Abstract
Several protocols for isolation of mycobacteria from water exist, but there is no established standard method. This study compared methods of processing potable water samples for the isolation of Mycobacterium avium and Mycobacterium intracellulare using spiked sterilized water and tap water decontaminated using 0.005% cetylpyridinium chloride (CPC). Samples were concentrated by centrifugation or filtration and inoculated onto Middlebrook 7H10 and 7H11 plates and Lowenstein-Jensen slants and into mycobacterial growth indicator tubes with or without polymyxin, azlocillin, nalidixic acid, trimethoprim, and amphotericin B. The solid media were incubated at 32°C, at 35°C, and at 35°C with CO2 and read weekly. The results suggest that filtration of water for the isolation of mycobacteria is a more sensitive method for concentration than centrifugation. The addition of sodium thiosulfate may not be necessary and may reduce the yield. Middlebrook M7H10 and 7H11 were equally sensitive culture media. CPC decontamination, while effective for reducing growth of contaminants, also significantly reduces mycobacterial numbers. There was no difference at 3 weeks between the different incubation temperatures.
The isolation of mycobacteria from both environmental and treated drinking water samples was first reported in the early 1900s. However, it has only been in the last three to four decades that these environmental mycobacteria have been recognized as pathogens of human disease. Compared to Mycobacterium tuberculosis, these organisms generally have low virulence and require a host defect for the establishment of disease (e.g., disseminated disease in AIDS patients or pulmonary disease in patients with underlying structural lung disease). However, there is a subset of patients who develop pulmonary disease without an obvious immune defect or a defect that is yet to be defined. Nontuberculous mycobacteria (NTM) have been demonstrated to be present in drinking water (1, 8, 11, 12, 15, 16, 18, 26, 35), drinking water distributions systems (17, 18, 23, 32-34), hot water systems (4), spas (6), and pools (14, 19). However, several authors have failed to identify NTM in water samples, often because of unsuitable isolation techniques. Variable growth rates, specific growth requirements, and different sources of water samples (e.g., treated, surface, or natural) are all variables that affect the choice of method used for identification. Because of the slow growth of these organisms, pretreatment methods are necessary to limit bacterial and fungal overgrowth and hence detect mycobacteria. However, the pretreatment method chosen may also prevent the detection of certain species of mycobacteria and reduce the rate of positive samples and the number of colonies seen. A number of different protocols have been described (29), but a standard protocol has not yet been established.
du Moulin and Stottmeier (5) first described the use of cetylpyridinium chloride (CPC) in 1978. They added 0.04% CPC to 1-liter samples of distilled water seeded with dilutions of 5-day-old cultures of mycobacteria grown in M7H9 broth and allowed them to stand for 24 h prior to filtration, rinsing, and application of membranes to M7H10 agar plates. The plates were incubated at 37°C (with 5 to 10% CO2) for 60 days. A control group of samples were processed in the same way but without CPC treatment. The level of survival of mycobacteria in spiked specimens varied from 1 to 100%, depending on the species (Mycobacterium kansasii, 18.4%; Mycobacterium gordonae, 8.4%; Mycobacterium intracellulare, 100%; Mycobacterium fortuitum, 1.1%; and Mycobacterium bovis, 39.9%). These authors actually reported greater survival of M. intracellulare in treated samples (7,400 viable units/liter) than in untreated samples (440 viable units /liter). Schulz-Robbecke et al. (27) compared CPC, sodium hydroxide (NaOH), and formaldehyde (HCHO) for their efficacy as decontamination substances for the isolation of mycobacteria from drinking water samples. They observed the highest level of recovery of mycobacteria and the lowest contamination rates with 0.005% CPC, using both spiked samples and environmental samples. These findings were confirmed by Neumann et al. (22). Glover et al. (10) found that decontamination of tap water samples with 0.04% CPC resulted in less contamination than decontamination with 1 to 3% NaOH or 5% oxalic acid but also the highest number of samples with no growth. CPC was applied at this concentration for 24 h to sterile water seeded with Mycobacterium avium complex at a final concentration of 1.5 × 103 CFU/500 ml. This resulted in an 89% reduction in the number of viable mycobacteria; 1% NaOH and 5% oxalic acid resulted in reductions of 64 and 59%, respectively.
Le Dantec et al. (18) used membrane filtration followed by decontamination with sodium dodecyl sulfate and NaOH, adjusting the pH with 40% phosphoric acid. Using M. gordonae-spiked sterile tap water, these authors showed that this decontamination method reduced the number of mycobacteria to 1% of the original number.
Falkinham et al. (7, 8) suggested that decontamination of drinking water may not be required. In a study published in 2001 (8) these authors initially processed samples without decontamination, but if plates were overgrown, they reprocessed them using CPC. Unfortunately, it is not stated in the paper how often decontamination was necessary. Only 15% of the samples contained slowly growing mycobacteria (3% M. avium and 1% M. intracellulare), and there were 2% rapid growers.
Other variables that may affect the yield of mycobacteria from environmental water samples include the sample volume, the use of sodium thiosulfate to neutralize chlorine-based disinfectants, the method of concentration (e.g., filtration versus centrifugation), the culture medium, and the incubation temperature.
In Queensland the main mycobacterial pathogen associated with pulmonary disease is M. intracellulare, followed by M. avium, M. abscessus, and M. kansasii. It has been postulated that patients acquire disease by inhaling aerosols containing mycobacteria from environmental water sources and water outlets in their homes (20). Patients may also aspirate contaminated water as a result of swallowing disorders or severe gastroesophageal reflux disease (31).
This pilot study was undertaken to try to identify the best method of processing water samples for the isolation of mycobacteria prior to a larger environmental survey. The aim of this study was to compare different methods of processing drinking water samples for the isolation of species of mycobacteria pathogenic to humans, particularly M. intracellulare and M. avium, with regard to the concentration method (centrifugation versus filtration), culture medium (Lowenstein-Jensen [LJ] slants, Middlebrook 7H10 and 7H11 plates, mycobacterial growth indicator tubes [MGIT], and MGIT with polymyxin, azlocillin, nalidixic acid, trimethoprim, and amphotericin B [PANTA]), and incubation temperature (32°C, 35°C, and 35°C with CO2).
MATERIALS AND METHODS
M. avium ATCC 35765 and M. intracellulare ATCC 13950 were inoculated into 7H9 broth (0.5 McFarland standard, correlating to a concentration of 1.5 × 108 CFU/ml) and diluted to a concentration of 100 CFU/500 ml water.
Control samples.
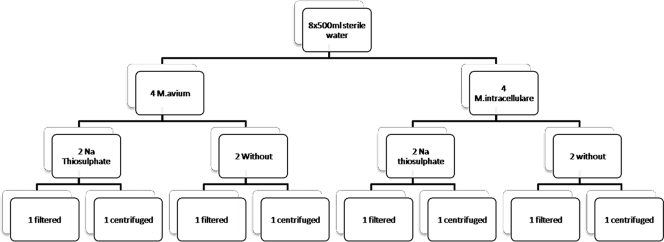
Organisms (M. avium and M. intracellulare separately) were added to eight 500-ml samples of sterile water (sterilized by filtration to preserve chlorination using a 0.2-μm MediaKap-2 hollow-fiber medium filter [Spectrum Laboratories Inc.]) to a final concentration of 100 CFU/500 ml. Sodium thiosulfate (0.5 ml of a 10% solution) was added to four of the samples (two M. avium samples and two M. intracellulare samples). One half of the samples were processed using filtration, and the other half were processed using centrifugation (Fig. 1).
FIG. 1.
Flow chart for the processing of sterile water samples.
Filtration.
Filtration was performed using 0.45-μm cellulose nitrate filters (Sartorius AG, Göttingen, Germany). The filters were then rinsed with 2 ml sterile distilled water (SDW) and macerated in 3 ml SDW. From the 3 ml of SDW, 0.1-ml aliquots were then transferred in triplicate to LJ slants and M7H10 and M7H11 plates and sealed in gas-permeable plastic bags for incubation at 32°C, at 35°C, and at 35°C with CO2. Aliquots (0.5 ml) were transferred to two MGIT tubes, one of which contained PANTA.
Centrifugation.
Four 500-ml samples (two samples containing sodium thiosulfate, one M. avium sample, and one M. intracellulare sample) were centrifuged in 250-ml sterile bottles at 5,000 × g for 20 min at 25°C. The pelleted cells were rinsed twice with phosphate-buffered saline (21). The resulting suspension was added to sterile diluent to obtain a 3-ml sample, and 0.1-ml aliquots were used to inoculate in triplicate each of the following: LJ slants and M7H10 and M7H11 plates. The plates were sealed in gas-permeable plastic bags and incubated as indicated above. Additional 0.5-ml aliquots were used to inoculate two MGIT tubes, one with PANTA and one without PANTA.
Tap water.
Tap water samples (four 500-ml samples) were collected after 2 min of flushing from a single tap in the laboratory. These tap water samples were spiked with M. avium (two samples) and M. intracellulare (two samples) to obtain a final concentration of 100 CFU/500 ml. The samples were then decontaminated with 0.005% CPC and incubated at room temperature for 30 min. Two samples (one M. avium sample and one M. intracellulare sample) were then processed using filtration, and two samples were processed using centrifugation, as described above for sterile samples.
All plates were read weekly. At 3 weeks all plates were photographed digitally and colonies were counted. Colonies from plates demonstrating growth were stained to confirm the presence of acid-fast bacilli, and morphologically different colonies were subcultured on M7H10 agar and incubated at 35°C. Subcultured organisms were then identified to the species level using multiplex PCR as described by Wilton and Cousins (36). All colonies grown from the tap water samples were treated similarly.
Data were analyzed using SPSS v12.0 for Windows 2003 (Apache Software Foundation). Tests of association were performed using Fisher's exact test for chi-square 2 × 2 tables. Statistical significance was defined as a two-sided P value of <0.05. Colony counts were also compared using the Mann-Whitney U test as the values were not normally distributed.
RESULTS
There were 88 spiked sterile water cultures and 44 spiked tap water samples. Mycobacteria grew in 83.3% of all filtered samples, compared to 12.1% for all centrifuged samples (P < 0.0001).
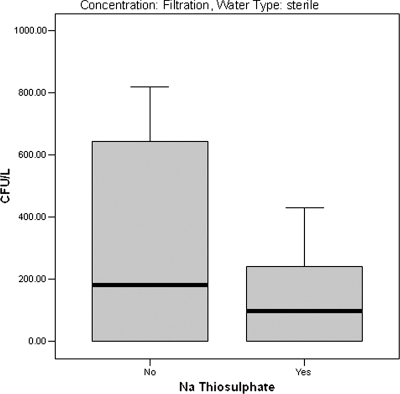
Mycobacteria grew in 52.3% of the spiked sterile samples not treated with sodium thiosulfate, compared to 43.2% for samples that were treated (P = 0.223). For filtered samples the addition of sodium thiosulfate did not affect recovery. However, for centrifuged samples, 4.5% of the treated samples were positive, compared to 22.7% of the untreated samples (P = 0.058). The colony counts were lower for filtered sterile samples to which sodium thiosulfate was added (151.7 ± 169.8 CFU/liter versus 259.0 ± 352.8 CFU/liter [means ± standard deviations]); however, the difference was not statistically significant (P = 0.178; Mann-Whitney U test, P = 0.709) (Fig. 2).
FIG. 2.
Box plot showing the median concentrations (CFU/liter) (solid bars), middle two quartiles (boxes), and ranges (error bars) for mycobacteria in spiked sterile water concentrated by filtration and processed with and without addition of sodium thiosulfate.
There was no overall difference between Middlebrook 7H10 and 7H11; 12 and 13 of 18 filtered samples, respectively, showed positive growth after 1 week. The LJ slants initially appeared to be less sensitive, but there was no difference between them and the Middlebrook media at 3 weeks (Table 1). There was no difference overall between the different incubation temperatures (Table 2).
TABLE 1.
Cultures positive for mycobacteria after concentration by filtration using different culture media after 1 and 6 weeks (n = 66)
| Culture medium | n | Week 1
|
Final culture results
|
||||
|---|---|---|---|---|---|---|---|
| No. contaminated | No. negative | No. positive | No. overgrown | No. negative | No. positive | ||
| 7H10 | 18 | 1 | 5 | 12 | 0 | 2 | 16 |
| 7H11 | 18 | 0 | 5 | 13 | 1 | 0 | 17 |
| LJ | 18 | 2 | 16 | 0 | 0 | 1 | 17 |
| MGIT | 6 | 0 | 6 | 0 | 0 | 5 | 1 |
| MGIT + PANTA | 6 | 0 | 3 | 3 | 0 | 2 | 4 |
TABLE 2.
Comparison of incubation temperatures for culture of mycobacteria in both spiked sterile and tap water samples processed by using centrifugation or filtration
| Processing method | Incubation conditions | No. contaminated | No. negative | No. positive | Total no. |
|---|---|---|---|---|---|
| Centrifugation | 32°C | 2 | 14 | 2 | 18 |
| 35°C | 2 | 14 | 2 | 18 | |
| 35°C + CO2 | 3 | 12 | 3 | 18 | |
| BACTEC | 0 | 11 | 1 | 12 | |
| Total | 7 | 51 | 8 | 66 | |
| Filtration | 32°C | 0 | 0 | 18 | 18 |
| 35°C | 0 | 0 | 18 | 18 | |
| 35°C + CO2 | 1 | 0 | 17 | 18 | |
| BACTEC | 0 | 7 | 5 | 12 | |
| Total | 1 | 7 | 58 | 66 |
For filtered samples CPC decontamination did not appear to affect the number of positive cultures at 3 weeks; 86.4% of the filtered samples treated with CPC were positive at the final reading, compared to 81.8% of the untreated samples. However, the colony counts were significantly reduced for spiked tap water samples (7.4 ± 8.5 CFU/liter [mean ± standard deviation]) compared to sterile samples (205.4 ± 262.4 CFU/liter; P = 0.0001). This equates to a mean reduction to 3.6% of the original numbers.
At 3 weeks, three samples not treated with CPC were overgrown, compared to none of the treated samples. In 9 of 88 (10.2%) spiked sterile samples contaminants grew in addition to mycobacteria, compared to 13/44 (29.5%) tap water samples (P = 0.012). The contaminants did not affect the ability to isolate mycobacteria. For the spiked sterile samples, on two of the plates there was fungal overgrowth at week 4, a week after they were photographed, and this was likely due to aerial contamination when the plates were inspected for photography. Of the remaining seven plates, four had single nonbuff colonies, two had two colonies, and one had three colonies. While the colonies were not formally identified, it is presumed that they entered the system during the processing of samples.
For a number of samples morphologically different colonies grew on Middlebrook medium plates. These colonies were subcultured and then identified to the species level using multiplex PCR, and they were found to be the same organism. Pulsed-field gel electrophoresis was not performed for these isolates. All colonies grown from the tap water samples were similarly processed. No mycobacteria other than the spiked organisms were identified in the tap water samples.
DISCUSSION
In this study we demonstrated that filtration is a more effective method than centrifugation for isolating mycobacteria from water samples. Apart from providing a far greater yield, filtration was also simpler and more time efficient. To our knowledge, there have been no direct comparative studies, but previous authors have been able to isolate mycobacteria from water samples processed using centrifugation. Perhaps it was our centrifugation technique, or alternatively, the success of previous authors may have been related to much higher concentrations of mycobacteria in the water sampled. In this study low concentrations of target organisms were used, like those that may be expected to exist in suburban, treated water supplies (3, 8, 9, 12, 18).
In the majority of reported studies the workers have processed samples with sodium thiosulfate to neutralize residual chlorine (2). It is not known whether neutralizing residual chlorine interferes with the ability to isolate mycobacteria by increasing bacterial overgrowth or whether the presence of residual chlorine reduces the yield and diversity of the species of mycobacteria subsequently isolated. As most opportunistic pathogenic NTM are relatively resistant to chlorine (18, 24, 25, 28, 30), the addition of sodium thiosulfate may not be necessary and may increase contamination rates.
The thiosulfate anion characteristically reacts with dilute acids to produce sulfur, sulfur dioxide, and water: S2O32−(aq) + 2H+(aq) → S(s) + SO2(g) + H2O(l), where aq, g, s, and l are aqueous, gas, solid, and liquid phases, respectively. Thiosulfate reduces the hypochlorite and in so doing is oxidized to sulfate. The complete reaction is 4NaClO + Na2S2O3 + 2NaOH → 4NaCl + 2Na2SO4 + H2O (13).
From our results it appears that sodium thiosulfate may have some antibacterial properties in water, perhaps due to generation of sulfur, as the contamination rates and mycobacterial colony counts were less in the treated samples. Although not statistically significant, this is an interesting observation. Importantly, it seems that for the purpose of isolating mycobacteria from water, the addition of sodium thiosulfate is unnecessary.
The addition of CPC to tap water samples spiked with M. avium and M. intracellulare resulted in survival of 3.6% of the organisms, but it did not affect the number of positive samples with the concentration of organisms used. The organisms used in our study were grown in 7H9 broth. It has been shown that antecedent growth conditions may affect susceptibility to chlorine-based disinfectants. Water-grown strains of M. avium were shown to be significantly more chlorine resistant than strains grown in medium (30). The magnitude of growth reduction that we observed may not necessarily apply to water-grown organisms from environmental or distribution system samples.
There were no differences between the temperatures tested or between the different solid media overall. However, the Middlebrook media were more sensitive at 1 week and had the advantage of quantitation of growth over LJ slants. The MGIT system has recently been introduced for the culture of clinical specimens and has not been used widely for the processing of water samples. Supplementation with PANTA is used to reduce contamination. A further study utilizing raw tap water samples (i.e., no decontamination) and the MGIT system (with PANTA to control contamination) is currently under way. The MGIT system without PANTA used in this study did not include oleic acid-albumin-dextrose-catalase enrichment, which may explain the lower yield using this system.
There have been a number of studies using different methods to isolate mycobacteria from water samples, and there is no established standard. We demonstrated that sodium thiosulfate may not be necessary and may interfere with growth. We confirmed the findings of previous authors that CPC controls contamination but also significantly reduces mycobacterial growth. While it would be appealing to process samples without decontamination, the utility of the method would depend on the origin of the samples.
This study added refinement to concentration and culture techniques for the isolation of mycobacteria from water; however, the major challenge remains the need for decontamination to reduce bacterial and fungal overgrowth. We and other workers have demonstrated that addition of CPC is effective for this purpose; however, we quantified the reduction in yield of M. intracellulare and M. avium, two of the main pathogens associated with lung disease, and found that it is significant. Given that the major environmental niche for M. intracellulare is biofilms (8) and only small numbers of this bacterium are found in water samples, the detection of low concentrations of organisms is important. Perhaps a metagenomic study may obviate the need for any decontamination and culture method, and developments in this area are awaited with interest.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Chang, C.-T., L.-Y. Wang, C.-Y. Liao, and S.-P. Huang. 2002. Identification of nontuberculous mycobacteria existing in tap water by PCR-restriction fragment length polymorphism. Appl. Environ. Microbiol. 68:3159-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 3.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.du Moulin, G., K. Stottmeier, P. Pelletier, A. Tsang, and J. Hedley-Whyte. 1988. Concentration of Mycobacterium avium by hospital hot water systems. JAMA 260:1599-1601. [DOI] [PubMed] [Google Scholar]
- 5.du Moulin, G. C., and K. D. Stottmeier. 1978. Use of cetylpyridinium chloride in the decontamination of water for culture of mycobacteria. Appl. Environ. Microbiol. 36:771-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embil, J., P. Warren, M. Yakrus, R. Stark, S. Corne, D. Forrest, and E. Hershfield. 1997. Pulmonary illness associated with exposure to Mycobacterium avium complex in hot tub water. Hypersensitivity pneumonitis or infection? Chest 111:813-816. [DOI] [PubMed] [Google Scholar]
- 7.Falkinham, J. 2004. Environmental sources of Mycobacterium avium linked to routes of exposure, p. 26-38. In S. Pedley, J. Bartram, G. Rees, A. Dufour, and J. A. Cotruvo (ed.), Pathogenic mycobacteria in water: a guide to public health consequences, monitoring and management. IWA Publishing, London, United Kingdom.
- 8.Falkinham, J., III, C. Norton, and M. Le Chavallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galassi, L., R. Donato, E. Tortoli, D. Burrini, D. Santianni, and R. Dei. 2003. Nontuberculous mycobacteria in hospital water systems: application of HPLC for identification of environmental mycobacteria. J. Water Health 1:133-139. [PubMed] [Google Scholar]
- 10.Glover, N., A. Holtzman, T. Aronson, S. Froman, O. Berlin, P. Dominguez, K. Kunkel, G. Overturf, G. Stelma, Jr., C. Smith, and M. Yakrus. 1994. The isolation and identification of Mycobacterium avium complex (MAC) recovered from Los Angeles potable water, a possible source of infection in AIDS patients. Int. J. Environ. Health Res. 4:63-72. [Google Scholar]
- 11.Goslee, S., and E. Wolinsky. 1976. Water as a source of potentially pathogenic mycobacteria. Am. Rev. Respir. Dis. 113:287-292. [DOI] [PubMed] [Google Scholar]
- 12.Hilborn, E. D., T. C. Covert, M. A. Yakrus, S. I. Harris, S. F. Donnelly, E. W. Rice, S. Toney, S. A. Bailey, and G. N. Stelma, Jr. 2006. Persistence of nontuberculous mycobacteria in a drinking water system after addition of filtration treatment. Appl. Environ. Microbiol. 72:5864-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holleman, A., and E. Wiberg. 2001. Inorganic chemistry. Academic Press, San Diego, CA.
- 14.Iivanainen, E., J. Northrup, R. D. Arbeit, M. Ristola, M.-L. Katila, and C. F. Von Reyn. 1999. Isolation of mycobacteria from indoor swimming pools in Finland. APMIS 107:193-200. [DOI] [PubMed] [Google Scholar]
- 15.Kubalek, I., and S. Komenda. 1995. Seasonal variations in the occurrence of environmental mycobacteria in potable water. APMIS 103:327-330. [DOI] [PubMed] [Google Scholar]
- 16.Kubalek, I., and J. Mysak. 1996. The prevalence of environmental mycobacteria in drinking water supply systems in a demarcated region in Czech Republic, in the period 1984-1989. Eur. J. Epidemiol. V 12:471-474. [DOI] [PubMed] [Google Scholar]
- 17.Le Dantec, C., J.-P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 68:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Dantec, C., J.-P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leoni, E., P. Legnani, M. T. Mucci, and R. Pirani. 1999. Prevalence of mycobacteria in a swimming pool environment. J. Appl. Microbiol. 87:683-688. [DOI] [PubMed] [Google Scholar]
- 20.Marras, T. K., R. J. Wallace, Jr., L. L. Koth, M. S. Stulbarg, C. T. Cowl, and C. L. Daley. 2005. Hypersensitivity pneumonitis reaction to Mycobacterium avium in household water. Chest 127:664-671. [DOI] [PubMed] [Google Scholar]
- 21.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 22.Neumann, M., R. Schulze-Robbecke, C. Hagenau, and K. Behringer. 1997. Comparison of methods for isolation of mycobacteria from water. Appl. Environ. Microbiol. 63:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norton, C. D., M. W. LeChevallier, and J. O. Falkinham III. 2004. Survival of Mycobacterium avium in a model distribution system. Water Res. 38:1457-1466. [DOI] [PubMed] [Google Scholar]
- 24.Payment, P. 1999. Poor efficacy of residual chlorine disinfectant in drinking water to inactivate water borne pathogens in distribution systems. Can. J. Microbiol. 45:709. [PubMed] [Google Scholar]
- 25.Pelletier, P. A., G. C. du Moulin, and K. D. Stottmeier. 1988. Mycobacteria in public water supplies: comparative resistance to chlorine. Microbiol. Sci. 5:147-148. [PubMed] [Google Scholar]
- 26.Pryor, M., S. Springthorpe, S. Riffard, T. Brooks, Y. Huo, G. Davis, and S. A. Sattar. 2004. Investigation of opportunistic pathogens in municipal drinking water under different supply and treatment regimes. Water Sci. Technol. 50:83-90. [PubMed] [Google Scholar]
- 27.Schulze-Robbecke, R., A. Weber, and R. Fischeder. 1991. Comparison of decontamination methods for the isolation of mycobacteria from drinking water samples. J. Microbiol. Methods 14:177-183. [Google Scholar]
- 28.Steed, K. A., and J. O. Falkinham III. 2006. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl. Environ. Microbiol. 72:4007-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stinear, T., T. Ford, and V. Vincent. 2004. Analytical methods for the detection of waterborne and environmental pathogenic mycobacteria, p. 55-73. In S. Pedley, J. Bartram, G. Rees, A. Dufour, and J. A. Cotruvo (ed.), Pathogenic mycobacteria in water: a guide to public health consequences, monitoring and management. IWA Publishing, London, United Kingdom.
- 30.Taylor, R. H., J. O. Falkinham III, C. D. Norton, and M. W. LeChevallier. 2000. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl. Environ. Microbiol. 66:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson, R. M., J. G. Armstrong, and D. F. Looke. 2007. Gastroesophageal reflux disease, acid suppression, and Mycobacterium avium complex pulmonary disease. Chest 131:1166-1172. [DOI] [PubMed] [Google Scholar]
- 32.Tobin-D'Angelo, M. J., M. A. Blass, C. del Rio, J. S. Halvosa, H. M. Blumberg, and C. R. Horsburgh, Jr. 2004. Hospital water as a source of Mycobacterium avium complex isolates in respiratory specimens. J. Infect. Dis. 189:98-104. [DOI] [PubMed] [Google Scholar]
- 33.Torvinen, E., S. Suomalainen, M. J. Lehtola, I. T. Miettinen, O. Zacheus, L. Paulin, M.-L. Katila, and P. J. Martikainen. 2004. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl. Environ. Microbiol. 70:1973-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaerewijck, M. J., G. Huys, J. C. Palomino, J. Swings, and F. Portaels. 2005. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol. Rev. 29:911-934. [DOI] [PubMed] [Google Scholar]
- 35.von Reyn, C. F., J. N. Marlow, R. D. Arbeit, T. W. Barber, and J. O. Falkinham. 1994. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137-1141. [DOI] [PubMed] [Google Scholar]
- 36.Wilton, S., and D. Cousins. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Applic. 4:269-273. [DOI] [PubMed] [Google Scholar]