Abstract
The abundance of potentially Microcystis aeruginosa-infectious cyanophages in freshwater was studied using g91 real-time PCR. A clear increase in cyanophage abundance was observed when M. aeruginosa numbers declined, showing that these factors were significantly negatively correlated. Furthermore, our data suggested that cyanophage dynamics may also affect shifts in microcystin-producing and non-microcystin-producing populations.
The major bloom-forming cyanobacterial species Microcystis aeruginosa forms noxious blooms in many eutrophic freshwater lakes, ponds, and reservoirs. A limited population (limited number of strains) of M. aeruginosa in the environment produces potent hepatotoxins called microcystins (7). These potent toxins in the M. aeruginosa blooms have caused many cases of animal and human poisoning (3, 8, 16).
Previously, most studies have focused on relationships among the cyanobacterial bloom dynamics and the changes in physicochemical factors (e.g., nutrient supply, light, and temperature) that influence cyanobacterial growth in the aquatic environment (28). Since the discovery that viruses are widespread in marine ecosystems (4), cyanophages that can infect cyanobacteria have been thought to be an alternative factor that may control the succession of cyanobacterial blooms (12, 14, 15, 18, 19). In addition, cyanophages can also influence the clonal composition of the host Synechococcus communities (14, 27) and could account for some of the cyanobacterial diversity observed in natural communities (22, 25, 30). Nevertheless, little is known about how freshwater cyanophages can affect the abundance and clonal composition of cyanobacterial blooms in lakes over time.
Our aim is to determine if the cyanophages have potential quantitative and qualitative effects on the M. aeruginosa communities in Lake Mikata in Japan. We performed two independent real-time PCR assays to monitor the dynamics of M. aeruginosa and its cyanophage communities. To quantify M. aeruginosa, we used the phycocyanin intergenic spacer (PC-IGS) that was previously used to examine total M. aeruginosa numbers (9, 32). A second real-time PCR assay was used to quantitatively detect potentially M. aeruginosa-infectious cyanophages using the primers targeting the viral sheath protein-encoding gene (g91) previously identified by Takashima et al. (21). To determine the effect of the cyanophages on the internal dynamics of the total M. aeruginosa communities, we examined the fluctuation in the abundance of potentially microcystin-producing M. aeruginosa populations using the real-time PCR and microcystin synthetase gene (mcyA)-specific primers (32) and monitored the relative size of the microcystin-producing subpopulation compared to the total population in relation to the cyanophage numbers using a field survey of M. aeruginosa blooms in a Japanese lake.
Water samples were collected from the surface layer (0.5 m) once per month from April to October in 2006 at a fixed point (35°33′N, 135°53′E) in Lake Mikata (Fig. 1). The cyanobacterial cells used for DNA extraction were sonicated gently and harvested by centrifugation at 14,400 × g for 10 min. DNA was extracted and purified using the previously described xanthogenate method (36). For phage DNA extraction, the samples were filtered through 0.2-μm polycarbonate filters (Advantec Toyo, Tokyo, Japan) and concentrated using ultracentrifugation at 111,000 × g for 1.5 h at 4°C; then the phage DNA was extracted as described previously (21). Each DNA extract was used as the PCR template for real-time PCR. The real-time PCR primer pairs 188F-254R, M1rF-M1rR, and SheathRTF-SheathRTR (Table 1) were used to amplify PC-IGS gene (66-bp), mcyA gene (107-bp), and g91 gene (132-bp) fragments, respectively. The mcyA primers M1rF and M1rR were designed to be M. aeruginosa specific based on a comparison of cyanobacterial mcyA sequences of M. aeruginosa strains NIES298 and NIES102, Planktothrix agardhii strain NIVA-CYA126/8, and Anabaena sp. strain 90 (DDBJ/EMBL/GenBank accession numbers AB092804, AB092805, AJ441056, and AJ536156, respectively) (32), using BLAST (1) and Clustal W (24). The PC-IGS and mcyA PCR were conducted as previously described by Yoshida et al. (32), and the g91 real-time PCR were conducted as previously described by Takashima et al. (21). Physicochemical parameters, temperature, the major dissolved inorganic nutrients, nitrate (NO3-N), orthophosphate (PO4-P), the number of cyanobacterial cells, and the total amounts of microcystins in the lake water samples were investigated as described previously (32, 33).
FIG. 1.
Sampling site in Lake Mikata (35°33′N, 135°53′E).
TABLE 1.
Primers used in this study
| Primer | Target gene | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| 188F | PC-IGS | GCT ACT TCG ACC GCG CC | 9 |
| 254R | PC-IGS | TCC TAC GGT TTA ATT GAG ACT AGC C | 9 |
| M1rF | mcyA | AGC GGT AGT CAT TGC ATC GG | 32 |
| M1rR | mcyA | GCC CTT TTT CTG AAG TCG CC | 32 |
| SheathRTF | g91 | ACA TCA GCG TTC GTT TCG G | 21 |
| SheathRTR | g91 | CAA TCT GGT TAG GTA GGT CG | 21 |
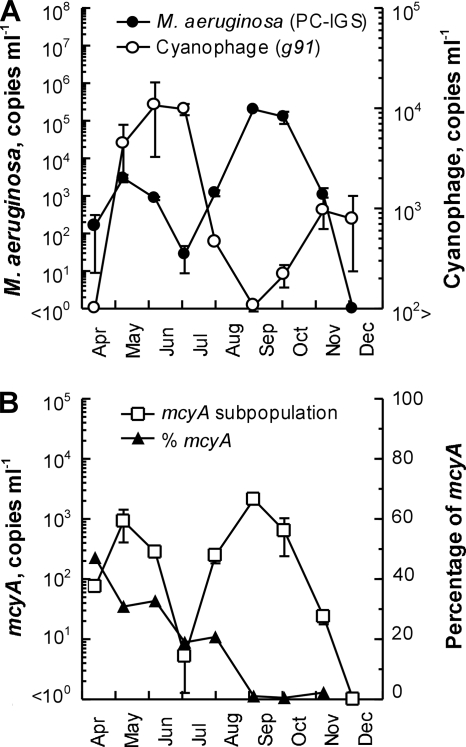
The abundance of M. aeruginosa and the cyanophages in Lake Mikata was monitored from the spring through the early winter in 2006. The PC gene copy numbers of M. aeruginosa were low at the beginning of the study (Fig. 2A), increased from April (1.6 × 102 copies ml−1) to May (3.0 × 103 copies ml−1), and then declined gradually through June (2.7 × 101 copies ml−1). During July, the number increased again and reached a maximum of 2.0 × 105 copies ml−1 on 5 September. The M. aeruginosa abundance decreased from then until November (1.0 × 103 copies ml−1), and M. aeruginosa was not detected on 5 December. According to the microscopic analysis, Planktothrix and Anabaena were dominant genera in Lake Mikata (Table 2). The numbers of M. aeruginosa cells showed a positive correlation (Spearman's r = 0.810; P = 0.015; n = 8) with the M. aeruginosa PC gene copy numbers (Table 2 and Fig. 2A). However, the M. aeruginosa PC gene copy numbers determined on all sampling dates except 4 July were 3 to >200 times higher than the cell numbers observed by microscopy. A similar case has been reported by Vaitomaa et al. (26). This could have been due to the microscopic counting method, which depends on morphological characteristics as the number of non-colony-forming M. aeruginosa cells might not be included in any cell count (33, 36). Another possible explanation is that high PC gene copy numbers detected by real-time PCR resulted from multiple genome copies in a cell. In fact, laboratory-based physiological data revealed that M. aeruginosa cells have several genome copies (maximum, 11 copies) during the transition from the logarithmic growth phase to the stationary phase (9), as observed in another cyanobacterium, Synechococcus (5, 6, 11).
FIG. 2.
(A) M. aeruginosa and cyanophage abundance in Lake Mikata from April to December 2006. The numbers of DNA copies per milliliter were determined using real-time PCR. The error bars indicate the standard deviations of three experiments. (B) mcyA subpopulation abundance and percentage of mcyA in Lake Mikata from April to December 2006. The numbers of mcyA gene copies per milliliter were determined using real-time PCR. The percentage of mcyA was determined by dividing the relative number of DNA copies in the mcyA subpopulation by the total number of copies in M. aeruginosa determined with the PC-IGS primer set.
TABLE 2.
Physical and chemical characteristics, numbers of cyanobacterial cells, and microcystin concentrations in Lake Mikata
| Sampling date | Temp (°C) | NO3-N concn (mg liter−1) | PO4-P concn (mg liter−1) | No. (%) of cells (cells ml−1) ofa:
|
Total cyanobacterium concn (cells ml−1)b | Microcystin concn (ng liter−1) | ||
|---|---|---|---|---|---|---|---|---|
| M. aeruginosa | Anabaena sp. | Planktothrix sp. | ||||||
| 13 April | 13.1 | 0.3636 | 0.0078 | 1.6 × 101 (0.3) | ND | 5.2 × 103 (99) | 5.2 × 103 | 0.15 |
| 9 May | 19.7 | 0.0032 | 0.0021 | 2.7 × 102 (0.4) | ND | 6.3 × 104 (99) | 6.3 × 104 | 3.25 |
| 6 June | 24.3 | 0.0016 | 0.0022 | 2.2 × 102 (0.05) | 2.7 × 105 (56) | 2.1 × 105 (43) | 4.8 × 105 | 1.34 |
| 4 July | 26.1 | 0.0025 | 0.0031 | 2.4 × 102 (0.07) | 1.5 × 105 (42) | 2.0 × 105 (57) | 3.5 × 105 | 0.75 |
| 1 August | 27.9 | 0.0507 | 0.0029 | 3.7 × 102 (0.2) | 2.1 × 104 (8) | 2.3 × 105 (91) | 2.5 × 105 | 3.34 |
| 5 September | 27.3 | 0.0020 | 0.0019 | 6.3 × 103 (2) | 2.6 × 105 (65) | 1.3 × 105 (33) | 4.0 × 105 | 5.03 |
| 3 October | 23.7 | 0.0232 | 0.0017 | 6.6 × 102 (1) | 4.7 × 102 (0.7) | 6.2 × 104 (98) | 6.3 × 104 | 1.86 |
| 8 November | 16.0 | 0.0030 | 0.0012 | 3.2 × 101 (0.3) | 1.0 × 104 (99) | ND | 1.0 × 104 | 0.44 |
| 4 December | 9.9 | 0.1602 | 0.0052 | ND | ND | 4.7 × 104 (100) | 4.7 × 104 | ND |
Percentages were determined as follows: number of cells of taxon/total number of cyanobacterial cells detected by microscopy × 100. ND, not detected.
The total cyanobacterium concentration is the sum of the M. aeruginosa, Anabaena, and Planktothrix concentrations.
The cyanophage g91 copy numbers detected were between 1.1 × 102 and 1.1 × 104 copies ml−1 throughout the sampling period, although the number was below the detection limit (<1.0 × 102 copies ml−1) on 13 April (Fig. 2A). The cyanophage abundance increased twice, with the first increase occurring from April (<1.0 × 102 copies ml−1) to June (1.1 × 104 copies ml−1) and the second increase occurring from September (1.1 × 102 copies ml−1) to November (9.5 × 102 copies ml−1). A clear increase in the phage abundance was observed when the host M. aeruginosa numbers declined. The cyanophage abundance was negatively correlated with the abundance of M. aeruginosa (Spearman's r = −0.857; P = 0.014; n = 7). The physicochemical factors, temperature, NO3-N, and PO4-P, which potentially controlled the growth of M. aeruginosa in the environment, were also considered in the correlation analysis (Table 2); however, we found no significant correlation (Spearman's r = 0.333, r = −0.095, and r = −0.571, respectively; n = 8). These observations suggest that the cyanophage is an important factor in determining the seasonal changes in M. aeruginosa abundance.
To evaluate the internal dynamics of M. aeruginosa blooms and to clarify the effect of the cyanophage on the community composition, the mcyA copy numbers of the potentially microcystin-producing populations were analyzed throughout the sampling period (Fig. 2B) and were found to be between 5.2 × 101 and 2.1 × 103 copies ml−1; copies were not detected only in December. The microcystin concentration in Lake Mikata (Table 2) had a statistically significant positive correlation with the copy number of the M. aeruginosa mcyA gene (Spearman's r = 0.762; P = 0.028; n = 8). The ratio of the mcyA-containing subpopulation to the total M. aeruginosa population was 0.50 to 47.1% (Fig. 2B). The relative abundance of the mcyA subpopulation in April, May, June, July, and August was greater (>18%) than that in September, October, and November (0.50 to 2.25%), and there were clear differences in the relative abundance of the mcyA subpopulation between bloom stages. The data indicate that the relative abundance of the microcystin-producing subpopulation of the species M. aeruginosa changes over time and that microcystin-producing populations were outcompeted by non-microcystin-producing populations during the summer. As mentioned above, in the summer we observed a temporal decline in the M. aeruginosa abundance when the cyanophage abundance increased, showing that temporal changes in the cyanophage numbers affected the host M. aeruginosa (Fig. 2A). Together, these data suggest that the cyanophages may have been responsible for the fact that the microcystin-producing subpopulation was outcompeted in the summer and may have influenced the seasonal shift in the composition of the different M. aeruginosa populations.
The regulation of the internal dynamics of M. aeruginosa populations is thought to be complicated by a broad range of environment conditions, including the intraspecific selective lysis caused by cyanophages. We reported previously that the dynamics of microcystin-producing and non-microcystin-producing M. aeruginosa populations showed a significant correlation with nitrate concentrations in lake water (32). In this study, our data also suggest that high NO3-N loading in the spring is a significant factor in increasing the growth of microcystin-producing M. aeruginosa populations at the beginning of the bloom (Table 2 and Fig. 2B).
The cyanophage communities in Lake Mikata determined using the g91 real-time PCR products from the samples obtained on 9 May, 6 June, 4 July, 8 November, and 4 December were sequenced after construction of the clone libraries (Table 3). Clone libraries could not be produced from four samples (13 April, 1 August, 5 September, and 3 October) because the amounts of the g91 PCR-amplified products were very small. A total of 108 sequences from five clone libraries (Table 3), which were almost identical at the amino acid level, were successfully generated (data not shown). At the nucleotide level, 10 different genotypes (designated genotypes a to j) were identified for the cyanophage isolates and also for the Lake Mikata sample. Genotype a was the genotype most frequently encountered in the Lake Mikata sample and was identical to sequences of Ma-LMM01, Ma-LMM02, Ma-LMM03, and Ma-HPM05 that specifically infect only a microcystin-producing M. aeruginosa strain (21, 34, 35). Genotypes b to j were detected only once in the PCR products from Lake Mikata. All of the genotypes had 98.9% identity to genotype a, as shown by a single base substitution at different nucleotide positions. The clonal sequence diversity of the cyanophage communities suggests that the phage particles detected by the g91 real-time PCR may be composed of different cyanophages (e.g., cyanophages having different host strain specificities) (2, 10, 13, 29, 37). A wide variety of host-phage systems in aquatic environments have been described by Sullivan et al. (17), Suttle and Chan (20), and Waterbury and Valois (27). We recently reported that M. aeruginosa populations have a high degree of genetic diversity at the intraspecies level (31), and their successional blooms revealed different dynamics during the bloom in Lake Mikata (33). We speculate there are various host-phage systems in the relationships between M. aeruginosa and its phages.
TABLE 3.
Sequence profiles from Lake Mikata clone libraries targeting the g91 genea
| Sampling date | Genotype | No. of clones detectedb |
|---|---|---|
| 9 May | a | 11 |
| 6 June | a | 25 |
| b | 1 | |
| c | 1 | |
| 4 July | a | 21 |
| d | 1 | |
| e | 1 | |
| 8 November | a | 23 |
| f | 1 | |
| g | 1 | |
| h | 1 | |
| 4 December | a | 19 |
| i | 1 | |
| j | 1 |
The PCR products from Lake Mikata samples obtained on 9 May, 6 June, 4 July, 8 November, and 4 December were sequenced.
Number of clones detected in the PCR products from Lake Mikata.
This report is the first report demonstrating that seasonal dynamics of the cyanophage community in freshwater was related to both the host M. aeruginosa abundance and shifts in the different populations (microcystin-producing and non-microcystin-producing populations). However, the results must be interpreted carefully. Our data also revealed the relatively high ratio of M. aeruginosa numbers to cyanophage numbers in late summer (Fig. 2A). This indicates that the cyanophage assemblage might have the ability to infect only a small percentage of the M. aeruginosa population present, and thus it is possible that the presence of the cyanophages results only in replacement of phage-sensitive populations by phage-insensitive populations (“killing the winner hypothesis”) (23) rather than having a quantitative impact on the M. aeruginosa abundance. To develop a better understanding of the ecological impact of cyanophages on M. aeruginosa blooms, further investigations of the multiple host-phage interactions at the population (strain) level are required.
Nucleotide sequence accession numbers.
The DDBJ/EMBL/GenBank accession numbers for the sequences reported here are AB361250 to AB361272 and AB375177 to AB375261.
Acknowledgments
This study was partially supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (JSPS Research Fellowships for Young Scientists) and by the Research Foundation of Fukui Prefecture for the Promotion of Science, Japan.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A. C., V. J. Goddard, J. Davy, D. C. Schroeder, D. G. Adams, and W. H. Wilson. 2006. Identification of a diagnostic marker to detect freshwater cyanophages of filamentous cyanobacteria. Appl. Environ. Microbiol. 72:5713-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley, V. R., W. O. Cook, A. M. Dahlem, S. B. Hooser, R. A. Lovell, and W. M. Valentine. 1989. Algae intoxication in livestock and waterfowl. Vet. Clin. N. Am. Food Anim. Pract. 5:345-361. [DOI] [PubMed] [Google Scholar]
- 4.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 5.Binder, B. J., and S. W. Chisholm. 1995. Cell cycle regulation in marine Synechococcus sp. strains. Appl. Environ. Microbiol. 61:708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder, B. J., and S. W. Chisholm. 1990. Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J. Bacteriol. 172:2313-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmichael, W. W. 1996. Toxic Microcystis and the environment, p. 1-11. In M. F. Watanabe, K.-I. Harada, W. W. Carmichael, and H. Fujiki (ed.), Toxic Microcystis. CRC Press, Boca Raton, FL.
- 8.Jochimsen, E. M., W. W. Carmichael, J. S. An, D. M. Cardo, S. T. Cookson, C. E. Holmes, M. B. Antunes, D. A. Melo Filho, T. M. Lyra, V. S. Barreto, S. M. Azevedo, and W. R. Jarvis. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 338:873-878. [DOI] [PubMed] [Google Scholar]
- 9.Kurmayer, R., and T. Kutzenberger. 2003. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl. Environ. Microbiol. 69:6723-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu, J., F. Chen, and R. E. Hodson. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl. Environ. Microbiol. 67:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann, N., and N. G. Carr. 1974. Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J. Gen. Microbiol. 83:399-405. [DOI] [PubMed] [Google Scholar]
- 12.Mann, N. H. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17-34. [DOI] [PubMed] [Google Scholar]
- 13.Marston, M. F., and J. L. Sallee. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69:4639-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mühling, M., N. J. Fuller, A. Millard, P. J. Somerfield, D. Marie, W. H. Wilson, D. J. Scanlan, A. F. Post, I. Joint, and N. H. Mann. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499-508. [DOI] [PubMed] [Google Scholar]
- 15.Okunishi, S., N. Yoshimura, H. Maeda, T. Yoshikawa, and T. Sakata. 2003. A virulent cyanophage affects the seasonal abundance of cyanobacterial picoplankton (Synechococcus sp.) in Kagoshima Bay. Microbes Environ. 18:10-15. [Google Scholar]
- 16.Pouria, S., A. Andrade, J. Barbosa, R. L. Cavalcanti, V. T. Barreto, C. J. Ward, W. Preiser, G. K. Poon, G. H. Neild, and G. A. Codd. 1998. Fatal microcystin intoxication in haemodialysis unit in Caruaru, Brazil. Lancet 352:21-26. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 18.Suttle, C. A. 2000. Cyanophage and their role in the ecology of cyanobacteria, p. 563-589. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Press, Dordrecht, The Netherlands.
- 19.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar. Ecol. Prog. Ser. 92:99-109. [Google Scholar]
- 21.Takashima, Y., T. Yoshida, M. Yoshida, Y. Shirai, Y. Tomaru, Y. Takao, S. Hiroishi, and K. Nagasaki. 2007. Development and application of quantitative detection of cyanophages phylogenetically related to cyanophage Ma-LMM01 infecting Microcystis aeruginosa in fresh water. Microbes Environ. 22:207-213. [Google Scholar]
- 22.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 23.Thingstad, T. F., and R. Lignell. 1997. A theoretical approach to the question of how trophic interactions control carbon demand, growth rate, abundance and diversity. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toledo, G., and B. Palenik. 1997. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63:4298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaitomaa, J., A. Rantala, K. Halinen, L. Rouhiainen, P. Tallberg, L. Mokelke, and K. Sivonen. 2003. Quantitative real-time PCR for determination of microcystin synthetase E copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 69:7289-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitton, B. A., and M. Potts (ed.). 2000. The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Press, Dordrecht, The Netherlands.
- 29.Wilson, W. H., I. R. Joint, N. G. Carr, and N. H. Mann. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl. Environ. Microbiol. 59:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, M., T. Yoshida, M. Satomi, Y. Takashima, N. Hosoda, and S. Hiroishi. Intra-specific phenotypic and genotypic variation in toxic cyanobacterial Microcystis strains. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 32.Yoshida, M., T. Yoshida, Y. Takashima, N. Hosoda, and S. Hiroishi. 2007. Dynamics of microcystin-producing and non-microcystin-producing Microcystis populations is correlated with nitrate concentration in a Japanese lake. FEMS Microbiol. Lett. 266:49-53. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, M., T. Yoshida, Y. Takashima, R. Kondo, and S. Hiroishi. 2005. Genetic diversity of the toxic cyanobacterium Microcystis in Lake Mikata. Environ. Toxicol. 20:229-234. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, T., K. Nagasaki, Y. Takashima, Y. Shirai, Y. Tomaru, Y. Takao, S. Sakamoto, S. Hiroishi, and H. Ogata. 2008. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J. Bacteriol. 190:1762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, T., Y. Takashima, Y. Tomaru, Y. Shirai, Y. Takao, S. Hiroishi, and K. Nagasaki. 2006. Isolation and characterization of a cyanophage infecting the toxic cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 72:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, T., Y. Yuki, S. Lei, H. Chinen, M. Yoshida, R. Kondo, and S. Hiroishi. 2003. Quantitative detection of toxic strains of the cyanobacterial genus Microcystis by competitive PCR. Microbes Environ. 18:16-23. [Google Scholar]
- 37.Zhong, Y., F. Chen, S. W. Wilhelm, L. Poorvin, and R. E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]