Abstract
Morphological differentiation in some arthropod-borne bacteria is correlated with increased bacterial virulence, transmission potential, and/or as a response to environmental stress. In the current study, we utilized an in vitro model to examine Rickettsia felis morphology and growth under various culture conditions and bacterial densities to identify potential factors that contribute to polymorphism in rickettsiae. We utilized microscopy (electron microscopy and immunofluorescence), genomic (PCR amplification and DNA sequencing of rickettsial genes), and proteomic (Western blotting and liquid chromatography-tandem mass spectrometry) techniques to identify and characterize morphologically distinct, long-form R. felis. Without exchange of host cell growth medium, polymorphic R. felis was detected at 12 days postinoculation when rickettsiae were seeded at a multiplicity of infection (MOI) of 5 and 50. Compared to short-form R. felis organisms, no change in membrane ultrastructure in long-form polymorphic rickettsiae was observed, and rickettsiae were up to six times the length of typical short-form rickettsiae. In vitro assays demonstrated that short-form R. felis entered into and replicated in host cells faster than long-form R. felis. However, when both short- and long-form R. felis organisms were maintained in cell-free medium for 12 days, the infectivity of short-form R. felis was decreased compared to long-form R. felis organisms, which were capable of entering host cells, suggesting that long-form R. felis is more stable outside the host cell. The relationship between rickettsial polymorphism and rickettsial survivorship should be examined further as the yet undetermined route of horizontal transmission of R. felis may utilize metabolically and morphologically distinct forms for successful transmission.
Rickettsia felis is an intracellular gram-negative bacterium transmitted primarily by the cat flea (Ctenocephalides felis). In laboratory colonies of cat fleas, R. felis is maintained via vertical transmission (1, 41). Horizontal transmission of viable R. felis from fleas to vertebrate hosts has not been demonstrated; however, mounting serological and molecular evidence suggests that this agent is infectious to humans (30). The transmission cycle of R. felis in nature involves small mammals, e.g., companion animals, rodents, and opossums, and their fleas (2, 6, 9, 39, 42); however, the mechanism by which R. felis moves from invertebrate to vertebrate host is not known.
Several genera of medically important obligate intracellular bacteria, including Chlamydia, Coxiella, Ehrlichia, and Anaplasma, have evolved the ability to produce morphologically distinct infectious forms (5, 10, 12, 18, 25, 33). Typical rickettsiae are short, rod-shaped organisms with an average size of 0.7 to 2.0 μm by 0.3 to 0.5 μm; however, atypical rickettsia-like organisms also have been reported in arthropod hosts and cell culture models. Within the tick host, wild-caught Dermacentor andersoni contained hemocyte-associated rickettsia-like organisms of three morphological types: coccobacillary, fine bacillary, and coarse bacillary (long form) (8, 31). The dynamic ultrastructure, including filamentous, irregular-pleomorphic, and spheroplast-like forms of Rickettsia prowazekii (strains Breinl and Madrid E), has been described in different tissue culture models (11, 15, 43). Filamentous Rickettsia bellii in cell culture has been reported as an adaptive form during nutrient exhaustion (22, 23, 32). Rickettsial biology is intimately coordinated with the metabolic activity of the arthropod host (27), and the rickettsiae maintained vertically within the arthropod host are subject to extreme variation in the nutrient availability associated with individual arthropod life cycle stages. The morphological characteristics of rickettsiae during the life cycle stages of the arthropod host will provide insight into the regulatory mechanisms for rickettsial propagation, dissemination, and subsequent transmission to vertebrate hosts.
Recently, we have isolated and propagated R. felis strain LSU from cat fleas in a tick-derived cell culture model, the ISE6 cell line (35). Interestingly, we identified morphologically distinct rickettsiae in R. felis-infected ISE6 cells. The long-form rickettsiae were identified as R. felis by PCR amplification of rickettsial genes. The infectivity and growth of rickettsiae have been assessed as a component of rickettsial virulence (21), but the contribution of polymorphic growth characteristics has not been examined. To examine the comparative infectious nature of morphologically distinct rickettsiae and further characterize the environmental factors contributing to the induction of a morphologically distinct form, we examined rickettsial growth and polymorphism under various environmental conditions, including nutrient availability and rickettsial density, and then compared virulence attributes of polymorphic R. felis organisms.
MATERIALS AND METHODS
Tick cells and rickettsial culture.
Ixodes scapularis-derived ISE6 cells, provided by T. Kurtti (University of Minnesota), were maintained in L15B growth medium supplemented with 10% heat-inactivated fetal bovine serum (HyClone) and 10% tryptose phosphate broth (Sigma) (26) at pH 6.8 to 7.0 in a humidified 5% CO2 incubator at 32°C. R. felis (LSU), originally isolated from C. felis, was maintained in ISE6 cells as previously described (35). Initial cultures of morphologically distinct R. felis (LSU) organisms were identified at passage 8. Nearly homogenous populations of morphologically distinct rickettsiae, consisting of >90% long-form organisms, were maintained independently; rickettsiae measuring at least three times the length of typical rickettsiae (0.25 to 0.3 μm in diameter by 0.8 to 1.0 μm) were considered long form. Populations of typical short-form R. felis were fed weekly by replacing half of the medium with new medium; long-form R. felis cultures were maintained by adding 1 ml of fresh medium into 5 ml of culture medium in a 25-cm2 tissue culture flask at weekly intervals. Both short-form and long-form R. felis organisms were subcultured to uninfected confluent ISE6 cells every 3 to 4 weeks. For all bioassays, rickettsial viability and enumeration were assessed by staining with a BacLight viability stain kit (Molecular Probes), and rickettsiae were counted in a Petroff-Hausser bacteria counting chamber (21) using a Leica microscope.
PCR amplification and DNA sequencing of rickettsial genes.
Genomic DNA of short- and long-form rickettsiae from infected ISE6 cells was extracted using a DNeasy tissue kit (Qiagen) according to the manufacturer's protocol. PCRs utilized gene-specific primers for the 17-kDa genus-specific antigen (primers Rr17.61p and Rr17.492n [42]) and the R. felis plasmids, pRF and pRFδ (primer sets pRFa-pRFb and pRFc-pRFd [28]). Reactions were performed using PCR master mix (Promega) and thermocycler conditions as previously described (24, 35). For each set of reactions, an environmental negative control for genomic DNA extraction (200 μl of phosphate-buffered saline [PBS]) and negative control for PCR (1 μl of water) were included. Amplified products were visualized on ethidium bromide-stained agarose gels. The PCR products were cloned into a pCR4-TOPO vector (Invitrogen) and sequenced by the dye terminator method on a 3130 genetic analyzer (Applied Biosystems) at the Louisiana State University School of Veterinary Medicine. Sequencing results were analyzed using the BioEdit sequence alignment editor (Ibis Biosciences); nucleotide similarity comparisons were made using the GenBank database.
Gel electrophoresis and Western immunoblotting.
Semipurified rickettsiae were recovered from R. felis-infected ISE6 cells via needle lysis of host cells and low- and high-speed centrifugation (40). Rickettsial pellets and an uninfected ISE6 cell pellet were resuspended in lysis buffer (PBS [pH 7.4], 1% NP-40, and 1× complete protease inhibitor cocktail [Roche]) and disrupted by an ultrasonic bath (Crest) for two 10-min intervals. Cell lysate was collected by centrifugation at 16,000 × g for 10 min at 4°C. Protein concentrations were determined using a detergent-compatible protein assay (Bio-Rad); 30 μg of each protein extract was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 4 to 12% Bis-Tris gels (Invitrogen). Separated proteins from companion gels were either stained with PageBlue protein staining solution (Fermentas) or transferred to a polyvinylidene fluoride membrane (Bio-Rad). The membrane was blocked with 5% skim milk in Tris-buffered saline-0.1% Tween 20 (TBST) for 1 h at room temperature. The membrane was then incubated with the anti-rickettsial outer membrane protein B (OmpB) monoclonal antibody (RC-9C2; Fuller Laboratories) for 2 h, washed with TBST, and then incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma). After the sample was washed with TBST, the signal was detected using a SuperSignal West Pico chemiluminescent substrate kit (Pierce).
Protein identification.
To identify protein in long-form R. felis, the protein band on the Coomassie-stained sodium dodecyl sulfate gel corresponding to the positive immunoblot signal was excised, using the ProteomeWorks Spot Cutter (Bio-Rad), and deposited into a 96-well plate before placement in the freezer (−22°C) until digestion. A MassPrep Station (Waters/Micromass) was used as the digestion robot to carry out an overnight digestion procedure. The gel plugs were automatically destained, reduced, alkylated, and digested with sequencing-grade modified trypsin. The peptides were automatically extracted from the gel plugs and transferred to another 96-well plate to be used for analysis. The peptide samples were then separated by liquid chromatography using an Atlantis dC18 column (75 μm by 100 mm; Waters). Mobile phase A consisted of 95% H2O-5% acetonitrile with 0.1% formic acid, and mobile phase B consisted of 5% acetonitrile-95% H2O with 0.1% formic acid. A Q-Tof (quadrupole time-of-flight) Micro (Waters/Micromass Corp) hybrid mass spectrometer (MS) was used for analysis. Electrospray analysis was carried out in positive mode. Data acquisition and analysis were executed using the ProteinLynx Global Server, version 2.0 (Waters/Micromass). A database search was performed using an online Mascot (Matrix Science) tandem MS (MS/MS) ion search against the NCBInr/Proteobacteria. Search parameters include peptide and MS/MS tolerances of 1 Da and 0.5 Da, 1 missed cleavage, oxidation of methionines, and carbamidomethylation of cysteines.
Microscopy.
For the immunofluorescence assay (IFA), cytospin preparations of R. felis-infected ISE6 cells were fixed in ice-cold acetone for 10 min; then they were incubated with blocking buffer (3% bovine serum albumin in PBS) for 1 h in a humidified chamber. Rickettsiae were labeled with polyclonal antibody against short-form R. felis organisms generated in mice. Briefly, a group of four mice were inoculated subcutaneously with a homogenate of purified, β-propiolactone (Sigma)-inactivated R. felis (25 to 50 μg) mixed 1:1 with Titermax. After two additional inoculations at 2-week intervals, anti-R. felis mouse serum was recovered and diluted 1:100 in blocking buffer for the assay. Fluorescein isothiocyanate-conjugated goat-anti-mouse IgG (KPL) was diluted 1:400 in blocking buffer and served as the secondary antibody; DAPI (4′, 6-diamidino-2-phenylindole; 1 μg/μl; Sigma) was used as a counter-stain. VectaShield (Vector Laboratories) was applied to the slides, and then slides were visualized using a Zeiss microscope. Slides in which no primary antibody was added served as a control for the fluorescein isothiocyanate-conjugated nonspecific binding.
For transmission electron microscopy, R. felis-infected ISE6 cells were prepared as described previously (14), with a modified procedure for epon/araldite embedding (35). For immunoelectron microscopy, R. felis-infected ISE6 cells were fixed in a mixture of 2.5% formaldehyde and 0.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3), washed in the same buffer, and then dehydrated in ethanol and embedded in LR White (SPI Supplies) as described previously (37). Ultrathin sections were cut by using an ultramicrotome and placed on nickel grids. Antigen grids were treated in blocking buffer (5% skim milk in PBS-0.1% Tween 20) at room temperature; then they were incubated with rabbit-anti-spotted fever group Rickettsia polyclonal antibody (NIH/RML-I7198; diluted 1:20 in blocking buffer) for 1.5 h at room temperature. The grids were washed in blocking buffer and incubated for 1 h at room temperature with goat-anti-rabbit IgG antibody labeled with 10-nm-diameter colloidal gold particles (Sigma) diluted 1:20 in blocking buffer. Preparations incubated with secondary antibody alone served as controls for nonspecific binding. Samples (for transmission electron microscopy and immunoelectron microscopy) were visualized on a JEM-1011 transmission electron microscope (JEOL) in the Microscopy Center at the Louisiana State University School of Veterinary Medicine.
Preparation of cell-free rickettsiae for bioassays.
Short-form R. felis was semipurified from R. felis-infected ISE6 cells by needle lysis and centrifugation as described above. To purify long-form R. felis, ISE6 cells were separated from rickettsiae by centrifugation at 100 × g at 4°C for 3 min; long-form R. felis organisms in the supernatant were concentrated by centrifugation. Semipurified short-form and long-form rickettsiae were counted, and comparable viability was confirmed for both forms prior to use in bioassays.
Polymorphic induction of rickettsiae.
To identify the culture parameters that facilitate induction of typical short-form R. felis into polymorphic, long-form R. felis, ISE6 cells (5 × 105 cells per well) were seeded into 12-well plates (NUNC) in 2 ml of L15B growth medium. After 2 days of culture at 32°C, ISE6 cells were exposed, in triplicate, to semipurified short-form R. felis at a multiplicity of infection (MOI) of 0.5, 5, or 50, and cells were incubated at 32°C. At 5, 12, and 19 days postinfection (dpi), the infected cells were detached by pipetting. Polymorphic, long-form development was assessed in 100 μl of cell suspension collected at 5, 12, and 19 dpi. Cell suspensions were disrupted by forcing the suspension through a 27-gauge needle 10 times, and the cells were collected by centrifugation at 16,000 × g at 4°C for 10 min, washed with 500 μl of 0.85% NaCl, and resuspended in 100 μl of 0.85% NaCl. Rickettsiae were assessed for viability and counted as described above.
Rickettsial infectivity and replication in ISE6 cells.
To compare infectivity and growth of polymorphic R. felis, ISE6 cells (5 × 105 cells per well) were seeded into 12-well plates (NUNC) in 2 ml of L15B growth medium and infected with R. felis at three different MOIs as described for the polymorphic induction assay. For the internalization assessment, 100 μl of cell suspension from 5, 12, and 19 dpi was stained with Diff-Quik (Dade Behring) according to the manufacturer's protocol. For each treatment (different MOIs and periods of infection), 200 ISE6 cells were counted by two individuals to estimate the percentage of R. felis-infected ISE6 cells. Only the intact ISE6 cells containing rickettsiae within the cell membrane were considered to be infected cells.
Rickettsiae in cell-free medium.
Conditioned L15B growth medium was collected from ISE6 cell culture flasks 3 days postseeding and filtered through a 2.0-μm-pore-size syringe filter (Whatman) to remove any ISE6 cells. Rickettsiae were assessed for viability and counted as described above, and serial quantities (2.5 × 105, 2.5 × 106, and 2.5 × 107 rickettsiae) of partially purified short-form and long-form R. felis were incubated in 1 ml of cell-free conditioned medium in a 24-well plate at 32°C. Rickettsial viability and quantity were assessed after 5, 12, and 19 days. To evaluate the infectivity of rickettsiae cultured in cell-free medium, uninfected ISE6 cells were exposed to 200 μl of rickettsial suspensions from day 12. After challenge with rickettsiae cultured in cell-free medium, ISE6 cells were incubated at 32°C until harvested at 5, 12, and 19 dpi, and then infectivity was assessed as described above.
Statistical analysis.
The SAS statistical package (version 9.1.3) general linear model procedure in an analysis of variance was used to examine potential differences in populations of short- and long-form rickettsiae. Data presented are from a single bioassay for the rickettsiae in a cell-free medium and two separate infectivity and growth assays; infectivity was assessed by two different individuals, and percentages represent the determined mean. When overall significance was identified, Tukey's honestly significant difference (HSD) post hoc test was used to examine pairwise differences of means of main effects. Pairwise t tests of least square means were performed for interaction effects to identify significant differences in rickettsial infectivity and growth. For all comparisons, a P value of ≤0.05 was considered significantly different.
RESULTS
PCR amplification and DNA sequencing.
The presence of rickettsiae in culture was confirmed using a PCR assay specific for portions of the 17-kDa antigen gene and pRF plasmid. An amplicon of 434 bp for the 17-kDa antigen gene was consistently obtained from homogenous populations of long-form R. felis. Likewise, primer sets pRFa-pRFb and pRFc-pRFd generated expected amplicons of 159 bp and 1.3 kb, respectively. The primer set pRFa-pRFd did not amplify DNA from short- or long-form R. felis samples. Representative amplicons for the Rr17.61p and Rr17.492n and pRFa-pRFb primer sets were cloned, and two clones were sequenced. Consistent with our previous reports (35), the 17-kDa antigen gene sequences of short- and long-form R. felis were 100% identical to each other and to the 17-kDa antigen gene of R. felis strain URRWXCal2 (GenBank accession number CP000053). The pRFa-pRFb sequences of both short and long forms were 100% identical to the R. felis plasmid, pRF (GenBank accession number CP000054). Amplification did not occur when the PCR templates used were an environmental control for genomic DNA extraction and water.
Western blot analysis and peptide identification.
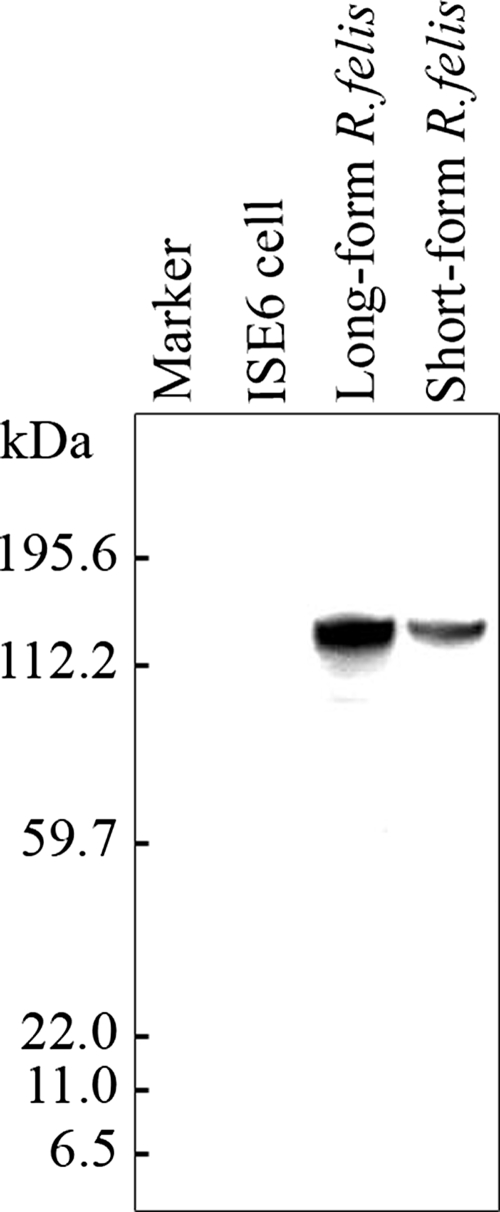
Western blot analysis was performed to assess the OmpB protein in long-form R. felis culture. Anti-OmpB monoclonal antibody strongly reacted with a 120-kDa protein, which also was present in short-form R. felis; no band was identified in the ISE6 cell extract not infected with R. felis (Fig. 1). For peptide identification, the corresponding protein band on a companion Coomassie-stained gel was excised after positions with the immmunoblot membrane were compared. The protein was digested and analyzed by using quadrupole time-of-flight micro MS. The data were dominantly matched to the OmpB of R. felis (GenBank accession number AAY61056) with a score of 236 using MASCOT MS/MS ion search software. The corresponding peptide and antigenic data confirm the expression of OmpB by long-form R. felis.
FIG. 1.
Western blot analysis of short- and long-form R. felis protein using anti-rickettsial OmpB monoclonal antibody. Proteins were extracted from semipurified short- and long-form R. felis bacteria. Marker, Kaleidoscope prestained standards (Bio-Rad). Other lanes contain protein extract from uninfected ISE6 cells and short- and long-form R. felis bacteria.
Microscopic analysis.
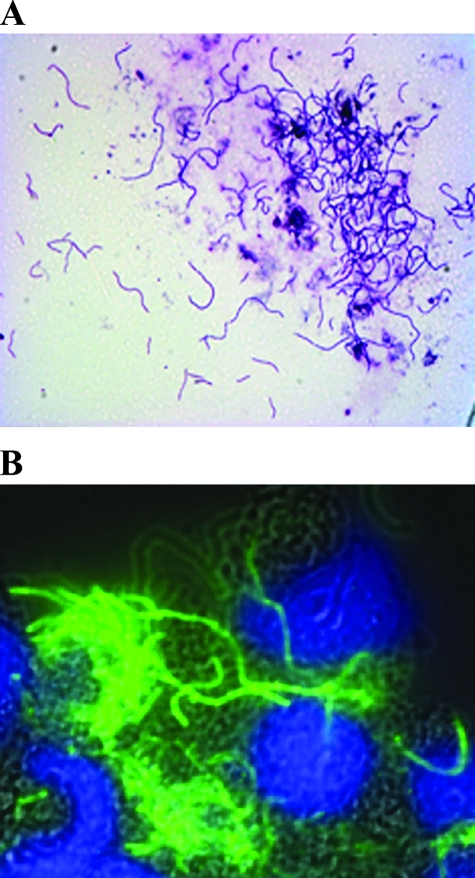
To study morphology, long-form R. felis organisms were stained by Diff-Quik and were characterized as bacillary bacteria, up to six times the length of short-form R. felis organisms, and present in the cytosol and outside the cells, with the majority of bacteria found outside the cells (Fig. 2A). While both short- and long-form R. felis induced a cytopathic effect in host cells, compared to short-form R. felis, the cytopathic effect in long-form R. felis-infected ISE6 cells was greater at 5 dpi. At an MOI of 50, long-form R. felis infection induced larger vacuoles than short-form R. felis and produced a sponge-like appearance in the host cell. Additional confirmation of the rickettsial origin of the long-form bacteria was accomplished with IFA utilizing polyclonal anti-short-form R. felis mouse serum. The antibody specifically bound the rickettsiae (Fig. 2B), while control slides lacking primary antibody were negative.
FIG. 2.
Long-form R. felis cultured in ISE6 cells. (A) Cytospin slide of long-form R. felis-infected ISE6 cells was stained using a Diff-Quik kit. (B) IFA of long-form R. felis-infected ISE6 cells using anti-R. felis mouse serum with DAPI counter-staining. A bright-field image was color merged with green fluorescence and DAPI images. The rickettsiae were visualized by green fluorescence, and the DNA was stained with DAPI. Both images are at a magnification of ×63.
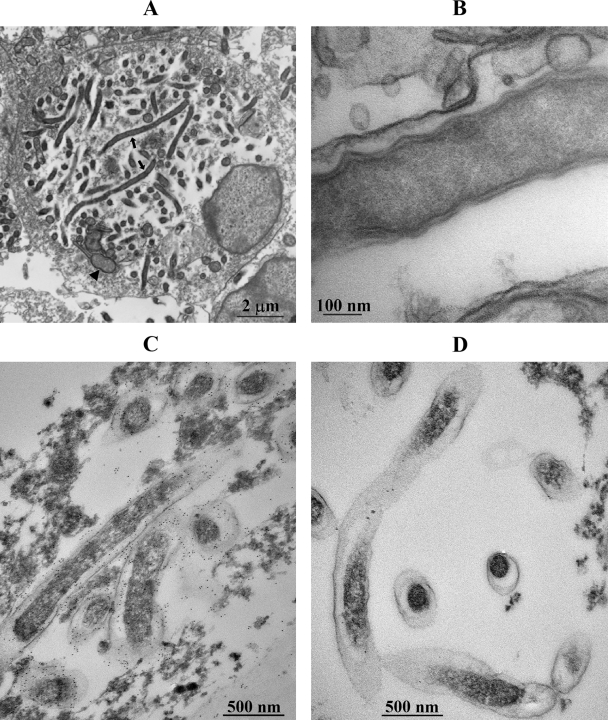
We utilized electron microscopy for further morphological characterization of long-form R. felis. In many ISE6 cells, the majority of the cytoplasm was occupied by long, filamentous rickettsiae up to 5 μm in length, free in their cytosol. Rickettsiae were usually found within electron-lucent spaces corresponding to their slime layers. Round rickettsiae 0.25 to 0.3 μm in diameter were most likely cross-sections of long-form R. felis organisms though some rickettsiae appeared to be of normal length of 0.8 to 1.0 μm. Additionally, Wolbachia-like organisms were identified in several cells (Fig. 3A). Long-form R. felis was surrounded by a typical envelope consisting of the cell wall membrane and cytoplasmic membrane separated by an unexpanded, narrow periplasmic space (Fig. 3B).
FIG. 3.
Electron micrographs of polymorphic R. felis in ISE6 cells. (A) Long-form R. felis organisms (arrows) associated with cytoplasmic clearance of ISE6 cells also infected with Wolbachia-like organisms (arrowhead). (B) Portion of long-form R. felis organisms within a cell with typical rickettsial envelope containing periplasmic space. (C) Immunolabeled long-form R. felis. (D) Secondary antibody alone; negative control.
In ultrathin sections of infected cells embedded in LR White and reacted with NIH/RML-I7198 antibody, the label was specifically localized at the surface of rickettsiae and specifically localized around the rickettsial membranes (Fig. 3C). Rickettsiae were not stained in control samples when NIH/RML-I7198 primary antibody was omitted and only secondary gold-labeled antibody was used (Fig. 3D).
Induction of rickettsial polymorphism.
The polymorphic long-form R. felis was first observed at 12 dpi at MOIs of 5 and 50; however, the number of long-form R. felis organisms was significantly increased by 19 dpi (P ≤ 0.05; Tukey's HSD test). Therefore, the proportion of polymorphic R. felis was dependent on the density of rickettsiae and the length of time in culture. Similar results were obtained using diluted (3:4) L15B growth medium with no significant difference between the two medium concentrations (data not shown).
Rickettsial infectivity and replication in ISE6 cells.
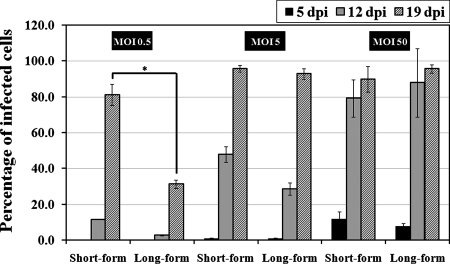
To assess the infectious nature of polymorphic R. felis for the ISE6 cell line, short- and long-form R. felis organisms immediately harvested from ISE6 cells or maintained in cell-free growth medium were inoculated at various MOIs onto previously R. felis-uninfected ISE6 cells. Cytospin preparations were stained with Diff-Quik, and the percentages of R. felis-infected ISE6 cells were counted at 5, 12, and 19 dpi. The ability to detect rickettsiae at the earliest assessment time point (5 dpi) was dependent on the MOI, with an increase in the percentage of infected cells correlating with an increased MOI and duration of infection. By 12 and 19 dpi, rickettsiae were detected in increased percentages of host cells, reaching nearly 100% infection of host cells by day 19 at MOIs of 5 and 50. A lower MOI correlated with a lower total number of cells infected by day 19; however, only at the lower MOI did we observe a significant difference between short- and long-form R. felis organisms, with the short form infecting a significantly higher percentage of ISE6 cells than the long form at the same MOI (Fig. 4).
FIG. 4.
Infectivity of short- and long-form R. felis bacteria for ISE6 cells. ISE6 cells were infected with freshly isolated short- and long-form R. felis organisms at MOIs of 0.5, 5, and 50. The infected ISE6 cells were counted at 5, 12, and 19 dpi. The results are means ± standard deviations (*, P ≤ 0.05; Tukey's studentized range HSD test).
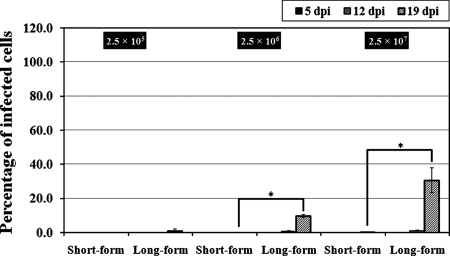
To examine environmental stability of polymorphic R. felis, short- and long-form R. felis organisms were recovered from ISE6 cells and maintained in filtered, conditioned cell-free L15B growth medium. While both short- and long-form R. felis organisms retained viability in cell-free L15B growth medium for up to 5 days, as assessed by BacLight staining, and replicated, as determined by an increase in the total number of rickettsiae, replication did not continue through 19 days of cell-free culture, despite the detection of viable rickettsiae (data not shown). To assess the infectious nature of cell-free rickettsiae, short- and long-form R. felis bacteria were maintained in cell-free L15B growth medium for 12 days and then passed onto uninfected ISE6 cells at various concentrations; infectivity and replication were assessed at 5, 12, and 19 dpi. While still viable, short-form R. felis was only infective for ISE6 cells at the highest concentration assessed, in contrast to the long-form R. felis, which was infectious at all concentrations (Fig. 5). At 19 dpi the infectivity of long-form R. felis (30.75%) was significantly higher than that of short-form R. felis (0.25%); this difference in infectivity was not observed in assays utilizing R. felis freshly isolated from ISE6 cells. The inability of short-form R. felis to infect new cells when maintained in cell-free L15B growth medium, in contrast to the long-form R. felis, which remained infectious after a prolonged extracellular period, suggests that polymorphism facilitates physiological changes in rickettsiae for survival outside the host cell.
FIG. 5.
Infection of ISE6 cells by short- and long-form R. felis bacteria. ISE6 cells were infected with short- and long-form R. felis organisms previously maintained for 12 days in cell-free L15B growth medium. The infected ISE6 cells were counted at 5, 12, and 19 dpi. The results are means ± standard errors of the means (*, P ≤ 0.05; Tukey's studentized range HSD test).
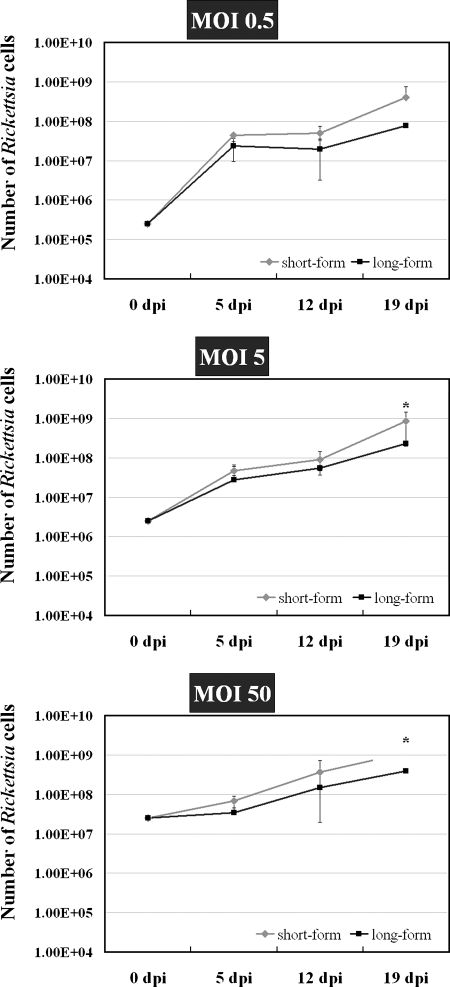
The rate of replication of short- and long-form R. felis organisms was determined by counting total numbers of viable rickettsiae during BacLight viability staining at 5, 12, and 19 dpi. Equal numbers of homogenous populations of short- or long-form R. felis were seeded onto uninfected ISE6 cells. At each time point assessed, the number of short-form rickettsiae were greater than the number of long-form R. felis organisms, with significant differences in the numbers of short-form R. felis at MOIs of 5 and 50 at 19 dpi (Fig. 6).
FIG. 6.
Growth of short- and long-form R. felis bacteria in ISE6 cells. ISE6 cells were infected with freshly isolated short- and long-form R. felis organisms at MOIs of 0.5, 5, and 50. Rickettsiae were separated from cells and counted at 5, 12, and 19 dpi. The results are means ± standard deviations (*, P ≤ 0.05; Tukey's studentized range HSD test).
DISCUSSION
In the present report, we characterize a homogenous population of polymorphic rickettsiae maintained in a tick-derived cell line. Molecular assays, including amplification of rickettsial DNA and protein characterization, confirmed that the long-form rickettsiae were R. felis organisms. Therefore, despite morphological differentiation, we did not identify a difference in the genetic composition of R. felis LSU bacteria. Although we were able to PCR amplify portions of the genus-specific 17-kDa antigen gene and the R. felis plasmid pRF, we were unable to detect plasmid pRFδ. The presence of two distinct plasmids (pRF and pRFδ) has been reported for R. felis strain URRWXCal2 and in wild-caught R. felis-infected fleas (28). Studies assessing our colonized fleas (24, 35) and the R. felis LSU isolate have been able to identify only one plasmid, pRF (3, 4).
The ultrastructural analysis of R. felis has been examined in the flea host (1, 7) without the report of long-form rickettsiae. However, the presence of elongated rickettsiae in arthropods (8, 31) suggests that this morphological differentiation is not simply an artifact of in vitro maintenance in various tissue culture models. In this study, microscopic analysis of long-form R. felis revealed the typical cell membrane consisting of inner and outer membranes separated by a peptidoglycan layer (29). Despite the absence of obvious physiological changes in the cell surface, the appearance of halted cell division could be observed in some organisms. Microbial interactions do influence the ecology of rickettsial transmission (8). The presence of Wolbachia-like organisms in some, but not all, cells is interesting. We have molecularly identified two different Wolbachia spp. in C. felis from the LSU colony (34), and the likelihood of coculture of these microorganisms with R. felis (LSU) warrants further investigation.
Although the specific mechanisms of induction of polymorphic rickettsiae were not identified, infectivity and growth studies suggest that both rickettsial density and nutrient availability contribute to the generation of morphologically distinct rickettsiae. Additionally, we identified decreased growth and infectivity in contrast to increased extracellular viability of long-form R. felis. In the current study, we did not directly correlate morphological changes with increased pathogenesis of R. felis; however, the morphologically distinct forms did demonstrate increased infectivity after prolonged extracellular phases. A study utilizing R. prowazekii described decreased infectivity of the rickettsiae after prolonged culture in cotton rat macrophage cells (15). Ultrastructural changes were not identified, and it was not clear if the cells were offered nutrients on a regular basis. In a study by Wisseman and Waddell (43), R. prowazekii on a second cycle of host cell infection underwent morphological differentiation and an increase in replication. Taken together, the data suggest that long-form rickettsiae represent an adaptive form that is more tolerant in nature, possibly contributing to transmission dynamics.
Recent isolation and partial molecular characterization of R. felis (LSU) cultivated in the tick-derived ISE6 cells demonstrated that, despite a strong flea association, R. felis will grow in alternate arthropod host cells (35). The growth of rickettsiae in atypical host cell environments has been examined (21), and the characterization of R. felis cultivated in mosquito (Aedes albopictus cell line C6/36) cell lines has been described for other R. felis isolates (13) and, more recently, in A. albopictus cell line Aa23 and Anopheles gambiae cell line Sua5B for R. felis (LSU) (38). Many rickettsial species are able to grow to large quantities in tick-derived cell lines in the absence of a cytopathic effect (21, 35). Accordingly, heavy infection by long-form R. felis appeared to have minimal effect on tick host cell survival; however, at an MOI 50 after 12 dpi there were increased cell clumping and lysis compared to short-form R. felis. Other investigators have reported this cytopathic effect for R. felis (LSU), as mosquito cell lines Aa23 and Sua5B supported fast-growth rickettsiae, resulting in clumping and cell lysis and subsequent release of rickettsiae into the extracellular environment. At 7 dpi, the number of infected cells approached 100%, coinciding with condensed and degraded nuclei of infected cells. The partially purified R. felis from the Aa23 and Sua5B cell lines also induced nuclear degradation and cell detachment in DAE100 (D. andersoni) and L929 (Mus musculus) cells (38). Cytopathology was not observed when R. felis (Marseille-URRWXCal2) was cultivated in Vero (Cercopithecus aethiops), L-929, and XTC-2 (Xenopus laevis) cell lines (36) or when R. felis (Pedreira) was maintained in mosquito cell lines (13). While the use of tick and other arthropod cell lines greatly facilitates the isolation and propagation of rickettsiae considered previously difficult to manipulate in vitro, caution should be exercised in the interpretation of the pathology associated with infection in these models.
The occurrence of morphologically distinct rickettsiae has been well described in other tick-rickettsiae systems, with the most extensive description concerning Anaplasma marginale, the causative agent of anaplasmosis in cattle. A. marginale development is closely coordinated with the tick feeding cycle (16, 17, 19) and is transmitted transstadially within the tick or intratransstadially by transfer of feeding male ticks among cattle (20). Upon infection of the tick, development of A. marginale begins when the erythrocytic stage invades the midgut epithelial cells. A reticulated form of A. marginale appears prior to the electron-dense form that survives extracellularly, and the electron-dense round form can exit and enter cells (16). Similar to observations about many spotted fever group rickettsiae and to our experience with R. felis in tick cells is a report that large numbers of A. marginale develop in ticks without injury to the host cell (16). Furthermore, rickettsial development in ticks and cell culture indicated that the development of A. marginale in various tick tissues appears to be dependent on physiological factors induced during tick feeding. Links between host cell physiology and rickettsial biology have been assessed in tick and mammalian cell culture systems, with the consistent realization that nutrient availability affects the morphology of rickettsiae.
Polymorphism has been described for a number of rickettsial species in concurrence with our data presented in this report. An early example of this was described by Gulevskaia et al. (11), who reported that polymorphic (filamentous) R. prowazekii 10 to 20 times longer than the regular form were observed at day 7 in guinea pig kidney cells without a change in medium composition. Likewise, analysis of R. prowazekii (Breinl and Madrid E strains) infection and growth in chicken embryo cells demonstrated large numbers of densely packed perinuclear rickettsiae. While inability to escape from a host cell resulted in a reduced number of spherical forms of rickettsiae, released rickettsiae that subsequently infected previously uninfected cells demonstrated morphological diversity. Similar to the R. felis in this study, elongated “spaghetti” forms with and without multiple division points were observed (43). A more recent example of rickettsial polymorphism is R. bellii (strain 369-C) identified by Philip et al. (32), which was described as a filamentous form or “long form” under undefined “suboptimal conditions of tissue culture” and measured 10 to 15 μm in length. Similarly, R. bellii (strain Ac25), recently isolated from Amblyomma ticks, demonstrated elongated forms (23), supporting the presence of polymorphism in several rickettsial species. In accordance with observations for other rickettsial species (11), we suspect that the long-form R. felis organisms in the current study are a result of unfavorable environmental conditions resulting in disrupted cell division rather than a distinct stage in a developmental cycle.
While polymorphism in Rickettsia is often observed with changes in medium composition or host cell type, it is probable that these regulations in rickettsial metabolism are critical to rickettsial survival within the arthropod host, which includes extended periods of limited nutrients. Further study of regulatory molecules in polymorphic rickettsiae will facilitate our understanding of rickettsial cell division and morphology. Likewise, the dynamic morphological changes in rickettsiae that occur when the organisms are subjected to various environmental settings, similar to conditions during transmission events, are ill defined, yet these changes are likely important to pathogen transmission. Using the complementary in vitro and in vivo R. felis hosts will allow us to decipher the exact mechanisms.
Acknowledgments
We thank Olga Borkhsenious for technical assistance with electron microscopy and Mike Kearney for his help with statistical analysis. We also thank Vsevold Popov, University of Texas Medical Branch Galveston, for helpful comments on the microscopy data and translation of papers. The monoclonal antibody RC-9C2 was provided by Lee Fuller, Fuller Laboratories.
This research was supported by the Louisiana Board of Regents (LEQSF), the National Institutes of Health (P20RR0201595), and the National Institute of Allergy and Infectious Diseases (K22AI60821 and R21AI070705).
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Adams, J. R., E. T. Schmidtmann, and A. F. Azad. 1990. Infection of colonized cat fleas, Ctenocephalides felis (Bouche), with a rickettsia-like microorganism. Am. J. Trop. Med. Hyg. 43:400-409. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. F., S. Radulovic, J. A. Higgins, B. H. Noden, and J. M. Troyer. 1997. Flea-borne rickettsioses: ecologic considerations. Emerg. Infect. Dis. 3:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldridge, G. D., N. Y. Burkhardt, R. F. Felsheim, T. J. Kurtti, and U. G. Munderloh. 2008. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl. Environ. Microbiol. 74:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldridge, G. D., N. Y. Burkhardt, R. F. Felsheim, T. J. Kurtti, and U. G. Munderloh. 2007. Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis. Appl. Environ. Microbiol. 73:4984-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blouin, E. F., and K. M. Kocan. 1998. Morphology and development of Anaplasma marginale (Rickettsiales: Anaplasmataceae) in cultured Ixodes scapularis (Acari: Ixodidae) cells. J. Med. Entomol. 35:788-797. [DOI] [PubMed] [Google Scholar]
- 6.Boostrom, A., M. S. Beier, J. A. Macaluso, K. R. Macaluso, D. Sprenger, J. Hayes, S. Radulovic, and A. F. Azad. 2002. Geographic association of Rickettsia felis-infected opossums with human murine typhus, Texas. Emerg. Infect. Dis. 8:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouyer, D. H., J. Stenos, P. Crocquet-Valdes, C. G. Moron, V. L. Popov, J. E. Zavala-Velazquez, L. D. Foil, D. R. Stothard, A. F. Azad, and D. H. Walker. 2001. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int. J. Syst. Evol. Microbiol. 51:339-347. [DOI] [PubMed] [Google Scholar]
- 8.Burgdorfer, W., S. F. Hayes, and A. J. Marvos. 1981. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii, p. 585-594. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY.
- 9.Case, J. B., B. Chomel, W. Nicholson, and J. E. Foley. 2006. Serological survey of vector-borne zoonotic pathogens in pet cats and cats from animal shelters and feral colonies. J. Feline Med. Surg. 8:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, S. A., E. R. Fischer, D. Howe, D. J. Mead, and R. A. Heinzen. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulevskaia, S. A., V. L. Popov, and V. F. Ignatovich. 1975. Current data on the polymorphism of Rickettsia prowazekii and burneti in cultured cells. Zh. Mikrobiol. Epidemiol. Immunobiol. 7:68-72. (In Russian.) [PubMed] [Google Scholar]
- 12.Heinzen, R. A., T. Hackstadt, and J. E. Samuel. 1999. Developmental biology of Coxiella burnettii. Trends Microbiol. 7:149-154. [DOI] [PubMed] [Google Scholar]
- 13.Horta, M. C., M. B. Labruna, E. L. Durigon, and T. T. Schumaker. 2006. Isolation of Rickettsia felis in the mosquito cell line C6/36. Appl. Environ. Microbiol. 72:1705-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, S., and Y. Rikihisa. 1981. Techniques for electron microscopy of rickettsiae, p. 213-240. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, Inc., New York, NY.
- 15.Kekcheeva, N. G., I. N. Kokorin, V. L. Popov, E. A. Chereshkova, N. S. Smirnova, O. A. Vovk, and E. M. Shirokova. 1992. Persistence of Rickettsia prowazekii in cotton rat macrophage cultures. Acta Virol. 36:103-110. [PubMed] [Google Scholar]
- 16.Kocan, K. M. 1986. Development of Anaplasma marginale Theiler in ixodid ticks: Coordinated development of a rickettsial organism and its tick host, p. 472-505. In J. R. Sauer and J. A. Hair (ed.), Morphology, physiology, and behavioral biology of ticks. John Wiley & Sons, New York, NY.
- 17.Kocan, K. M. 1992. Recent advances in the biology of Anaplasma spp. in Dermacentor andersoni ticks. Ann. N. Y. Acad. Sci. 653:26-32. [DOI] [PubMed] [Google Scholar]
- 18.Kocan, K. M., S. A. Ewing, D. Holbert, and J. A. Hair. 1982. Morphologic characteristics of colonies of Anaplasma marginale Theiler in midgut epithelial cells of Dermacentor andersoni Stiles. Am. J. Vet. Res. 43:586-593. [PubMed] [Google Scholar]
- 19.Kocan, K. M., W. L. Goff, D. Stiller, P. L. Claypool, W. Edwards, S. A. Ewing, J. A. Hair, and S. J. Barron. 1992. Persistence of Anaplasma marginale (Rickettsiales: Anaplasmataceae) in male Dermacentor andersoni (Acari: Ixodidae) transferred successively from infected to susceptible calves. J. Med. Entomol. 29:657-668. [DOI] [PubMed] [Google Scholar]
- 20.Kocan, K. M., D. Holbert, W. Edwards, S. A. Ewing, S. J. Barron, and J. A. Hair. 1986. Longevity of colonies of Anaplasma marginale in midgut epithelial cells of Dermacentor andersoni. Am. J. Vet. Res. 47:1657-1661. [PubMed] [Google Scholar]
- 21.Kurtti, T. J., J. A. Simser, G. D. Baldridge, A. T. Palmer, and U. G. Munderloh. 2005. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J. Invertebr. Pathol. 90:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labruna, M. B., R. C. Pacheco, L. J. Richtzenhain, and M. P. Szabo. 2007. Isolation of Rickettsia rhipicephali and Rickettsia bellii from Haemaphysalis juxtakochi ticks in the state of Sao Paulo, Brazil. Appl. Environ. Microbiol. 73:869-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labruna, M. B., T. Whitworth, M. C. Horta, D. H. Bouyer, J. W. McBride, A. Pinter, V. Popov, S. M. Gennari, and D. H. Walker. 2004. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 42:90-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macaluso, K. R., W. Pornwiroon, V. Popov, and L. D. Foil. 2008. Identification of Rickettsia felis in the salivary glands of cat fleas. Vector Borne Zoonotic Dis., in press. [DOI] [PMC free article] [PubMed]
- 25.Munderloh, U. G., S. D. Jauron, V. Fingerle, L. Leitritz, S. F. Hayes, J. M. Hautman, C. M. Nelson, B. W. Huberty, T. J. Kurtti, G. G. Ahlstrand, B. Greig, M. A. Mellencamp, and J. L. Goodman. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munderloh, U. G., and T. J. Kurtti. 1989. Formulation of medium for tick cell culture. Exp. Appl. Acarol. 7:219-229. [DOI] [PubMed] [Google Scholar]
- 27.Munderloh, U. G., and T. J. Kurtti. 1995. Cellular and molecular interrelationships between ticks and prokaryotic tick-borne pathogens. Annu. Rev. Entomol. 40:221-243. [DOI] [PubMed] [Google Scholar]
- 28.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS. Biol. 3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang, H., and H. H. Winkler. 1994. Analysis of the peptidoglycan of Rickettsia prowazekii. J. Bacteriol. 176:923-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parola, P., B. Davoust, and D. Raoult. 2005. Tick- and flea-borne rickettsial emerging zoonoses. Vet. Res. 36:469-492. [DOI] [PubMed] [Google Scholar]
- 31.Philip, R. N., and E. A. Casper. 1981. Serotypes of spotted fever group rickettsiae isolated from Dermacentor andersoni (Stiles) ticks in western Montana. Am. J. Trop. Med. Hyg. 30:230-238. [DOI] [PubMed] [Google Scholar]
- 32.Philip, R. N., E. A. Casper, R. L. Anacker, J. Cory, S. F. Hayes, W. Burgdorfer, and C. E. Yunker. 1983. Rickettsia bellii sp. nov.: a tick-borne rickettsia, widely distributed in the United States, that is distinct from the spotted fever and typhus biogroups. Int. J. Syst. Bacteriol. 33:94-106. [Google Scholar]
- 33.Popov, V. L., V. C. Han, S. M. Chen, J. S. Dumler, H. M. Feng, T. G. Andreadis, R. B. Tesh, and D. H. Walker. 1998. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 47:235-251. [DOI] [PubMed] [Google Scholar]
- 34.Pornwiroon, W., M. T. Kearney, C. Husseneder, L. D. Foil, and K. R. Macaluso. 2007. Comparative microbiota of Rickettsia felis-uninfected and -infected colonized cat fleas, Ctenocephalides felis. ISME J. 1:394-402. [DOI] [PubMed] [Google Scholar]
- 35.Pornwiroon, W., S. S. Pourciau, L. D. Foil, and K. R. Macaluso. 2006. Rickettsia felis from cat fleas: isolation and culture in a tick-derived cell line. Appl. Environ. Microbiol. 72:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raoult, D., S. B. La, M. Enea, P. E. Fournier, V. Roux, F. Fenollar, M. A. Galvao, and X. L. de. 2001. A flea-associated Rickettsia pathogenic for humans. Emerg. Infect. Dis. 7:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rikihisa, Y., and S. Ito. 1980. Localization of electron-dense tracers during entry of Rickettsia tsutsugamushi into polymorphonuclear leukocytes. Infect. Immun. 30:231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakamoto, J. M., and A. F. Azad. 2007. Propagation of arthropod-borne Rickettsia spp. in two mosquito cell lines. Appl. Environ. Microbiol. 73:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schriefer, M. E., J. B. Sacci, Jr., J. P. Taylor, J. A. Higgins, and A. F. Azad. 1994. Murine typhus: updated roles of multiple urban components and a second typhuslike rickettsia. J. Med. Entomol. 31:681-685. [DOI] [PubMed] [Google Scholar]
- 40.Simser, J. A., A. T. Palmer, U. G. Munderloh, and T. J. Kurtti. 2001. Isolation of a spotted fever group Rickettsia, Rickettsia peacockii, in a Rocky Mountain wood tick, Dermacentor andersoni, cell line. Appl. Environ. Microbiol. 67:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wedincamp, J., Jr., and L. D. Foil. 2002. Vertical transmission of Rickettsia felis in the cat flea (Ctenocephalides felis Bouche). J. Vector Ecol. 27:96-101. [PubMed] [Google Scholar]
- 42.Williams, S. G., J. B. Sacci, Jr., M. E. Schriefer, E. M. Andersen, K. K. Fujioka, F. J. Sorvillo, A. R. Barr, and A. F. Azad. 1992. Typhus and typhuslike rickettsiae associated with opossums and their fleas in Los Angeles County, California. J. Clin. Microbiol. 30:1758-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisseman, C. L., Jr., and A. D. Waddell. 1975. In vitro studies on rickettsia-host cell interactions: intracellular growth cycle of virulent and attenuated Rickettsia prowazeki in chicken embryo cells in slide chamber cultures. Infect. Immun. 11:1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]