43). Methanomicrobiales were represented by the 85-bp TRF and partly by the 394-bp TRF in the present study. The organisms represented by the 85-bp TRF showed only a low abundance (less than 8%) throughout the incubations (Fig. 4), while the organisms represented by the 394-bp TRF were abundant at 30°C (Table 1). However, Methanomicrobiales were hardly detected at 45°C, particularly in the later stages. Therefore, the members of Methanomicrobiales in our rice field soil were apparently not adapted to high temperatures (45°C) but possibly contributed to CH4 production at 30°C.
Acknowledgments
We thank Ralf Conrad for discussion and critical reading of the manuscript. We also thank the anonymous reviewers for the valuable comments and suggestions that improved the manuscript.
This study was supported by the Natural Science Foundation of China (grants 40571080 and 40625003) and the Max-Planck Society as a partner group.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Akasaka, H., T. Izawa, K. Ueki, and A. Ueki. 2003. Phylogeny of numerically abundant culturable anaerobic bacteria associated with degradation of rice plant residue in Japanese paddy field soil. FEMS Microbiol. Ecol. 43:149-161. [DOI] [PubMed] [Google Scholar]
- 2.Chin, K. J., T. Lukow, S. Stubner, and R. Conrad. 1999a. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30°C). FEMS Microbiol. Ecol. 30:313-326. [DOI] [PubMed] [Google Scholar]
- 3.Chin, K. J., T. Lukow, and R. Conrad. 1999b. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin, K. J., T. Lueders, M. W. Friedrich, M. Klose, and R. Conrad. 2004. Archaeal community structure and pathway of methane formation on rice roots. Microbiol. Ecol. 47:59-67. [DOI] [PubMed] [Google Scholar]
- 5.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediment. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 6.Conrad, R. 2002. Control of microbial methane production in wetland rice fields. Nutr. Cycl. Agroecosys. 64:59-69. [Google Scholar]
- 7.Conrad, R., and M. Klose. 2006. Dynamics of the methanogenic archaeal community in anoxic rice soil upon addition of straw. Eur. J. Soil. Sci. 57:476-484. [Google Scholar]
- 8.Denier Van Der Gon, H. A. C., and H. U. Neue. 1995. Influence of organic matter incorporation on the methane emission from a wetland rice field. Global Biogeochem. Cycles 9:11-22. [Google Scholar]
- 9.Erkel, C., D. Kemnitz, M. Kube, P. Ricke, K. J. Chin, S. Dedysh, R. Reinhardt, R. Conrad, and W. Liesack. 2005. Retrieval of first genome data for rice cluster I methanogens by a combination of cultivation and molecular techniques. FEMS Microbiol. Ecol. 53:187-204. [DOI] [PubMed] [Google Scholar]
- 10.Fey, A., and R. Conrad. 2000. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl. Environ. Microbiol. 66:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fey, A., K. J. Chin, and R. Conrad. 2001. Thermophilic methanogens in rice field soil. Environ. Microbiol. 3:295-303. [DOI] [PubMed] [Google Scholar]
- 12.Fey, A., P. Claus, and R. Conrad. 2004. Temporal change of 13C-isotope signatures and methanogenic pathways in rice field soil incubated anoxically at different temperatures. Geochim. Cosmochim. Acta 68:293-306. [Google Scholar]
- 13.Glissmann, K., and R. Conrad. 2000. Fermentation pattern of methanogenic degradation of rice straw in anoxic paddy soil. FEMS Microbiol. Ecol. 31:117-126. [DOI] [PubMed] [Google Scholar]
- 14.Glissmann, K., S. Weber, and R. Conrad. 2001. Localization of processes involved in methanogenic degradation of rice straw in anoxic paddy soil. Environ. Microbiol. 8:502-511. [DOI] [PubMed] [Google Scholar]
- 15.Glissmann, K., K. J. Chin, P. Casper, and R. Conrad. 2004. Methanogenic pathway and archaeal community structure in the sediment of eutrophic Lake Dagow: effect of temperature. Microb. Ecol. 48:389-399. [DOI] [PubMed] [Google Scholar]
- 16.Großkopf, R., P. H. Janssen, and W. Liesack. 1998a. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Großkopf, R., S. Stubner, and W. Liesack. 1998b. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heal, O. W., J. M. Anderson, and M. J. Swift. 1997. Plant litter quality and decomposition: a historical overview, p. 3-30. In G. Cadish and K. E. Giller (ed.), Driven by nature: plant litter quality and decomposition. CAB International, Oxon, United Kingdom.
- 19.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1990. Acetate threshold values and acetate activating enzymes in methanogenic bacteria. FEMS Microbiol. Ecol. 73:339-344. [Google Scholar]
- 20.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88:181-198. [Google Scholar]
- 21.Jurgens, G., F. O. Glöckner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Münster. 2000. Identification of novel archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 22.Khalil, M. A., R. A. Rasmussen, M. J. Shearer, R. W. Dalluge, L. Ren, and C. H. Duan. 1998. Factors affecting methane emissions from rice fields. J. Geophys. Res. 103:25219-25231. [Google Scholar]
- 23.Kimura, M., and C. C. Tun. 1999. Microscopic observation of the decomposition process of leaf sheath of rice straw and colonizing microorganisms during the cultivation period of paddy rice. Soil. Sci. Plant Nutr. 45:427-437. [Google Scholar]
- 24.Kruger, M., P. Frenzel, D. Kemnitz, and R. Conrad. 2005. Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51:323-331. [DOI] [PubMed] [Google Scholar]
- 25.Lu, Y., R. Wassmann, H. U. Neue, C. Huang, and C. S. Bueno. 2000. Methanogenic responses to exogenous substrates in anaerobic rice soils. Soil Biol. Biochem. 32:1683-1690. [Google Scholar]
- 26.Lu, Y., A. Watanabe, and M. Kimura. 2003. Carbon dynamics of rhizodeposits, root- and shoot-residues in rice soil. Soil Biol. Biochem. 35:1223-1230. [Google Scholar]
- 27.Lu, Y., T. Lueders, W. Michael, M. W. Friedrich, and R. Conrad. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326-336. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Y., and R. Conrad. 2005. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309:1088-1090. [DOI] [PubMed] [Google Scholar]
- 29.Lueders, T., and M. W. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 31.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 32.Page, A. L., R. H. Miller, and D. R. Keeney. 1982. Methods of soil analysis. Part 2: chemical and microbiological properties, 2nd ed. American Society of Agronomy, Madison, WI.
- 33.Puget, P., and L. E. Drinkwater. 2001. Short-term dynamics of root- and shoot derived carbon from a leguminous green manure. Soil Sci. Soc. Am. J. 65:771-779. [Google Scholar]
- 34.Ramakrishnan, B., T. Lueders, P. F. Dunfield, R. Conrad, and M. W. Friedrich. 2001. Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37:175-186. [Google Scholar]
- 35.Sakai, S., H. Imachi, Y. J. Sekiguchi, A. Ohashi, H. Harada, and Y. Kamagata. 2007. Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster rice cluster I. Appl. Environ. Microbiol. 73:4326-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsuki, K., and F. N. Ponnamperuma. 1987. Behavior of anaerobic decomposition products in submerged soils. Effects of organic material amendment, soil properties, and temperature. Soil Sci. Plant Nutr. 33:13-33. [Google Scholar]
- 37.Watanabe, A., K. Katoh, and M. Kimura. 1993. Effect of rice straw application on CH4 emission from paddy fields. 2. Contribution of organic constituents in rice straw. Soil Sci. Plant Nutr. 39:707-712. [Google Scholar]
- 38.Watanabe, A., M. Yoshida, and M. Kimura. 1998. Contribution of rice straw carbon to CH4 emission from rice paddies using 13C-enriched rice straw. J. Geophys. Res. 103:8237-8242. [Google Scholar]
- 39.Watanabe, A., T. Takeda, and M. Kimura. 1999. Evaluation of origins of CH4 carbon emitted from rice paddies. J. Geophys. Res. 104:23623-23629. [Google Scholar]
- 40.Watanabe, T., S. Asakawa, A. Nakamura, K. Nagaoka, and M. Kimura. 2004. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS. Microbiol. Lett. 232:153-163. [DOI] [PubMed] [Google Scholar]
- 41.Wiegel, J. 1990. Temperature spans for growth: hypothesis and discussion. FEMS Microbiol. Rev. 75:155-170. [Google Scholar]
- 42.Wu, X. L., K. J. Chin, S. Stubner, and R. Conrad. 2001. Functional patterns and temperature response of cellulose-fermenting microbial cultures containing different methanogenic communities. Appl. Microbiol. Biotechnol. 56:212-219. [DOI] [PubMed] [Google Scholar]
- 43.Wu, X. L., M. W. Friedrich, and R. Conrad. 2006. Diversity and ubiquity of thermophilic methanogenic archaea in temperate anoxic soils. Environ. Microbiol. 8:394-404. [DOI] [PubMed] [Google Scholar]
- 44.Zeikus, J. G., and M. R. Winfrey. 1976. Temperature limitation of methanogenesis in aquatic sediments. Appl. Environ. Microbiol. 31:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]