Abstract
Seven methods of soil DNA extraction and purification were tested in a set of 14 soils differing in bedrock, texture, pH, salinity, moisture, organic matter content, and vegetation cover. The methods introduced in this study included pretreatment of soil with CaCO3 or purification of extracted DNA by CaCl2. The performance of innovated methods was compared to that of the commercial kit Mo Bio PowerSoil and the phenol-chloroform-based method of D. N. Miller, J. E. Bryant, E. L. Madsen, and W. C. Ghiorse (Appl. Environ. Microbiol. 65:4715-4724, 1999). This study demonstrated significant differences between the tested methods in terms of DNA yield, PCR performance, and recovered bacterial diversity. The differences in DNA yields were correlated to vegetation cover, soil pH, and clay content. The differences in PCR performances were correlated to vegetation cover and soil pH. The innovative methods improved PCR performance in our set of soils, in particular for forest acidic soils. PCR was successful in 95% of cases by the method using CaCl2 purification and in 93% of cases by the method based on CaCO3 pretreatment, but only in 79% by Mo Bio PowerSoil, for our range of soils. Also, the innovative methods recovered a higher percentage of actinomycete diversity from a subset of three soils. Recommendations include the assessment of soil characteristics prior to selecting the optimal protocol for soil DNA extraction and purification.
DNA purification is a critical step in soil DNA extraction, once the problems with lysis are overcome (23). Over time, bead beating has been recommended as the most effective technique in soil aggregate and cell disruption (13, 14, 17, 22) and is also used in commercial kits. So, all the methods tested here included bead beating for the disruption step to ensure a high yield of DNA. Yet, harsh direct lysis is known to recover DNA contaminated with humic acids and unknown amounts of eukaryotic DNA (22).
DNA contamination with humic acids has resulted in PCR inhibition and microbial diversity bias (16, 17, 27). Solutions were found in the dilution of the template DNA, which improved both PCR performance and recovered diversity (5) in some soils, or, in contrast, by increasing the amount of template DNA, which was effective in other soils (4, 10). However, it seems that further purification of template DNA is preferred to DNA dilution because a loss of diversity is more likely with dilution (27).
DNA purification is often difficult because humic acids seem to make complexes with the extracted DNA that are not easily separable. Purification by washing DNA embedded in low-melting-point agarose, desalting on Sephadex columns, treating with polyvinylpolypyrrolidone, and using polyethylene glycol 8000, commercial glass milk, Ionex, and membrane-based purification kits did improve the soil DNA quality and subsequent PCR performance in some soils but not in others (14, 17, 18, 19). In many soils, DNA extraction was negatively influenced by clay content (10, 20, 28). Recently, most difficulties have been attributed to acidic forest soils (12).
Consequently, in this study, two innovative methods were developed to improve the quality of extracted DNA in acidic soils. The first method included pretreatment of soils with CaCO3, while in the second method, separation of humic acids from soil DNA by CaCl2 (buffered to pH 8) was added after extraction. The aims of the study were (i) to compare the performance of the innovative methods with other widely used soil DNA extraction methods and (ii) to assess the importance of soil characteristics, low pH and clay content in particular, with regard to DNA yield and PCR performance in all tested methods.
MATERIALS AND METHODS
Soil samples.
Fourteen sites were selected containing soils with a wide spectrum of soil characteristics, including pH, conductivity, moisture, organic matter content, vegetation cover, base rock, particle size, and soil type. Sites were located in the Czech Republic and France.
Soil samples were collected in the spring and summer of 2005. Approximately 1 kg of soil was cut from the top 20 cm using a small spade and knife. Samples were placed in plastic bags and cooled for transport. Soil was homogenized manually by thorough mixing, and subsamples of 250 g were set aside for soil analysis. Soil for DNA analysis was frozen to −20°C. All tools and materials used were washed and sterilized.
Soil analysis.
Particle size was determined by the standard method at the accredited soil laboratory Geologie and Geotechnica, a.s. (Prague, Czech Republic) (8). Soil pH and conductivity were measured in soil water extract (20 g of soil and 50 ml of water were mixed and left to stand overnight at room temperature). Moisture was assessed by drying soil at 80°C to constant weight. Organic matter content was estimated by combustion in an oven at 550°C to constant weight. Humic substances were extracted according to Rezacova et al. (21) and quantified spectrophotometrically at 465 nm. Cations of Ca, Mg, Al, and Fe were determined by atomic absorption spectrophotometry after extraction with 1 M ammonium acetate by Gematest, s.r.o. (Prague, Czech Republic). Phytosociological relevés were performed at all sites, and the vegetation type was ascertained. Direct bacterial counts were assessed using the method of Bakken and Lindahl (3); 1 g of soil was suspended in 7 ml of SET buffer (100 mM NaCl, 10 mM Tris, and 1 mM EDTA, pH 8) and vortexed for 2 min, 3 ml of Nycodenz AG (Axis/Shield, Oslo, Norway; 1.3 g in 1 ml water) was added, and the mixture was centrifuged at 8,000 × g for 40 min. The supernatant was diluted 1,000× for counting. Bacteria were stained with DAPI (4′,6′-diamidino-2-phenylindole) and enumerated under an epifluorescence microscope (Olympus BX 60), with the number of fields counted always sufficient to reach at least 500 counted cells.
Experimental design.
Two methods of DNA extraction, three methods of sample pretreatment, and one method of DNA purification, seven combinations in total, were compared according to the design shown in Table 1.
TABLE 1.
Experimental design for comparison of soil DNA extraction and purification methods
| Method | Soil pretreatment | DNA extraction | DNA purification | Reference or source |
|---|---|---|---|---|
| M | Mo Bio PowerSoil | Included in the kit | ||
| MV | Incubation with suspended CaCO3 | Mo Bio PowerSoil | Included in the kit | This work |
| S | Modified method of Miller et al. (17, 27) | 17 | ||
| SA | AlNH4(SO4)2, 50 mM in extraction buffer | Modified method of Miller et al. (17, 27) | 9 | |
| SC | CaCl2, 50 mM in extraction buffer | Modified method of Miller et al. (17, 27) | 4 | |
| SV | Incubation with suspended CaCO3 | Modified method of Miller et al. (17, 27) | This work | |
| SK | Modified method of Miller et al. (17, 27) | Incubation with CaCl2, GeneClean Turbo DNA kit | This work |
The two DNA extraction methods were (i) the Mo Bio PowerSoil DNA isolation kit (M) (Carlsbad, CA), used according to the manufacturer's protocol, and (ii) our modification of the method of Miller et al. (S) (17, 27), for which the procedure was as follows. Soil (0.5 g) was homogenized in a mini-bead beater (BioSpec Products, Bartlesville, OK) for 90 s, at 2,500 rpm with 600 μl of extraction buffer (50 mM Na-phosphate buffer [pH 8], 50 mM NaCl, 500 mM Tris-HCl [pH 8], and 5% sodium dodecyl sulfate) and 300 μl of phenol-chloroform-isoamyl alcohol (25:24:1) and 0.5 g sterile glass beads (0.25 mg of 0.1-mm diameter and 0.25 mg of 0.5-mm diameter). The homogenate was centrifuged at 16,000 × g for 2 min. The supernatant was mixed with the same volume of phenol-chloroform-isoamyl alcohol (25:24:1) and centrifuged at 6,000 × g for 5 min. (In this step, half the volume of phenol can be used for obtaining larger DNA fragments.) The supernatant was mixed with an equal volume of chloroform-isoamyl alcohol (24:1) and centrifuged at 16,000 × g for 5 min. To the supernatant, NaCl was added to a final concentration of 1.5 M, and CTAB was added to 1% and incubated at 65°C for 30 min. The incubated solution was cooled, mixed with an equal volume of chloroform-isoamyl alcohol (24:1), and centrifuged at 3,400 × g for 20 min. The supernatant was then precipitated with isopropanol.
Sample pretreatment included one method from the literature, SA; two methods developed in our laboratory, SV and MV; and one modified in our laboratory, SC. For the SA pretreatment method, soil was treated with AlNH4(SO4)2 [a stock solution of 200 mM AlNH4(SO4)2 in 100 mM Tris-HCl, pH 7], added to the extraction buffer to a final concentration of 50 mM as described by Dong et al. (9), and extracted by the S method. Alternatively, a 1 M suspension of CaCO3 was added to the soil sample (5 g) in a 1:1 ratio by volume, and the mixture was left at room temperature for 1 h. Then, DNA was extracted by method M (for the MV pretreatment method) or S (for the SV pretreatment method). For the SC pretreatment method, soil was treated with CaCl2 (a stock solution of 200 mM CaCl2 in 100 mM Tris-HCl, pH 7), added to the extraction buffer to a final concentration of 50 mM, and extracted by the S method.
SK, a newly developed method, was used for further purification of extracted DNA. DNA extracted by the S method was treated with CaCl2 (1 M CaCl2 in 1 M HEPES-NaOH, pH 7), which was added to the DNA dissolved in water in a 1:1 ratio by volume and left to stand for 30 min at room temperature. Then, the mixture was purified with a GeneClean Turbo DNA kit (Qbiogene, Irvine, CA).
DNA was extracted three times from separate aliquots of each soil by each method (294 extractions in total). The yields of isolated DNA were determined from 1% agarose gel using AIDA software (Raytest, Straubenhardt, Germany). DNA yields from the more concentrated samples were determined after dilutions. The estimations from the gels were then correlated to the measurements made by the NanoDrop ND-1000 spectrophotometer at 260 nm for a comparison of accuracy.
PCR.
To test the purity of DNA extracted by the tested methods, PCRs were conducted for all samples with universal eubacterial primers for the 16S rRNA gene. A PCR was performed in 50 μl total volume, according to the following protocol: 50 to 100 ng of DNA, 1× Taq buffer, 0.4 μM primers, 0.4 mM deoxynucleoside triphosphate mixture, 1.5 mM MgCl2, 0.6 mg ml−1 bovine serum albumin, and 1 U Taq DNA polymerase. The program on the PCR thermocycler Biometra T1 included a hot start at 94°C for 5 min, 35 cycles of 94°C for 45 s, 57°C for 45 s, 72°C for 1 min 30 s, and a final extension at 72°C for 5 min. The primers were 27f Hex, 5′-AGA GTT TGA TCM TGG CKC AG-3′, and 783r (a mixture of 783a [5′-CTA CCA GGG TAT CTA ATC CTG-3′], 783b [5′-CTA CCG GGG TAT CTA ATC CCG-3′], and 783c [5′-CTA CCC GGG TAT CTA ATC CGG-3′], as described by Sakai et al. (24). A PCR was performed once for each DNA extraction; so, three PCRs were performed for each soil sample. The set of PCRs always included negative and positive controls. A PCR was considered successful if a specific product was visible in one-tenth of a reaction mixture on the gel.
To test the DNA extracted by each tested method for recovered bacterial diversity, terminal restriction fragment length polymorphism (T-RFLP) analysis was performed. DNA sufficiently pure to produce a successful PCR was extracted by all methods from only 3 out of the 14 soils: Oblik, Saline Giraud, and Srbsko. So, those samples were selected for a comparison between methods of the levels of recovered diversity. For the T-RFLP analysis, PCRs were performed twice for each sample, once with the universal eubacterial primers and once with the actinomycete-specific primers, according to the same protocol. The actinomycete-specific reverse primer, 623r (5′-ACA CCA GGA ATT CCA GTC TC-3′), was designed using Primrose software (2). Altogether, three T-RFLP analyses were performed in one run for all samples, and those with the highest signal were selected for statistical analysis.
T-RFLP.
PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and cleaved by restriction endonuclease AluI at 37°C for 4 h. Inactivation at 65°C for 20 min was followed by purification with Sigma spin postreaction cleanup columns (Sigma-Aldrich, St. Louis, MO). Fragment analysis was performed at Genomac International, s.r.o. (Prague, Czech Republic) with a 96-capillary sequencer (Applied Biosystems, Foster City, CA).
Statistical analysis.
Soil characteristics were correlated using the Spearman rank-order correlation coefficient (r). DNA yields among the tested methods were compared using the Friedman test (26). Soil characteristics were related to DNA yields using stepwise linear regression maximizing the Akaike information criterion, with logarithmic transformation of DNA yields as the response and soil characteristics as regressors (11, 25). The logarithmic transformation proved to be necessary to achieve approximate normality in the response. A linear regression was used to predict the influence of soil clay content on DNA yields. Soil characteristics were related to PCR performance using stepwise logistic regression maximizing the Akaike information criterion, with logarithmic transformation of DNA yields as the response and soil characteristics as regressors. T-RFLPs were treated using the approach of Abdo et al. (1) and statistically compared using Friedman's test.
RESULTS
Sites distinguished by three types of vegetation cover, forest, meadow, and scattered vegetation, and combinations of base rock, soil water and humic acid content, pH, or particle size (Table 2) were included in our set of soils.
TABLE 2.
Soil and site characteristics
| Site | pH | Conductivity (μS) | Organic matter (%) | Water content (%) | Humic acids (A465) | Ca (mg·kg−1) | Mg (mg·kg−1) | Al (mg·kg−1) | Fe (mg·kg−1) | Clay (%) | Silt (%) | Sand (%) | Soil type | Vegetation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpilles | 7.5 | 256 | 16.1 | 25.6 | 0.046 | 5,900 | 184 | <3.0 | <1.0 | 5 | 22 | 73 | Rendzina | Pine forest |
| Kotyz | 7.5 | 310 | 26.6 | 20.4 | 0.074 | 7,600 | 191 | <3.0 | 1.2 | 2 | 11 | 87 | Rendzina | Pine forest |
| Meluzina | 5.0 | 86 | 7.6 | 66.0 | 0.370 | 2,700 | 137 | 10.4 | 2.5 | 4 | 51 | 45 | Cryptopodzol | Mixed mountain |
| Nechranice | 6.1 | 64 | 11.6 | 24.4 | 0.143 | 3,470 | 618 | <3.0 | <1.0 | 31 | 39 | 30 | Vertisol | Hard wood |
| Podyji | 5.6 | 75 | 10.2 | 41.5 | 0.063 | 2,010 | 404 | 4.1 | 2.9 | 3 | 37 | 60 | Fluvisol | Riparian forest |
| Srbsko | 7.7 | 141 | 8.2 | 20.2 | 0.009 | 4,740 | 141 | <3.0 | <1.0 | 26 | 57 | 17 | Cambisol | Mixed forest |
| Trebon | 4.0 | 80 | 9.1 | 30.2 | 0.077 | 1,160 | 289 | 3.4 | 4.4 | 2 | 2 | 96 | Podzol | Mixed forest |
| Bozi Dar | 3.3 | 47 | 95.7 | 75.3 | 0.812 | 1,290 | 106 | 5.3 | 1.4 | 0 | 0 | 0 | Histosol | Peat bog |
| Devin | 7.9 | 200 | 12.0 | 27.1 | 0.028 | 6,210 | 163 | <3.0 | <1.0 | 2 | 21 | 77 | Rendzina | Steppe |
| Oblik | 7.9 | 200 | 21.5 | 50.2 | 0.010 | 6,090 | 307 | <3.0 | <1.0 | 4 | 29 | 67 | Cambisol | Steppe |
| Rynholec | 6.3 | 53 | 7.2 | 19.3 | 0.002 | 2,840 | 83 | 6.5 | <1.0 | 37 | 43 | 20 | Technosol | Wet meadow |
| Slanisko Nesyt | 8.0 | 537 | 6.6 | 22.4 | 0.010 | 3,390 | 1,250 | <3.0 | <1.0 | 10 | 19 | 71 | Phaeosol | Marsh |
| Saline Giraud | 8.1 | 21,350 | 3.3 | 10.9 | 0.001 | 3,150 | 67 | <3.0 | <1.0 | 9 | 15 | 76 | Arenosol | Scattered |
| Slepici vrch | 4.6 | 24 | 1.5 | 3.1 | 0.015 | 100 | 29 | <3.0 | <1.0 | 2 | 2 | 96 | Arenosol | Scattered |
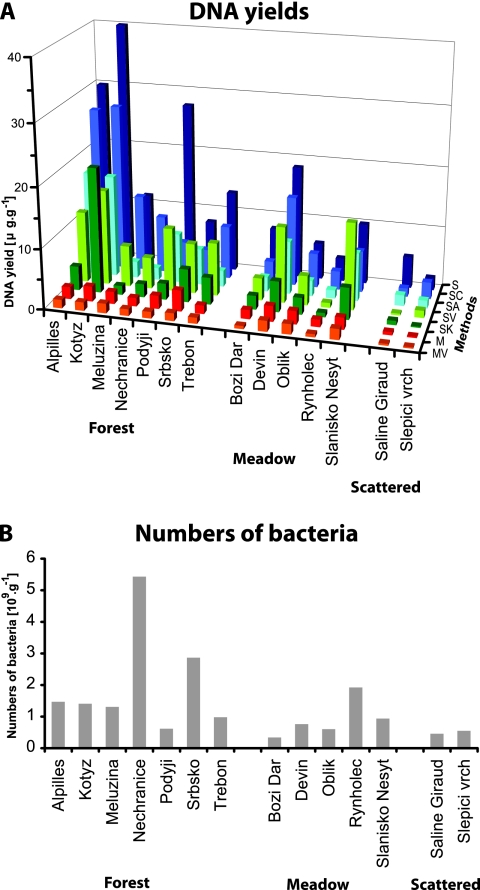
DNA yields reached values from 1 μg per g soil from sandy soils with scattered vegetation to tens of μg per g soil from forest soils (Fig. 1A; see Table S1 in the supplemental material). DNA yields were decreased by all purification procedures. The order of the methods according to the average DNA yield over all sites was S, SC, SV, SA, SK, M, and MV. Differences in DNA yields among the tested methods were significant (χ2 = 65.4, df = 6, and P < 0.001). However, the DNA yields at several sites did not follow the overall efficiency of the respective method. For example, the highest DNA yields at the Slanisko Nesyt site were reached by the method SV and at the sites Meluzina, Nechranice, and Slepici Vrch by the method SC, rather than by the method S, the method most efficient for other sites. The methods based on the Mo Bio PowerSoil kit gave low yields, and the method MV gave the lowest yields of all sites.
FIG. 1.
(A) DNA yield averages (μg·g−1) (dry weight) of soil from sites with different vegetation cover. (For respective standard deviations of the three measurements, see Table S1 in the supplemental material.) Abbreviations (Methods) stand for DNA extraction methods (see Materials and Methods). (B) Numbers of bacteria in different types of vegetation cover. Data are from direct counts under a microscope.
The influence of soil characteristics on DNA yields with respect to the tested methods was analyzed by linear models. As nothing was known about the significance of any of the soil characteristics prior to data analysis, a stepwise regression was used to show that method (compared to method M, P = 0.04 for MV, P = 2e-16 for S, P = 4.1e-10 for SA, P = 0.001 for SK, P = 1.8 e-14 for SC, and P = 1.1e-14 for SV), vegetation cover (compared to forest, P = 1.2e-05 for meadow and P = 2e-16 for scattered vegetation), clay (P = 8.2e-13), pH (P = 2e-05), and water content (P = 0.01) might affect the DNA yields. The final model explained over 75% of the variability of DNA yields. However, these findings should be considered only a suggestion for further study due to the multiplicity testing problem of stepwise methods.
The DNA yields were compared with counted numbers of bacteria, and no correlation was observed for the averages of DNA yields recovered by all tested methods (Fig. 1B). Since vegetation cover and clay content were assigned as the most significant soil characteristics influencing the DNA yield by the linear model, the relationship between DNA yields and number of bacteria was further analyzed with respect to these soil characteristics. First, DNA yields were highest for forest sites, intermediate for meadow, and lowest for scattered vegetation for all tested methods as expected (see Fig. S1A in the supplemental material). However, DNA yields recovered by the methods derived by S (phenol-chloroform extraction) from the meadow and forest soils often exceeded the amount expected from the numbers of bacteria. Second, the yields of DNA were correlated to the numbers of bacteria for all methods with respect to the clay content in the soil. A linear model showed that DNA yields were significantly lower than expected from the numbers of bacteria in high-clay soils (>20%; three soils) than in low-clay soils (<20%; 11 soils) (P < 0.001) (see Fig. S1B in the supplemental material). The model explained over 50% of the variability in DNA yields.
The tested methods differed significantly in the purity of extracted DNA, when the levels of purity were compared by PCR performance (χ2 = 22.07, df = 6, and P < 0.001). As with the DNA yields, a stepwise logistic regression was used to find the influencing factors. The model suggested that method (P = 1.2e-10), pH (P = 5.1e-07), and vegetation cover (P = 0.02) might be most closely related to PCR performance. The final model explained over 75% of the variability in PCR performance.
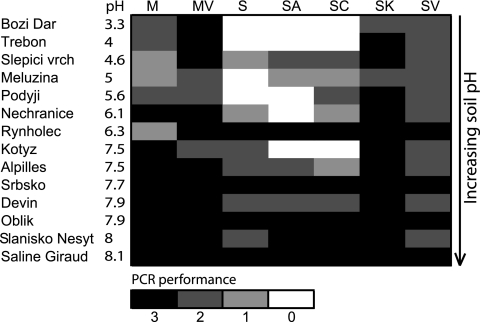
The method SK was the most successful when evaluated by PCR performance (40 successful PCRs out of 52), followed by MV (39 out of 52), M (33 out of 52), SV (32 out of 52), SC (25 out of 52), SA (23 out of 52), and S (22 out of 52). PCRs were less dependent on the method when the soil pH was higher. With pH levels higher than 7.5, almost no difference in PCR performances was found among the tested methods (Fig. 2). The most successful PCR performance was achieved with template DNA prepared by the methods that produced the purest DNA. The correlation between the DNA solution absorbance at a wavelength of 320 nm, which represents humic substances, and the number of successful PCRs, was significant (P < 0.001).
FIG. 2.
PCR performance. Evaluation of PCR success from three independent reactions. Sites are ordered according to pH (low pH at the top and high pH at the bottom). Abbreviations stand for DNA extraction methods (see Materials and Methods).
The methods differed in the numbers of recovered T-RF lengths, which represent bacterial phylotypes. The differences were found to be significant at three selected sites, Oblik, Saline de Giraud, and Srbsko, for both eubacteria (χ2 = 17.5, df = 6, and P = 0.008) and actinomycetes (χ2 = 14.9, df = 6, and P = 0.02) (see Table S2 in the supplemental material). All pretreatment and purification methods improved the quality of extracted DNA with respect to the number of detected T-RFs, particularly for eubacteria. The method MV improved the method M by 25% for eubacteria. The method SK improved the method S by 20% for eubacteria and by 30% for actinomycetes. The method SK was more efficient than all the other tested methods in recovering actinomycete diversity at the three sites (see Table S2 in the supplemental material).
DISCUSSION
DNA yields.
DNA yields obtained from our soils reached values similar to those found in other studies (5, 17). Bacteria numbers have regularly been used for evaluation of DNA yields (7, 17, 18), but we did not find a clear correlation between numbers of bacteria and DNA yields in respective soils; rather, the DNA recovery was influenced by soil characteristics, namely vegetation cover and clay and water content.
Different factors influencing DNA yields have already been suggested. Organic matter correlation with DNA yields has been suggested by Zhou et al. (29), Miller et al. (17), and Frostegård et al. (10). Soil particle size and water content were correlated to DNA yields in a study by Burgmann et al. (5). Such disagreements in assigning different factors responsibility for the inconsistency in DNA yields can be explained by their close correlation or similar mechanisms of influence.
Another point of view explaining various DNA yields can be given by the sensitivity, or rather the “aggressiveness,” of individual methods in assessing both the microbial community and the already-freed DNA. First, different methods may recover different parts of the bacterial community, and because bacteria contain different amounts of DNA and DNA originating from diverse species might be released differently (14), the yields may be affected by a method's specificity. Second, differences in DNA yields can be explained by the presence of organisms other than bacteria and again the different sensitivity levels of the respective methods in their DNA recovery. Finally, the decrease in DNA yields might be attributed to DNA binding to particles. Clay, but also humus, particles are negatively charged and bind and exchange cations (6), which lead to an explanation of lowered DNA yields because of the adsorption of free DNA on clay (10, 12, 20, 29), but also on organic matter particles (23). Often, separation of the two influences, clay and humus, becomes difficult, which was the case in our study, in which the samples high in clay were also relatively high in organic matter content.
PCR performance.
The tested methods differed in their PCR performances, particularly in forest soils of low pH. In agreement with our findings, Burgmann et al. (5) concluded that the low-pH soil from a forest site needed further purification because a sample of a forest soil with pH 3.4 strongly inhibited PCR. Also, Zhou et al. (29) reported two boreal temperate forest soils of pH 4.8 and 6.1 needed more purification than other soils with higher pH. Both of our purification approaches, pretreatment of soil with CaCO3 (MV and SV) and crude DNA purification with buffered CaCl2 followed by a cleanup with glass milk columns (SK), proved more efficient in recovering purer DNA and consequently had higher PCR success. Inhibition of PCRs was often ascribed to DNA contaminated with humic acids coextracted from the soil (10, 22, 27, 28). Also, soil organic C was negatively correlated to PCR performance, also possibly due to increased humic substances in DNA from more-organic soils (20, 29). In our study, vegetation cover and pH influenced DNA purity and consequently PCR performance. Both vegetation cover and pH are related to organic matter (see above), and at low pH, larger amounts of humic acids can be released to the extracted DNA.
The reason why the methods taken from the literature have already proved to be optimal, but yet were not successful with our soils, is most probably the restricted range of studied soil types. Also, differing soil characteristics assigned to the changes in PCR performance might be due to their intercorrelation. The numbers of tested soils in various studies were, for example, one soil type (27), three soils (28), four soils (9), six soils (5), and eight soils (29). Vegetation cover, one of the most important factors influencing PCR performance in our study, was not considered in many studies (4, 7, 10, 15) and only generally described in others (5, 12, 27, 29). The other important factor, pH, was more often included in the studies, but in some cases studied soils were in narrow pH ranges, such as pH 4.3 to 5.8 (10), pH 5.9 to 7.1 (28), pH 6.2 to 6.3 (15), and pH 5.7 to 6.5 (17), or included just one sample of extremely low pH, such as pH 4.8 (29) or pH 4 (27). Finally, in a comprehensive study by Braid et al. (4), who tested 20 soils, no data on pH were presented.
In conclusion, the commonly used methods can be successfully applied to higher-pH soils with limited vegetation biomass or organic matter content. We believe our innovative methods have a wider range of application.
Bacterial diversity.
The comparison of recovered bacterial diversity using T-RFLP was limited to three soil samples, which were the only ones yielding template DNA suitable for subsequent PCR amplification by all methods. Consequently, the compared soils were of similar characteristics, high pH and average humic acid and Ca content. Our purification methods (MV, SV, and SK) were more efficient than the Mo Bio PowerSoil kit (M) in recovering the diversity of eubacteria. The innovative method SK was more efficient than any other studied method in recovering the diversity of actinomycetes.
Conclusions and perspectives.
The introduced purification methods were successfully applied to our set of soils, in which the innovative methods proved more efficient in PCR performance and description of bacterial communities, particularly with regard to actinomycete diversity. The innovative method SK is rather time-consuming; it can therefore be used when precision is required, for extreme soils or when actinomycetes are targeted. In other cases, the treatment of soil by CaCO3 followed by the Mo Bio PowerSoil kit (M) might be a proper choice.
The attempt was made to find relationships between soil characteristics, DNA yield, and PCR performance (Table 3). We suggest that soil pH and clay content, but possibly also organic matter or water content or Ca, be assessed at a site prior to deciding on an extraction and purification protocol. Particular attention should be paid to soils high in clay, in which DNA yields can be very low.
TABLE 3.
Soil characteristics important for choice of the DNA extraction method
| Decisive soil characteristic | DNA extraction method(s) for:
|
|||
|---|---|---|---|---|
| PCR
|
PCR + DNA yield
|
|||
| Recommended | Not recommended | Recommended | Not recommended | |
| pH <7.5 | MV, SV, SK | SV | MV | |
| Clay, >20% | M | S, SC | MV, SK, SV | |
| Vegetation cover | ||||
| Forest | MV, SK, SV | M, S, SC, SA | SK, SV | |
| Scattered | MV, SK | M, S | SA, SC, SV | |
| pH >7.5 | ||||
| Organic matter, >20% | SC, SA | |||
| Vegetation cover, scattered | SC, SA | S, SA, SC | M, MV, SK, SV | |
Supplementary Material
Acknowledgments
This work was supported by the Grant Agency of the Academy of Sciences of the Czech Republic, grant no. IAA6020410 and IAA600200519 and Institutional Research Concept no. AV0Z50200510.
We thank Tomas Cajthaml for helpful discussion and Steve Ridgill for English revisions.
Footnotes
Published ahead of print on 14 March 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdo, Z., U. M. E. Schüette, S. J. Bent, C. J. Williams, L. J. Forney, and P. Joyce. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929-938. [DOI] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken, L. R., and V. Lindahl. 1995. Recovery of bacterial cells from soil, p. 9-27. In J. D. van Elsas and J. T. Trevors (ed.), Nucleic acids in the environment: methods and applications. Springer-Verlag, Heidelberg, Germany.
- 4.Braid, M. D., L. M. Daniels, and C. L. Kitts. 2003. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 52:389-393. [DOI] [PubMed] [Google Scholar]
- 5.Burgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 6.Buscot, F. 2005. What are soils?, p. 3-18. In F. Buscot and A. Varma (ed.), Microorganisms in soils: roles in genesis and functions. Springer-Verlag, Berlin, Germany.
- 7.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993. [Google Scholar]
- 8.DIN, ISSMGE. 1998. Recommendations of the ISSMGE for geotechnical laboratory testing. Beuth Verlag, Berlin, Germany.
- 9.Dong, D., A. Yan, H. Liu, X. Zhang, and Y. Xu. 2006. Removal of humic substances from soil DNA using aluminium sulfate. J. Microbiol. Methods 66:217-222. [DOI] [PubMed] [Google Scholar]
- 10.Frostegård, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie, T. J., and D. Pregibon. 1992. Generalized linear models, p. 195-248. In J. M. Chambers and T. J. Hastie (ed.), Statistical models in S. Chapman & Hall/CRC, Boca Raton, FL.
- 12.He, J., Z. Xua, and J. Hughes. 2005. Pre-lysis washing improves DNA extraction from a forest soil. Soil Biol. Biochem. 37:2337-2341. [Google Scholar]
- 13.Heuer, H., and K. Smalla. 1997. Application of denaturation gradient electrophoresis and temperature gradient electrophoresis for studying soil microbial communities, p. 353-373. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, NY.
- 14.Kauffmann, I. M., J. Schmitt, and R. D. Schmid. 2004. DNA isolation from soil samples for cloning in different hosts. Appl. Microbiol. Biotechnol. 64:665-670. [DOI] [PubMed] [Google Scholar]
- 15.Lakay, F. M., A. Botha, and B. A. Prior. 2007. Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. J. Appl. Microbiol. 102:265-273. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreira, D. 1998. Efficient removal of PCR inhibitors using agarose embedded DNA preparations. Nucleic Acids Res. 26:3309-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranjard, L., F. Poly, J. Combrisson, A. Richaume, and S. Nazaret. 1998. A single procedure to recover DNA from the surface or inside aggregates and in various size fractions of soil suitable for PCR-based assays of bacterial communities. Eur. J. Soil Biol. 34:89-97. [Google Scholar]
- 21.Rezacova, V., H. Hrselova, H. Gryndlerova, I. Miksik, and M. Gryndler. 2006. Modifications of degradation-resistant soil organic matter by soil saprobic fungi. Soil Biol. Biochem. 38:2292-2299. [Google Scholar]
- 22.Robe, P., R. Nalin, C. Capellano, T. Vogel, and P. Simonet. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39:183-190. [Google Scholar]
- 23.Roose-Amsaleg, C. L., E. Garnier-Sillam, and M. Harry. 2001. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 18:47-60. [Google Scholar]
- 24.Sakai, M., A. Matsuka, T. Komura, and S. Kanazawa. 2004. Application of a new PCR primer for terminal restriction fragment length polymorphism analysis of the bacterial communities in plant roots. J. Microbiol. Methods 59:81-89. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto, Y., M. Ishiguro, and G. Kitagawa. 1986. Akaike information criterion statistics. D. Reidel Publishing Co., Dordrecht, Holland.
- 26.Sokal, R. R., and F. J. Rohlf. 1995. Biometry: the principles and practice of statistics in biological research. W. H. Freeman and Company, New York, NY.
- 27.Stach, J. E., S. Bathe, J. P. Clapp, and R. G. Burns. 2001. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 36:139-151. [DOI] [PubMed] [Google Scholar]
- 28.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.