Abstract
A novel type of mutanase (termed mutanase RM1) was isolated from Paenibacillus sp. strain RM1. The purified enzyme specifically hydrolyzed α-1,3-glucan (mutan) and effectively degraded biofilms formed by Streptococcus mutans, a major etiologic agent in the progression of dental caries, even following brief incubation. The nucleotide sequence of the gene for this protein contains a 3,873-bp open reading frame encoding 1,291 amino acids with a calculated molecular mass of 135 kDa. The protein contains two major domains, the N-terminal domain (277 residues) and the C-terminal domain (937 residues), separated by a characteristic sequence composed of proline and threonine repeats. The characterization of the recombinant proteins for each domain which were expressed in Escherichia coli demonstrated that the N-terminal domain had strong mutan-binding activity but no mutanase activity whereas the C-terminal domain was responsible for mutanase activity but had mutan-binding activity significantly lower than that of the intact protein. Importantly, the biofilm-degrading activity observed with the intact protein was not exhibited by either domain alone or in combination with the other. Therefore, these results indicate that the structural integrity of mutanase RM1 containing the N-terminal mutan-binding domain is required for the biofilm-degrading activity.
Streptococcus mutans has been strongly implicated in the etiology of dental caries (28). The pathogenicity of S. mutans is closely related to its ability to synthesize insoluble glucans in an extracellular matrix (51). These molecules are characterized as unique polysaccharides composed of α-1,6-glucan (dextran) and α-1,3-glucan (mutan) chains (50). The dextrans exhibit water solubility, while the mutans are water insoluble. Therefore, the insolubility of the glucans is thought to be attributable to their high mutan contents (30). S. mutans strains have strong potential to adhere to tooth surfaces, thereby contributing to the formation of dental plaque (biofilms) (2, 53). In addition, it is generally thought that biofilms formed by S. mutans promote the adherence and further accumulation of other bacteria into dental plaque (26). Organic acids produced by S. mutans, as well as other bacteria, accumulate within the biofilms, most likely due to limitations in acid diffusion, leading to demineralization of enamel or dentin tissues (dental caries) (54). Biofilms are also known to play a central role in the initiation and development of periodontitis (44) and likely also in the maintenance of oral bacterial resistance to antimicrobial agents. Therefore, biofilms formed by S. mutans together with other bacteria are thought to be important factors in dental caries as well as in periodontitis. Thus, agents which inhibit the formation of biofilms or promote their destruction should be useful tools to facilitate the development of new therapeutic approaches against dental caries.
Previous in vitro and in vivo studies showed that the enzyme mutanase has potential for inhibiting dental plaque formation by suppressing the formation of mutan and/or by promoting the degradation of mutan into low-molecular-weight glucans (46). The low-molecular-weight forms of glucans are known to increase in water solubility (12). In addition, dental plaque formation is regulated by a balance between positive and negative factors involved in biofilm formation. Given that endogenous mutanase activity is barely detectable in the oral cavity, the hydrolysis of mutan should be less than its rate of formation. Simonson et al. (41) indicated that biofilms formed by S. mutans OMZ 176 and NTCT 10449 were extensively degraded by treatment with Pseudomonas sp. mutanase at 37°C for 90 min. Inoue et al. (20) also reported that insoluble glucan formed by cell-free S. mutans OMZ 176 glucosyltransferases (GTFs) was degraded by incubation with Pseudomonas sp. mutanase at 37°C for 24 h. To our knowledge, however, little is known about mutanases having the ability to degrade biofilms following short-term treatments. Given the short duration of tooth-brushing and oral rinsing by humans, it is of therapeutic importance to identify or develop molecules possessing the ability to degrade biofilms following brief manipulation and also to be retained in the oral cavity.
In this study, we describe a novel type of mutanase, termed mutanase RM1, in Paenibacillus sp. strain RM1 which expresses the ability to effectively degrade in vitro biofilms formed by S. mutans. We also demonstrate the mechanism for destruction of the biofilms by this enzyme.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Streptococcus downei MFe28 and S. mutans ATCC 25175 (NTCT 10449) were obtained from ATCC (Manassas, VA), Escherichia coli JM109 and E. coli NV522 were obtained from Takara Bio (Shiga, Japan), and E. coli BL21 was obtained from Novagen (Madison, WI). Nutrient broth (NB), peptone, yeast extract, and brain heart infusion (BHI) were purchased from Kyokuto Pharmaceutical Industrial (Tokyo, Japan), Nissui Pharmaceutical (Tokyo, Japan), Oxoid (Cambridge, United Kingdom), and Becton Dickinson (Franklin Lakes, NJ), respectively. The vectors Charomid9-36, pUC118, and pColdI as well as pT7Blue were from Nippon Gene (Tokyo, Japan), Takara, and Novagen, respectively. Restriction enzymes were from Takara, Toyobo (Osaka, Japan), or New England Biolabs (Ipswich, MA). AccuPrime PCR enzyme and Taq polymerase were from Invitrogen (Carlsbad, CA).
Preparation of mutan.
The gtfI gene of S. downei MFe28 (7) was cloned in the plasmid pUC118, and E. coli JM109 was transformed with this plasmid. The transformants were then cultured in BHI medium including 50 μg/ml ampicillin at 37°C for 18 h, and the bacterial cells were collected by centrifugation at 8,000 × g for 20 min at 4°C. The cells were suspended in 0.1 M Tris-HCl buffer, 6 M urea, 10 mM EDTA, pH 6.8, and incubated for 1 h at 25°C. The supernatant including GTF-I encoded by gtfI was obtained by centrifugation at 8,000 × g for 20 min at 4°C. The GTF-I preparation and 20% sucrose were then incubated in 0.1 M potassium phosphate buffer, pH 6.5, at 37°C for 16 h. The synthesized mutan was collected by centrifugation at 8,000 × g for 20 min at 4°C followed by washing with distilled water, and the mutan was recovered by lyophilization. Our supplementary study by nuclear magnetic resonance confirmed that the mutan consisted of 99.5% α-1,3-glucosidic linkages.
Screening and identification for mutanase-producing bacteria.
Soil samples collected in Japan were suspended in saline solution, and aliquots of the suspension were plated on NB agar medium containing 0.2% (wt/vol) mutan and cultured at 40°C for 2 days. The microorganisms which exhibited halos on the plates were then isolated. Emergence of a halo was indicative of mutan-degrading activity. The positive microorganisms were cultured in a medium containing 0.1% mutan, 1% inositol, 0.5% peptone, 0.3% yeast extract, 0.2% KH2PO4, 0.2% NH4NO3, and 0.03% MgSO4·7H2O, pH 7 (MIPY), at 40°C for 24 h. The crude enzymes were obtained in the supernatant of the cultures. Mutanase-positive organisms with the highest activities were selected following assays of mutanase activity and assessment of the degradation of biofilms described below. One microorganism which was found to produce a novel mutanase was analyzed bacteriologically according to Bergey's Manual of Systematic Bacteriology (25). Furthermore, genomic DNA of the microorganism was prepared. The 16S rRNA was amplified by the primers described by Fox et al. (8). The amplified 16S rRNA was purified with a QIAquick Spin PCR purification kit (Qiagen, Hilden, Germany) and sequenced with an ABI 373A automatic DNA sequencer (Perkin-Elmer, Waltham, MA). The analyzed sequence was compared to bacterial sequences in the GenBank database using BLAST and classified as Paenibacillus sp. strain RM1.
Assay of mutanase activity.
Mutan (3%, wt/vol) in 0.1 M acetate buffer, pH 5.0, was treated with a Hiscotron homogenizer (Microtec, Chiba, Japan) for 15 min. One hundred microliters of the mutan solution which was preincubated at 35°C and 100 μl of the enzyme solution were mixed and incubated at 35°C for 10 min. The reaction was stopped by adding 0.4 ml of DNS solution (29), and 0.5 ml of the supernatant was boiled for 10 min followed by cooling in cold water for 10 min. Five milliliters of distilled water was added to the reaction mixture, and the absorbance at 530 nm was measured. One unit of mutanase activity was defined as the amount of enzyme that released 1 μmol of reducing sugar per min under standard conditions. To verify substrate specificity of the mutanase, nigeran (Sigma, St. Louis, MO), dextran T2000 (Pharmacia, Uppsala, Sweden), amylose (Wako Pure Chemical Industries, Osaka, Japan), pullulan (Seikagaku Kogyo, Tokyo, Japan), laminariheptaose (Seikagaku Kogyo), or cellohexaose (Seikagaku Kogyo) was tested in addition to mutan.
Purification of mutanase from Paenibacillus sp. strain RM1.
Paenibacillus sp. strain RM1 was cultured in MIPY medium at 40°C for 24 h. The supernatant of the culture was concentrated by ultrafiltration and then precipitated with 90% saturated ammonium sulfate. The precipitate was dissolved in 50 mM Tris-HCl buffer, pH 8.0, and dialyzed against the same buffer. The crude enzyme solution was loaded onto a DE52 anion-exchange chromatography column (Whatman, Maidstone, United Kingdom) equilibrated with 50 mM Tris-HCl buffer, pH 8.0, and the flowthrough fractions were pooled. The pooled fractions were then dialyzed against 50 mM Tris-HCl buffer, pH 8.5. The resultant solution was loaded onto a DE52 column equilibrated with 50 mM Tris-HCl buffer, pH 8.5, and the adsorbed mutanase was eluted with a linear gradient of NaCl from 0 to 200 mM. Active fractions were pooled and dialyzed against 10 mM sodium phosphate buffer, pH 7.0.
Assessment of degradation of biofilms.
S. mutans ATCC 25175 precultured in BHI medium was inoculated in a glass test tube containing 3 ml of BHI medium with 1% sucrose. The glass test tube was inclined at an angle of 30° and cultured in an anaerobic culture apparatus at 37°C for 18 h. During the cultivation, S. mutans adhered to the wall of the glass test tube, forming biofilms with an insoluble glucan matrix. Following two rinses with 4 ml of distilled water, 3 ml of 0.1 M sodium phosphate buffer, pH 6.0, was added, and the test tube was preincubated at 37°C for 10 min. One milliliter of mutanase solution was added to the tube and incubated for 3 min at 37°C. After 3 min of mutanase treatment, the test tube was rinsed three times with 4 ml distilled water. Subsequently, 4 ml of 0.1 M sodium phosphate buffer, pH 6.0, was added into the test tube and the contents were incubated at 37°C for 6 h. After the buffer was discarded, the test tube was rinsed twice with 4 ml of distilled water. After addition of an additional 4 ml of distilled water, residual biofilm remaining on the wall of the test tube was dispersed by sonication for 3 s (ultrasonic homogenizer US-50; Nissei, Tokyo, Japan). Absorbance at 550 nm was then measured for the sonicated suspension (colorimeter ANA-7S; Tokyo-Koden, Saitama, Japan), and the degradation rate of the biofilms was calculated with the following equation: degradation (%) = [1 − (absorbance at 550 nm after mutanase treatment/absorbance at 550 nm for the untreated control)] × 100. One hundred percent degradation was defined as the absorbance intensity when distilled water was measured at 550 nm, i.e., zero, which should be equivalent to complete degradation of the biofilm. The assays were performed in triplicate.
Cloning of the mutanase RM1 gene.
Genomic DNA of Paenibacillus sp. strain RM1 was prepared by standard methods (36). The DNA was partially digested with Sau3AI, and the resulting fragments were inserted into the BamHI cloning site of Charomid9-36. Transformant colonies of E. coli NV522 were cultured in one-half M9 agar containing 0.2% mutan at 37°C for 5 days, and the colonies forming halos were selected. The plasmid of the colony carrying the mutanase gene was digested by SacI and KpnI and ligated into the cloning sites of pUC118. The DNA sequence of the insert was determined with an ALFred DNA sequencer (Amersham Pharmacia Biotech, Uppsala, Sweden) and registered as GenBank accession number E16590 (23).
Preparation of recombinant N- and C-terminal protein domains of mutanase RM1.
PCRs using the AccuPrimePCR enzyme were carried out with a template of the mutanase-cloned plasmid and the indicated primer sets (Table 1). The primers were designed to contain NaeI sites in the 5′-terminal regions of amplified cDNA and XbaI sites in the 3′-terminal regions. Amplified cDNA was modified with dATP at the 3′ terminus with Taq polymerase and inserted into the pT7Blue cloning vector. These plasmids were digested with NaeI and XbaI, and each cDNA was inserted into the same cloning sites of the pColdI vector. E. coli BL21 was transformed with these plasmids and cultured in L broth at 37°C until the absorbance at 600 nm reached about 0.5. After the culture was cooled at 16°C for 30 min, 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture, which was further cultured at 16°C for 24 h. After centrifugation at 15,000 × g for 10 min at 4°C, the pellets were suspended in 10 mM sodium phosphate buffer, pH 7.4. Recombinant proteins were extracted by sonication treatment, i.e., 10 s of irradiation six times and pausing under the condition of iced water (ultrasonic homogenizer US-50) and purified by His-affinity chromatography (GE Healthcare, Uppsala, Sweden) with a buffer change to 10 mM sodium phosphate buffer, pH 7.0, on a PD10 column (GE Healthcare).
TABLE 1.
Primers used in this study
| Recombinant protein | Direction | Oligonucleotide sequencea |
|---|---|---|
| Mature region | Forward | 5′-CATATGGCGGGAGGACCGAATC-3′ |
| Reverse | 5′-TCTAGACTAATTATTGATGATCAGATTGAACCC-3′ | |
| N-terminal domain | Forward | 5′-CATATGGCGGGAGGACCGAATC-3′ |
| Reverse | 5′-TCTAGATGAAGTCGACGCTTCGACC-3′ | |
| C-terminal domain | Forward | 5′-CATATGGGCGGCAACATCGCC-3′ |
| Reverse | 5′-TCTAGACTAATTATTGATGATCAGATTGAACCC-3′ |
Restriction sites in the oligonucleotides are underlined.
Mutan binding assay.
One hundred microliters of 500 μg/ml protein solution and 100 μl of 3% (wt/vol) mutan in 0.1 M sodium acetate buffer, pH 5.0, were mixed and incubated for 10 min at 35°C. After centrifugation at 15,000 × g for 10 min at 4°C, the precipitated mutan (containing the mutan-binding protein) and supernatant (the fraction of unbound proteins) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Novex 4 to 12% Bis-Tris gel, morpholinepropanesulfonic acid [MOPS] buffer system; Invitrogen). Intensity of the bands was quantified by densitometry (UVP, Upland, Sweden). The mutan-binding activity was calculated by the following equation: mutan binding (%) = (density for mutan-binding proteins/density for the sum of mutan-binding and unbound proteins) × 100.
Protein assay.
Protein concentration was determined by the Bradford method (Bio-Rad Laboratories, Hercules, CA) using immunoglobulin G as the protein standard.
Analysis of N-terminal amino acids.
Mutanase RM1 was blotted onto a polyvinylidene difluoride membrane following SDS-PAGE. Coomassie brilliant blue-stained bands were cut out and analyzed using a Porton PI-2020 protein sequencer (Beckman Coulter, Fullerton, CA).
Molecular weight analysis by MALDI-TOF mass spectrometry.
Mutanase RM1 and matrix (sinapinic acid) were mixed and spotted onto silicon plates. Mass spectra were collected with the matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry system (Biflex III; Bruker Daltonics, Billerica, MA).
Nucleotide sequence accession number.
The DNA sequence of mutanase RM1 was determined and deposited as GenBank accession number E16590 (23).
RESULTS
Biochemical properties of mutanase RM1.
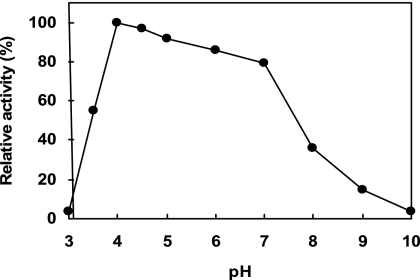
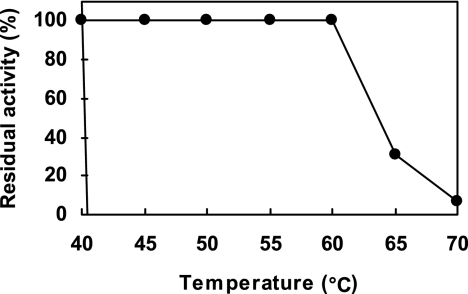
Considering the relatively short duration for tooth-brushing for removing human dental plaque, we first established new screening methods to search for novel types of mutanases which can efficiently degrade biofilms formed by S. mutans in relatively short time frames. Those bacteria exhibiting strong mutanase activity, which produce halos on culture plates, were selected under the conditions described in Materials and Methods. Of the microorganisms exhibiting halos in NB medium containing 0.2% mutan, we identified a novel type of mutanase in a new species of Paenibacillus (named Paenibacillus sp. strain RM1), thus termed “mutanase RM1,” and purified it from the culture medium of this bacterium by chromatographic procedures using sensitive differences in adsorption behavior between pH 8.0 and pH 8.5. The final preparation of the enzyme obtained by DE52 anion-exchange chromatography at pH 8.5 revealed a single major protein band by SDS-PAGE. The purified mutanase RM1 degraded only α-1,3-glucan (mutan) but no other glucan substrates with different types of glucosidic linkages and displayed a specific activity of 5.6 U/mg. When mutan was incubated with mutanase RM1 in a wide range of buffers (pH 3 to 10) at 35°C for 10 min, the pH optimum was approximately pH 4.0 (Fig. 1). At pH 4.0 to 7.0, mutanase RM1 showed more than 80% of the maximal activity at pH 4.0. The enzyme was stable following 10 min of incubation at temperatures below 60°C and unstable at temperatures above 65°C (Fig. 2). The apparent molecular mass of the enzyme by MALDI-TOF mass spectrometry was estimated to be 132 kDa. The N-terminal amino acid sequence was revealed as Ala-Gly-Gly-Pro-Asn-Leu-Thr-Pro-Gly-Lys-Pro-Ile-Thr-Ala-Ser-Gly-Gln.
FIG. 1.
pH dependence of the hydrolysis of α-1,3-glucan (mutan) by purified mutanase RM1 (0.28 U/ml). The buffers used were citrate-phosphate (pH 3 to 7), sodium phosphate (pH 6 to 8), and glycine-NaOH (pH 9 to 10). Mean values are expressed at each pH. The optimal pH for maximal activity, pH 4.0, was set at 100%.
FIG. 2.
Thermostability of mutanase RM1. After a 10-min incubation of mutanase RM1 in 20 mM acetate buffer, pH 5 (1.7 U/ml), at the indicated temperatures, the hydrolytic activity against mutan was measured. The activity before incubation was set at 100%.
Degradation of S. mutans biofilms by mutanase RM1.
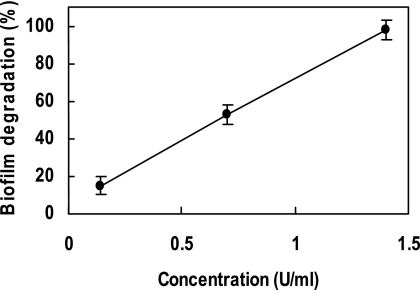
Purified mutanase RM1 strongly degraded biofilms formed by S. mutans in vitro in a dose-dependent manner. The complete degradation of the biofilm was observed at 1.4 U/ml of mutanase RM1 (Fig. 3).
FIG. 3.
Degradation of S. mutans biofilms by mutanase RM1. Increasing concentrations of mutanase RM1 were incubated for 3 min with S. mutans biofilms formed in test tubes. Following removal of the enzyme, 0.1 M phosphate buffer, pH 6.0, was added and further incubation was carried out for 6 h. Biofilm degradation was expressed as the relative values (%) compared to the absorbance intensity when distilled water was measured that should be equivalent to complete degradation of the biofilm, 100%. Error bars represent standard deviations.
DNA sequencing and structural analysis.
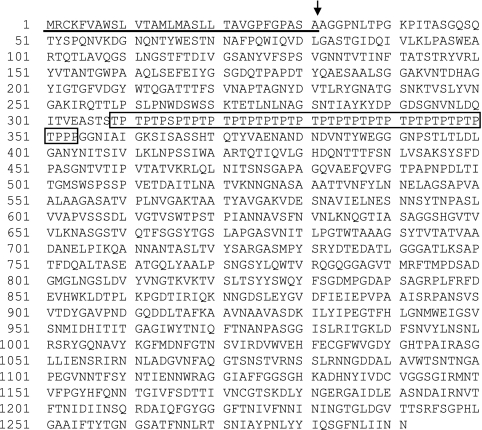
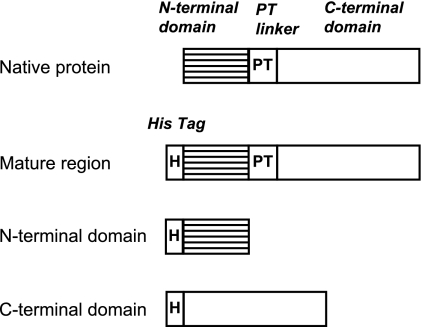
The DNA sequence of mutanase RM1 showed an open reading flame of 3,873 base pairs (GenBank accession no. E16590) (23). Figure 4 shows the deduced amino acid sequence comprising 1,291 amino acids. The estimated molecular mass was 135 kDa. The results of the deduced amino acid sequence and the analysis of the N-terminal amino acid sequence suggested that the first 308 amino acid residues contain a signal peptide (31 amino acid residues) and N-terminal mature domain (277 amino acid residues). The next 46 amino acid residues represented a characteristic repeated sequence composed of proline and threonine (40). The remaining C-terminal domain comprising 937 amino acid residues is thought to be the mature C-terminal domain. The primary structure of mutanase RM1 was thus characterized by two different domains divided by a unique repeated proline-threonine sequence (PT linker) (Fig. 4). In order to explore the physiological significance of each domain, we generated recombinant proteins for both N-terminal and C-terminal domains, as well as the mature region, in which both domains were connected with a linker. We initially constructed His-tagged fusion proteins (Fig. 5) for expression in E. coli and subsequent purification by His-tag affinity chromatography. SDS-PAGE revealed that each recombinant protein was purified to near homogeneity.
FIG. 4.
Deduced amino acid sequence of mutanase RM1. The signal peptide region is underlined, and the linker region is boxed. The arrow indicates the cleavage site for the N-terminal domain of the protein. The DNA sequence was registered as GenBank accession number E16590 (23).
FIG. 5.
Recombinant proteins of the mutanase RM1 derivatives. PT, proline-threonine.
We first examined the ability of each purified protein to bind mutan. Mutan binding of the native protein was 55% under our standard conditions. The recombinant protein for the N-terminal domain strongly bound to mutan (100%), more than did the mature protein (45%) and much more than did the C-terminal domain (28%). We next determined the ability of each protein to degrade mutan. The recombinant proteins for the C-terminal domain and the mature region degraded mutan (mutanase activities of 3.9 and 4.7 U/mg, respectively), whereas that for the N-terminal domain was devoid of mutan-hydrolyzing activity (mutanase activity of 0). (The native protein showed mutanase activity of 5.6 U/mg.) These results thus indicate that the N- and C-terminal domains of mutanase RM1 are essential for mutan-binding and mutan-hydrolyzing activities, respectively.
Degradation of S. mutans biofilms by each recombinant protein.
Next, to determine to what extent each recombinant protein could degrade biofilms, we treated the biofilms of S. mutans formed in glass test tubes with each protein at 37°C and pH 6.0 for 3 min. Protein concentrations were 1.4 U/ml. After being washed with distilled water, the tubes were further incubated at 37°C and pH 6.0 for 6 h, and then the amounts of biofilm retained on the tubes were measured. The recombinant protein for the mature region exhibited strong biofilm-degrading activity (94%) comparable to that of the native mutanase RM1 (98%). In contrast, the recombinant proteins for N- and C-terminal domains and the combination of these proteins showed little or no biofilm-degrading activity, indicating that the structural integrity of mutanase RM1, linked between the N-terminal mutan-binding and the C-terminal mutan-hydrolyzing domains, is essential for efficient degradation of S. mutans biofilms following short-term treatment.
DISCUSSION
This study was initiated to isolate a novel mutanase for possible application as an oral hygiene product, and we were successful in isolating mutanase RM1 derived from Paenibacillus sp. strain RM1. We registered the DNA sequence of mutanase RM1 in GenBank as accession number E16590 in 1998 (23). This was the first registration of a DNA sequence for a bacterial mutanase. Recently, mutanases similar to mutanase RM1 have been identified. In 2006, Yano et al. reported on a mutanase derived from Bacillus circulans KA-304 (52). It was confirmed that this mutanase is approximately 80% similar to mutanase RM1. However, the functional properties of the B. circulans KA-304 mutanase, such as the functions of the N-terminal or C-terminal domain, were not reported. Moreover, very recently, Sumitomo et al. described a mutanase derived from Paenibacillus sp. strain KSM-M86 which has 65.6% similarity to mutanase RM1 (43). The Paenibacillus sp. strain KSM-M86 mutanase contains sequences similar to the C-terminal catalytic domain of mutanase RM1 but different from the N-terminal region of the latter, which we showed to be important for mutanase RM1 to effectively degrade biofilms. However, relative to the catalytic domain, B. circulans KA-304 and the Paenibacillus sp. strain KSM-M86 mutanases could be classified in the same family as the RM1 enzyme.
Many other types of mutanases derived from different microorganisms have been reported. Mutanase-producing fungi (1, 9, 13, 16, 38, 48, 49, 55), yeasts (10, 32), and bacteria (6, 18, 33, 34, 42, 43, 45, 47, 52) were described. Mutanases were isolated and characterized biochemically, and some of them were tested regarding caries-preventive efficacy (14, 15, 19, 21, 22, 35). However, mutanase RM1 can be readily differentiated from all of these previously reported enzymes as documented in the present study.
Mutanase RM1 exhibited biochemical properties which suggest some practical applications. For example, it was observed that the enzyme had higher thermostability (Fig. 2). Moreover, the effective pH range of the mutanase RM1 was quite broad (pH 4.0 to 7.0) (Fig. 1). In the oral cavity, the salivary pH is between 6.0 and 7.0 and the acids produced by plaque bacteria which ferment carbohydrates can lower the pH to around 4.0 (39). The acidic pH causes demineralization of enamel and dentin tissues, ultimately leading to dental cavities. Therefore, mutanase RM1 is expected to be active in the environment of the oral cavity.
We provided evidence for the modular structure of mutanase RM1 as well as the function of each domain in the structure (Fig. 4). The N-terminal domain has mutan-binding activity while the C-terminal domain expresses mutan-hydrolytic functions (see Results). This is the first demonstration of a mutan-binding domain in a bacterial mutanase.
Why does the mutanase RM1 contain a mutan-binding domain? The catalytic domain of mutanase RM1 (C-terminal domain) alone can hydrolyze mutan in vitro (see Results) but has no detectable biofilm-degrading activity. Under in vitro mutanase assay conditions, the concentration of the substrate mutan is in excess relative to the enzyme concentration, and accessibility and reactivity of the enzyme for the substrate mutan are thought to be greater than those for the mutan in the biofilm due to use of the homogenized substrate mutan by the Hiscotron homogenizer. Mutans exist only in trace amounts in the oral cavity. Also, mutans in natural environments likely exist tightly associated with the cell walls of certain basidiomycetes (37), yeasts (24), and other microorganisms. The absence of a mutan-binding domain in a mutanase may result in low affinity for biofilms or other species of environmental mutans. If the affinity of mutanases for mutan is increased, the enzymes could more efficiently hydrolyze mutans in natural environments. Therefore, mutanase-producing microorganisms could then efficiently utilize mutan as a carbohydrate source.
In addition, it is of interest to consider why the mutan-binding domain bound to mutan more strongly than the intact enzyme. While the mutan-binding domain was completely bound to mutan, the intact enzyme was only partially bound under these experimental conditions (see Results). It was suggested that, to degrade insoluble mutan efficiently, the intact enzyme must dissociate from the mutan after initial attack by the catalytic domain following binding to its substrate mediated by the mutan-binding domain. Following initial cleavage of the mutan, the enzyme must move to another undigested region of mutan. Therefore, the intact enzyme might require both the binding and dissociation functions within a single molecule. It is suggested that the hydrolytic domain may also exhibit dissociating activity from mutan based upon the present data showing lower binding activity of the C-terminal domain (see Results). On the other hand, it may be that strong binding for mutan by the mutan-binding domain is required, especially in the first step of the enzymatic reaction.
Mutans are also water insoluble and, therefore, are of lower reactivity than are water-soluble substrates. To degrade water-insoluble substrates efficiently, many enzymes possess substrate-binding domains. There are many examples regarding modular structures in glucanases such as those found in cellulases (17), xylanases (4), chitinases (11), mannanases (3), amylases (5), etc. For example, regarding cellulose, cellulose-binding domains are important for degrading insoluble cellulose. The loss of cellulose-binding domains decreased the hydrolytic activity for insoluble substrates (i.e., crystalline cellulose and amorphous cellulose) but did not change the hydrolysis of soluble substrates (i.e., carboxymethyl cellulose) (31). Similarly, mutanases might have evolved a modular structure to hydrolyze insoluble substrates more efficiently. Therefore, the role of the mutan-binding domain might be to concentrate the catalytic domains on mutans and thereby increase the biofilm-degrading activity of the enzyme.
Mutanase RM1 possesses excellent biofilm-degrading properties. Enzymes for use in oral hygiene products should act relatively quickly to be effective. Therefore, it is important to characterize the kinetics of activity for potential oral hygiene products. For this reason, it was important to demonstrate that mutanase RM1 degraded S. mutans biofilms effectively in a relatively brief treatment. Until now, very few studies have reported the efficacy of degrading “preformed” biofilms. Simonson et al. indicated that it took a 90-min incubation for Pseudomonas sp. mutanase to degrade biofilms of S. mutans OMZ 176 by 70% and those of S. mutans NTCT 10449 (ATCC 25175) by 42% (41). Similarly, Inoue et al. indicated that it took 24 h for the Pseudomonas sp. mutanase to dissociate insoluble glucan by cell-free GTFs (20). To our knowledge, there have been no reports which assessed the efficacy of mutanases when the biofilms were treated with enzymes for relatively short periods.
We were also interested in the mechanism of mutanase RM1 activity. Therefore, we examined biofilm degradation relative to the modular structure of the enzyme. Data in Results clearly demonstrate that the absence of the mutan-binding domain from mutanase RM1 resulted in the complete loss of biofilm-degrading activity. In addition, the presence of a mixture of the individual functional domains (mutan-binding and catalytic domains) did not lead to biofilm degradation. These results suggest that the mutan-binding domain is essential for biofilm degradation during short-term treatment. Therefore, we propose a novel mechanism for mutanase removal of S. mutans-induced biofilms. According to this model, biofilms formed by S. mutans contain an extracellular matrix primarily composed of insoluble glucans which contain α-1,6- and α-1,3-linked glucose residues. Within the first 3 min of contact, the mutan-binding domain allows adherence of the intact mutanase to the mutan regions of the biofilm matrix and allows access of this substrate to the catalytic domain of mutanase RM1. Even after washing, mutanase RM1 could continue to degrade the mutan, which leads to weakening of the biofilm matrix. In addition, the mutan-binding domain would allow the catalytic domain to act at relatively high concentrations on the glucan matrix, thus increasing the frequency of cleavage of the mutan. Therefore, in the oral cavity, the mutan-binding ability of the RM1 mutanase would be expected to enhance the degradation of cariogenic biofilms formed by S. mutans. This study is the first to demonstrate the relationship between the structure (modular structure) and the biofilm-degrading activity of a mutanase.
For biotechnological applications, the mutan-binding domain may be coupled to functional components of proteins for novel biofilm control. In this regard, we have demonstrated that the mutan-binding domain of mutanase RM1 has high affinity for mutan. Recent advances in protein engineering are expected to allow for the rational design of artificial proteins. Therefore, mutanase RM1 or its mutan-binding protein could be fused with other functional proteins, such as dextranases, oxidases, antibodies, or antimicrobial peptides. The mutan-binding domain could be used as a targeting agent against biofilms. Moreover, the mature mutanase RM1 could degrade or penetrate the biofilms and thereby could allow penetration of other fused agents. Previously, Lis and Kuramitsu suggested that a fusion protein composed of galactose oxidase and a glucan-binding domain of a GTF might be useful for controlling dental plaque formation (27). In the future, rationally designed proteins could be used in oral hygiene products as novel therapeutic agents for controlling dental plaque formation.
Acknowledgments
We thank Kenji Yamamoto and Yoshihisa Yamashita, Faculty of Dental Science, Kyushu University, for kind guidance and support.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Ait-Lahsen, H., A. Soler, M. Rey, J. de La Cruz, E. Monte, and A. Llobell. 2001. An antifungal exo-α-1,3-glucanase (AGN13.1) from the biocontrol fungus Trichoderma harzianum. Appl. Environ. Microbiol. 67:5833-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, H., T. Shiroza, M. Hayakawa, S. Sato, and H. K. Kuramitsu. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 53:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cann, I. K., S. Kocherginskaya, M. R. King, B. A. White, and R. I. Mackie. 1999. Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J. Bacteriol. 181:1643-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cazemier, A. E., J. C. Verdoes, A. J. van Ooyen, and H. J. Op den Camp. 1999. Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae. Appl. Environ. Microbiol. 65:4099-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desseaux, V., C. Seigner, Y. Pierron, M. L. Grisoni, and G. Marchis-Mouren. 1988. Porcine pancreatic alpha-amylase: a model for structure-function studies of homodepolymerases. Biochimie 70:1163-1170. [DOI] [PubMed] [Google Scholar]
- 6.Ebisu, S., K. Kato, S. Kotani, and A. Misaki. 1975. Isolation and purification of Flavobacterium alpha-1,3-glucanase-hydrolyzing, insoluble, sticky glucan of Streptococcus mutans. J. Bacteriol. 124:1489-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti, J. J., M. L. Gilpin, and R. R. B. Russell. 1987. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 69:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, G. E., J. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 9.Fuglsang, C. C., R. M. Berka, J. A. Wahleithner, S. Kauppinen, J. R. Shuster, G. Rasmussen, T. Halkier, H. Dalboge, and B. Henrissat. 2000. Biochemical analysis of recombinant fungal mutanases. A new family of α1,3-glucanases with novel carbohydrate-binding domains. J. Biol. Chem. 275:2009-2018. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, I., D. Jimenez, V. Martin, A. Duran, and Y. Sanchez. 2005. The alpha-glucanase Agn1p is required for cell separation in Schizosaccharomyces pombe. Biol. Cell 97:569-576. [DOI] [PubMed] [Google Scholar]
- 11.Gleave, A. P., R. K. Taylor, B. A. Morris, and D. R. Greenwood. 1995. Cloning and sequencing of a gene encoding the 69-kDa extracellular chitinase of Janthinobacterium lividum. FEMS Microbiol. Lett. 131:279-288. [DOI] [PubMed] [Google Scholar]
- 12.Guggenheim, B. 1970. Enzymatic hydrolysis and structure of water-insoluble glucan produced by glucosyltransferases from a strain of Streptococcus mutans. Helv. Odontol. Acta 14(Suppl. 5):89. [PubMed] [Google Scholar]
- 13.Guggenheim, B., and R. Haller. 1972. Purification and properties of an alpha-(1-3) glucanohydrolase from Trichoderma harzianum. J. Dent. Res. 51:394-402. [DOI] [PubMed] [Google Scholar]
- 14.Guggenheim, B., B. Regolati, and H. R. Muhlemann. 1972. Caries and plaque inhibition by mutanase in rats. Caries Res. 6:289-297. [DOI] [PubMed] [Google Scholar]
- 15.Guggenheim, B., B. Regolati, R. Schmid, and H. R. Muhlemann. 1980. Effects of the topical application of mutanase on rat caries. Caries Res. 14:128-135. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa, S., and J. H. Nordin. 1969. Enzymes that hydrolyze fungal cell wall polysaccharides. I. Purification and properties of an endo-alpha-d-(1-3)-glucanase from Trichoderma viride. J. Biol. Chem. 244:5460-5470. [PubMed] [Google Scholar]
- 17.Hazlewood, G. P., and H. J. Gilbert. 1998. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic Acid Res. Mol. Biol. 61:211-241. [DOI] [PubMed] [Google Scholar]
- 18.Imai, K., T. Kikuta, M. Kobayashi, and K. Matsuda. 1977. An α-1,3-glucanase from Streptomyces sp. KI-8. Production and purification. Agric. Biol. Chem. 41:1339-1346. [Google Scholar]
- 19.Inoue, M., T. Yakushiji, J. Mizuno, Y. Yamamoto, and S. Tanii. 1990. Inhibition of dental plaque formation by mouthwash containing an endo-alpha-1,3 glucanase. Clin. Prev. Dent. 12:10-14. [PubMed] [Google Scholar]
- 20.Inoue, M., T. Yakushiji, M. Katsuki, N. Kudo, and T. Koga. 1988. Reduction of the adherence of Streptococcus sobrinus insoluble alpha-d-glucan by endo-(1-3)-alpha-d-glucanase. Carbohydr. Res. 182:277-286. [DOI] [PubMed] [Google Scholar]
- 21.Kelstrup, J., P. Holm-Pedersen, and S. Poulsen. 1978. Reduction of the formation of dental plaque and gingivitis in humans by crude mutanase. Scand. J. Dent. Res. 86:93-102. [DOI] [PubMed] [Google Scholar]
- 22.Kelstrup, J., T. D. Funder-Nielsen, and E. N. Moller. 1973. Enzymatic reduction of the colonization of Streptococcus mutans in human dental plaque. Acta Odontol. Scand. 31:249-253. [DOI] [PubMed] [Google Scholar]
- 23.Kigawa, H., I. Shimotsuura, Y. Yokobori, Y. Asai, and M. Ohdera. November 1996. Mutanase gene. Japan patent 1998201483-A/1.
- 24.Kopecká, M., G. H. Fleet, and H. J. Phaff. 1995. Ultrastructure of the cell wall of Schizosaccharomyces pombe following treatment with various glucanases. J. Struct. Biol. 114:140-152. [DOI] [PubMed] [Google Scholar]
- 25.Krieg, N. R., and J. G. Holt. 1986. Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, MD.
- 26.Li, Y. H., and G. H. Bowden. 1994. Characteristics of accumulation of oral gram-positive bacteria on mucin-conditioned glass surfaces in a model system. Oral Microbiol. Immunol. 9:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Lis, M., and H. K. Kuramitsu. 1997. Galactose oxidase-glucan binding domain fusion proteins as targeting inhibitors of dental plaque bacteria. Antimicrob. Agents Chemother. 41:999-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luchsinger, W. W., and R. A. Cornesky. 1962. Reducing power by the dinitrosalicylic acid method. Anal. Biochem. 4:346-347. [DOI] [PubMed] [Google Scholar]
- 30.Masumoto, K., K. Yamashita, A. Yoshida, S. Hayashi, Y. Machida, and T. Nagai. 1987. Production and physicochemical properties of water-insoluble glucan from Streptococcus mutans. Chem. Pharm. Bull. 35:3813-3821. [DOI] [PubMed] [Google Scholar]
- 31.McGavin, M., and C. W. Forsberg. 1989. Catalytic and substrate-binding domains of endoglucanase 2 from Bacteroides succinogenes. J. Bacteriol. 171:3310-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, M. T., and H. J. Phaff. 1977. Survey for α-(1 → 3)-glucanase activity among yeasts. J. Bacteriol. 131:702-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer, M. T., and H. J. Phaff. 1980. Purification and properties of (1-3)-α-glucanases from Bacillus circulans WL-12. J. Gen. Microbiol. 118:197-208. [Google Scholar]
- 34.Pleszczynska, M., M. Marek-Kozaczuk, A. Wiater, and J. Szczodrak. 2007. Paenibacillus strain MP-1: a new source of mutanase. Biotechnol. Lett. 29:755-759. [DOI] [PubMed] [Google Scholar]
- 35.Regolati, B., and B. Guggenheim. 1974. Effects of protease activity in crude mutanase on caries and plaque in rats. Helv. Odontol. Acta 18:97-100. [PubMed] [Google Scholar]
- 36.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 37.Sanchez, H. M. E., M. C. Garcia, and M. Novaes-Ledieu. 1993. Two different alkali-soluble alpha-glucans in hyphal walls of the basidiomycete Armillaria mellea. Microbiologia 9:34-42. [PubMed] [Google Scholar]
- 38.Sanz, L., M. Montero, J. Redondo, A. Llobell, and E. Monte. 2005. Expression of an α-1,3-glucanase during mycoparasitic interaction of Trichoderma asperellum. FEBS J. 272:493-499. [DOI] [PubMed] [Google Scholar]
- 39.Schachtele, C. F., and M. E. Jensen. 1982. Comparison of methods for monitoring changes in the pH of human dental plaque. J. Dent. Res. 61:1117-1125. [DOI] [PubMed] [Google Scholar]
- 40.Shen, H., M. Schmuck, I. Pilz, N. R. Gilkes, D. G. Kilburn, R. C. Miller, Jr., and R. A. J. Warren. 1991. Deletion of the linker connecting the catalytic and cellulose-binding domains of endoglucanase A (CenA) of Cellulomonas fimi alters its conformation and catalytic activity. J. Biol. Chem. 266:11335-11340. [PubMed] [Google Scholar]
- 41.Simonson, L. G., R. W. Gaugler, B. L. Lamberts, and D. A. Reiher. 1983. Glucanohydrolases and the control of glucans, p. 211-221. In R. J. Doyle and J. G. Ciardi (ed.), Glucosyltransferases, glucans, sucrose and dental caries. IRL Press, Washington, DC.
- 42.Simonson, L. G., R. W. Gaugler, B. L. Lamberts, and D. A. Reiher. 1982. Purification and properties of endo-1,3-alpha-d-glucanase from Pseudomonas. Biochim. Biophys. Acta 715:189-195. [DOI] [PubMed] [Google Scholar]
- 43.Sumitomo, N., K. Saeki, K. Ozaki, S. Ito, and T. Kobayashi. 2007. Mutanase from a Paenibacillus isolate: nucleotide sequence of the gene and properties of recombinant enzymes. Biochim. Biophys. Acta 1770:716-724. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, N., A. Yoshida, and Y. Nakano. 2005. Quantitative analysis of multi-species oral biofilms by TaqMan real-time PCR. Clin. Med. Res. 3:176-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, N., Y. Sato, and K. Takamori. 1983. Isolation and characterization of an oral bacterial alpha-1,3-glucanase from Bacteroides oralis Ig4a. J. Dent. Res. 62:471. [Google Scholar]
- 46.Takehara, T., and M. Inoue. 1981. Inhibitory effects of endo-alpha-1,3-glucanase on glucan film formation and glucan synthesis by the glucosyltransferase of the oral bacterium Streptococcus mutans. Arch. Oral Biol. 26:217-222. [DOI] [PubMed] [Google Scholar]
- 47.Takehara, T., M. Inoue, T. Morioka, and K. Yokogawa. 1981. Purification and properties of endo-alpha-1,3-glucanase from a Streptomyces chartreusis strain. J. Bacteriol. 145:729-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsunoda, A., T. Nagai, Y. Sakano, and T. Kobayashi. 1997. Purification and properties of an exo-α-1,3-glucanase from Trichoderma viride. Agric. Biol. Chem. 41:939-943. [Google Scholar]
- 49.Walker, G. J., and M. D. Hare. 1977. Metabolism of the polysaccharides of human dental plaque. Part II. Purification and properties of Cladosporium resinae (1→3)-alpha-d-glucanase, and the enzymic hydrolysis of glucans synthesised by extracellular d-glucosyltransferases of oral streptococci. Carbohydr. Res. 58:415-432. [DOI] [PubMed] [Google Scholar]
- 50.Yakushiji, T., M. Inoue, and T. Koga. 1984. Inter-serotype comparison of polysaccharides produced by extracellular enzymes from Streptococcus mutans. Carbohydr. Res. 127:253-266. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yano, S., M. Wakayama, and T. Tachiki. 2006. Cloning and expression of an alpha-1,3-glucanase gene from Bacillus circulans KA-304: the enzyme participates in protoplast formation of Schizophyllum commune. Biosci. Biotechnol. Biochem. 70:1754-1763. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zero, D. T., J. van Houte, and J. Russo. 1986. The intra-oral effect on enamel demineralization of extracellular matrix material synthesized from sucrose by Streptococcus mutans. J. Dent. Res. 65:918-923. [DOI] [PubMed] [Google Scholar]
- 55.Zonneveld, B. J. 1972. A new type of enzyme, an exo-splitting 1,3-glucanase from non-induced cultures of Aspergillus nidulans. Biochim. Biophys. Acta 258:541-547. [DOI] [PubMed] [Google Scholar]