Abstract
Vibrio nigripulchritudo, the etiological agent of Litopenaeus stylirostris summer syndrome, is responsible for mass mortalities of shrimp in New Caledonia. Epidemiological studies led to the suggestion that this disease is caused by an emergent group of pathogenic strains. Genomic subtractive hybridization was carried out between two isolates exhibiting low and high virulence. Our subtraction library was constituted of 521 specific fragments; 55 of these were detected in all virulent isolates from our collection (n = 32), and 13 were detected only in the isolates demonstrating the highest pathogenicity (n = 19), suggesting that they could be used as genetic markers for high virulence capacity. Interestingly, 10 of these markers are carried by a replicon of 11.2 kbp that contains sequences highly similar to those of a plasmid detected in Vibrio shilonii, a coral pathogen. The detection of this plasmid was correlated with the highest pathogenicity status of the isolates from our collection. The origin and consequence of this plasmid acquisition are discussed.
Vibrioses are important diseases threatening the sustainable development of the penaeid shrimp aquaculture industry (6, 7, 20, 21). However, little research attention has been paid to these diseases, mostly because viruses are considered the most significant diseases in crustacean aquaculture (22). As a result, solutions to control vibriosis remain scarce. The massive use of antibiotics is certainly not a sustainable control strategy, since it favors the emergence of antibiotic-resistant strains (17). Moreover, antibiotics are often banned from aquaculture ponds for commercial reasons and because of the increasing concern about residue issues. Additionally, vaccination is not possible in invertebrates, limiting the ways to reduce disease impact in shrimp culture (2). Therefore, there is a need to understand the environmental, physiological, and bacterial conditions leading to the expression of the disease in order to determine ecological or zootechnical methods that could in the end control these diseases (16). One step consists of diagnosing and quantifying the etiologic agent of the disease, both in the cultivated animal and in its environment.
It is recognized that strains belonging to the same Vibrio species can have different virulence patterns ranging from highly pathogenic (HP) strains to nonvirulent ones (8, 13, 19). Therefore, the diagnosis often needs to be infraspecific, i.e., based on epidemiologically relevant sequence polymorphisms that can be regarded as genetic markers of virulence (9, 25).
Suppressive subtractive hybridization (SSH) has been successfully and extensively used in a wide range of bacterial species to identify strain-specific genes (14, 32, 33). In comparisons of virulent and nonvirulent strains, the different genes evidenced may encode virulence factors. Moreover, identifying these strain-specific regions can help to bring to light the traces of horizontal gene transfers, which are known to provide selective advantages and to be implicated in the emergence of new pathogens (4, 10, 24).
In New Caledonia (South Pacific; 19°S, 23°W), a disease called “summer syndrome” has occurred seasonally in penaeid shrimp farms since 1997 and causes severe epizootic mortality. A multidisciplinary research program aiming at a global understanding of this disease was set up, bringing together rearing technology, pond ecosystem studies, shrimp physiology and immunology, nutrition and genetics, pathology, and bacteriology approaches (11). Epidemiological studies have revealed that this disease is a vibriosis due to HP Vibrio nigripulchritudo isolates (7, 8). To the best of our knowledge, this is the first reported disease associated with this Vibrio species. Because the New Caledonian shrimp production is also affected by another vibriosis, namely, syndrome 93, occurring during the cool season (6), the spreading of the summer syndrome to other shrimp farms would undoubtedly threaten the sustainable development of the New Caledonian shrimp industry. Preliminary studies based on a collection of V. nigripulchritudo isolates have brought to light different virulence levels, according to experimental-infection results (7). The genetic structures of 58 selected V. nigripulchritudo isolates were studied using arbitrarily primed PCR (AP-PCR) and multilocus sequence typing (MLST) (8). These two typing methods gave congruent results, revealing a clustering of HP and moderately pathogenic isolates (MP). None of the nonpathogenic (NP) isolates was present in this cluster. The hypothesis that this particular cluster of pathogenic V. nigripulchritudo isolates emerged within a shrimp farm environment has been proposed. This emergence could be linked to the recent acquisition of one or several genetic elements, leading a moderately virulent isolate to become HP.
Our study was aimed at identifying and characterizing genetic markers of V. nigripulchritudo virulence by an SSH performed between the genomes of an HP isolate and a genetically close NP isolate. In a second step, the distribution of the screened SSH fragments was studied in a selection of both virulent (either HP or MP) and NP V. nigripulchritudo isolates by macroarray. This allowed us to determine more precisely which DNA fragments were constantly associated with virulence and could possibly be part of the virulence determinants. Lastly, the discovery of a replicon detected only in HP V. nigripulchritudo isolates led to a discussion of the role of mobile genetic elements in the emergence of pathogenicity in V. nigripulchritudo.
MATERIALS AND METHODS
Bacterial strains, media, and DNA extraction.
The V. nigripulchritudo isolates used in this study have been described previously (8) and are presented in Table 1. The Vibrio shilonii strain AK1 was purchased from the Pasteur Institute collection (CIP107136T). Vibrio strains were grown in marine broth or marine agar at 30°C. Escherichia coli strains were routinely grown in Luria-Bertani medium at 37°C. All media were from Difco. When necessary, the media were supplemented with ampicillin (100 μg/ml). Total genomic DNA from Vibrio strains was prepared as described previously (30).
TABLE 1.
V. nigripulchritudo strains and field isolates used in the present study
| Strain namea | Context | Virulence for L. stylirostris |
|---|---|---|
| CIP 103195T | V. nigripulchritudo type strain | NP |
| SFn1 | Summer syndrome, moribund shrimp hemoculture | HP |
| SFn2 | Summer syndrome, moribund shrimp hemoculture | HP |
| SFn27 | Sediment pore water, diseased pond | HP |
| SFn48 | Summer syndrome, moribund shrimp hemoculture | HP |
| SFn49 | Grow-out pond water, diseased pond | HP |
| SFn105 | Grow-out pond water, diseased pond | HP |
| SFn106 | Summer syndrome, moribund shrimp hemoculture | HP |
| SFn111 | Carapace of a healthy crab (Portunus pelagicus), diseased farm | NP |
| SFn115 | Lagoon water in front of pumps, diseased farm | NP |
| SFn118 | Lagoon water in front of pumps, diseased farm | NP |
| SFn127 | Healthy shrimp hemoculture, before disease outbreak | HP |
| SFn128 | Summer syndrome, moribund shrimp hemoculture | HP |
| SFn135 | Grow-out pond water, diseased pond | HP |
| AgMn1 | NP | |
| AgMn2 | Healthy shrimp hemoculture, before disease outbreak (same animal) | NP |
| AgMn3 | NP | |
| AgMn7 | Healthy shrimp hemoculture, before disease outbreak | HP |
| AgMn8 | Summer syndrome, moribund shrimp hemoculture | HP |
| AgMn9 | Grow-out pond water, diseased pond | HP |
| AgMn10 | Summer syndrome, moribund shrimp hemoculture | HP |
| AgMn12 | Sediment pore water, diseased pond | HP |
| AgMn13 | Sediment pore water, diseased pond | HP |
| POn2 | Healthy shrimp hemoculture, healthy pond 2, healthy farm | HP |
| POn3 | Healthy shrimp hemoculture, healthy pond 3, same healthy farm | HP |
| POn4 | Healthy shrimp hemoculture, healthy pond 6, same healthy farm | NP |
| POn10 | Moribund shrimp hemoculture, no vibriosis, healthy pond 5, same healthy farm | NP |
| POn12 | Healthy shrimp hemoculture, healthy pond 4, same healthy farm | NP |
| POn13 | Healthy shrimp hemoculture, same healthy pond 4, same healthy farm | NP |
| POn19 | Healthy shrimp hemoculture, same healthy pond 4, same healthy farm | HP |
| SOn1 | Moribund shrimp hemoculture, no vibriosis | NP |
| SOn2 | Healthy shrimp hemoculture, healthy pond, healthy farm | NP |
| FTn1 | Moribund shrimp hemoculture, no vibriosis | NP |
| SBn2 | Healthy shrimp hemoculture, healthy pond, healthy farm | NP |
| Wn1 | Moribund shrimp hemoculture, opportunistic vibriosis | MP |
| Wn3 | Moribund shrimp hemoculture, opportunistic vibriosis | MP |
| Wn13 | Moribund shrimp hemoculture, opportunistic vibriosis (same animal) | MP |
| Wn14 | MP | |
| BDn1 | Healthy shrimp hemoculture, healthy pond, healthy farm | MP |
| BDn2 | Healthy shrimp hemoculture, healthy pond, healthy farm | MP |
| Fn1 | Healthy shrimp hemoculture, healthy pond, healthy farm | MP |
| Fn2 | Healthy shrimp hemoculture, healthy pond, healthy farm | NP |
| AQn1 | Healthy shrimp hemoculture, healthy pond, healthy farm | MP |
| AQn2 | Healthy shrimp hemoculture, healthy pond, healthy farm | MP |
| MT1 | Moribund shrimp hemoculture, opportunistic vibriosis, brood stock | MP |
| BLFn1 | Moribund shrimp hemoculture, opportunistic vibriosis | MP |
| BLFn2 | Moribund shrimp hemoculture, opportunistic vibriosis | MP |
| ENn1 | Healthy shrimp hemoculture, healthy brood stock | NP |
| ENn2 | Healthy shrimp hemoculture, healthy brood stock | MP |
| SVn2 | Moribund shrimp hemoculture, no vibriosis | NP |
| SVn3 | Healthy shrimp hemoculture, healthy farm | NP |
| ESn2 | Healthy shrimp hemoculture, healthy brood stock | NP |
Isolates from farms that were affected by summer syndrome are in boldface. Isolates collected during surveys specifically dedicated to the isolation of V. nigripulchritudo strains are in italics.
PCR.
Long-range PCR was performed using Herculase DNA polymerase fusion II to amplify the entire plasmid of V. nigripulchritudo or V. shilonii following the manufacturer's instructions (Stratagene). Other PCRs were performed using the Bioline Taq polymerase according to the manufacturer's instructions. Conditions for amplification were as follows: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, melting temperature minus 10°C for 30 s, and 72°C for 60 s per kbp.
SSH and macroarrays.
SSH was carried out using the PCR-Select bacterial genomic-subtraction kit (Clontech) essentially following the manufacturer's instructions. Isolate SFn1 (HP) genomic DNA was used as the tester, and SFn118 (NP) genomic DNA was used as the driver DNA. During SSH, a high annealing temperature of 63°C was used to enrich for the recovery of SFn1-specific unique sequences. The PCR products obtained from SSH, representing tester-specific sequences, were cloned into pCRII-TOPO (Invitrogen) and transformed into E. coli strain TOP10.
Recombinant clones were screened with macroarrays. Briefly, inserts were PCR amplified and spotted in duplicate onto nylon membranes (Millipore). Genomic DNAs were labeled and used as probes in hybridization experiments using the Dig labeling and detection kit according to the manufacturer's instruction (Roche diagnostics).
DNA sequencing and sequence analysis.
DNAs for sequencing were amplified using a Templi Phi amplification kit (Amersham) and sequenced on an ABI Prism 3100 genetic analyzer (Applied Biosystems) following the manufacturer's instructions. DNA sequences were blasted on public databases using the BlastX algorithm (1). Similarities with an E value smaller than 10−5 were considered significant. Sequencing and contig assembly were performed by using SEQMAN (Lasergene Software).
Plasmid extraction and characterization.
Plasmid DNA extraction trials were conducted using different commercial kits, namely, the Qiafilter Plasmid Midi kit (Qiagen), the Plasmid Midiprep kit (Sigma Aldrich), and the Wizard DNA purification system (Promega), following the instructions of the manufacturers.
Plasmids were digested with the restriction enzymes EcoRI and XhoI, size fractionated by 1% agarose electrophoresis, and analyzed by Southern blotting (30). For this, a fragment of the plasmid pSFn1 (SSH clone 16) was PCR amplified and labeled using the Dig labeling system (Roche).
Complete sequences of pSFn1 and pAK1 were obtained by shotgun sequencing. After SauIIIa partial digestion, the purified 3-kbp to 8-kbp restriction DNA fragments were ligated in a pUC18 vector predigested with BamHI (Amersham) and transformed into TOP10 competent cells (Invitrogen). open reading frame (ORF) annotation was performed using GeneMark software, and synteny analysis was performed using Arthemis software.
Nucleotide sequence accession numbers.
The DNA sequences of the plasmids pSFn1 and pAK1 have been assigned accession numbers EU156059 and EU159455, respectively.
RESULTS
Genomic subtraction between V. nigripulchritudo isolates.
An SSH was carried out between the HP isolate SFn1 and the NP isolate SFn118. In order to check the specificity of the technique, a total of 1,112 inserts from the SSH library were screened by macroarray using SFn1 or SFn118 genomic DNA as a probe. Six hundred twenty-two SFn1-specific fragments were selected, revealing 44.1% nonspecific DNA fragments. Sequencing of the SFn1-specific fragments resulted in 521 DNA sequences, 143 of which (27.4%) showed no significant matches with entries in the public database. The remaining 378 predicted ORFs showed homology to proteins described in other bacterial species, and among them, 43 (18.9%) corresponded to conserved hypothetical proteins.
Correlation between macroarrays and virulence status.
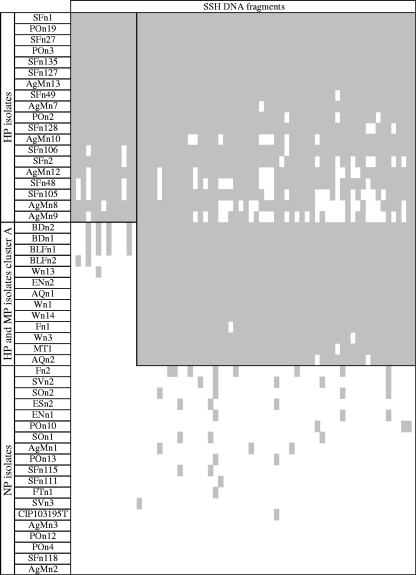
In a first set of experiments, macroarray analyses were performed using the 521 SFn1-specific DNA fragments as targets and a collection of 19 V. nigripulchritudo genomic DNAs as probes. This allowed the selection of 68 DNA fragments: 13 were found only in the DNAs of the HP isolates, whereas 55 were present in both HP and MP isolates (n = 5 and 6 isolates, respectively). The genomic DNAs of the eight NP isolates hybridized with almost none of the 68 selected DNA fragments. In a second set of experiments, the 68 fragments specific to the pathogenic isolates (both HP and MP) were spotted on membranes that were then hybridized with genomic DNAs extracted from 33 additional V. nigripulchritudo isolates. Hybridization profiles were correlated with virulence status, i.e., 13 fragments were found to be specific to the 19 HP isolates, 55 fragments were found in HP and MP isolates, and only a few fragments (n = 23) were found sporadically in NP isolates (Fig. 1).
FIG. 1.
Correlation between subtracted fragments of genomic DNA from V. nigripulchritudo SFn1 and virulence statuses; summary of macro-array results. Hybridizations were performed using the 68 SFn1-specific DNA fragments as targets and a collection of 51 V. nigripulchritudo DNAs as probes. The names and virulence statuses of the strains are indicated in the ordinate, and DNA-subtracted fragments are indicated in the abscissa in the same order as in Table 1. Positive signals are highlighted in gray.
The homology search suggested that some of the 68 putative inferred ORFs could play roles in the virulence process (Table 2). Clone 106 showed similarity to a vulnibactin outer membrane receptor precursor, clone 198 to an RTX protein or autotransporter adhesin, clone 458 to cyanobacterial toxins, and clone 486 to a capsule biosynthesis protein, CapA. Nine putative ORFs (13%) showed homology to transposase, integrase, or other proteins implicated in recombination, suggesting a role of mobile elements in the SFn1 genome specificity.
TABLE 2.
Summary of sequence analysis of clone inserts specific to pathogenic strains of V. nigripulchritudo and absent from NP strains
| Specific DNA fragments | SSH clone | GenBank accession no. | SSH DNA fragment size (bp) | Predicted protein size (bp) | Homologya | BLAST type | E value | Identity (%) | Homologue accession no. |
|---|---|---|---|---|---|---|---|---|---|
| HP strains | 16; ORF pSFn1 no. 6 | ET024018 | 459 | 990 | 13.5-kbp plasmid sequence Z2Z3, putative serine peptidase S49 (V. shilonii AK1) | n | 9E−175 | 92 | AAB65791 |
| 64 | ET023962 | 427 | 1,380 | Hypothetical protein VV0144 (V. vulnificus YJ016) | x | 6E−76 | 97 | ZP_932937 | |
| 68; ORF pSFn1 no. 7 | ET024019 | 250 | 553 | 13.5-kbp plasmid sequence Z8 | n | 7E−57 | 94 | AF009903 | |
| 104 | ET023965 | 415 | 2,691 | Hypothetical protein Neut_2547 (Nitrosomonas eutropha C71) | x | 2E−48 | 72 | ABI60750 | |
| 155; ORF pSFn1 no. 7 | ET024020 | 312 | 553 | 13.5-kbp plasmid sequence Z8 (V. shilonii AK1) | n | 5E−86 | 88 | AF009903 | |
| 191; ORF pSFn1 no. 4 | ET024021 | 323 | 1,788 | Predicted phage tail protein (V. vulnificus) | x | 2E−05 | 45 | ABB90701 | |
| 284; ORF pSFn1 no. 8 | ET024022 | 262 | 1,035 | Hypothetical protein R2601_22861 (Roseovarius sp. strain HTCC2601) | x | 5E−04 | 55 | ZP_01444696 | |
| 302; ORF pSFn1 no. 5 | ET024023 | 367 | 2,280 | Phage tail tape measure protein TP901, core region (Thiomicrospira crunogena XCL-2) | x | 4E−16 | 40 | YP_390968 | |
| 378 | ET024024 | 277 | Unknown COG | x | |||||
| 414 | ET024025 | 537 | Unknown COG | x | |||||
| 506 | ET024007 | 405 | 909 | Predicted transcriptional regulator (V. alginolyticus 12G01) | x | 2E−05 | 47 | ZP_01259885 | |
| 522; ORF pSFn1 no. 5 | ET024026 | 405 | 1,815 | Putative tail length determinant (bacteriophage K139) | x | 2E−13 | 35 | ZP_536663 | |
| 535; ORF pSFn1 no. 3 | ET024027 | 365 | 489 | Hypothetical protein pC46022_21, V. vulnificus | x | 3.00E−33 | 53 | YP_001393180 | |
| All strains of cluster A | 39 | ET023960 | 187 | Unknown COG | x | ||||
| 55 | ET023961 | 416 | 765 | tsaC, RSc2351; probable toluenesulfonate zinc-independent alcohol dehydrogenase oxidoreductase protein (Xanthobacter autotrophicus Py2) | x | 6E−15 | 63 | ZP_01196427 | |
| 73 | ET023963 | 429 | 1,080 | Putative signal peptide protein (Marinomonas sp. strain MED121) | x | 9E−42 | 59 | ZP_01077543 | |
| 86 | ET023964 | 331 | 1,227 | Probable tartrate dehydrogenase/3-isopropylmalate dehydrogenase (Rhodococcus sp. strain RHA1) | x | 1E−18 | 60 | YP_708005 | |
| 106 | ET023966 | 255 | 2,061 | Vulnibactin outer membrane receptor precursor (V. vulnificus) | x | 3E−15 | 56 | AAF28471 | |
| 116 | ET023967 | 322 | 1,143 | Hypothetical protein VP1567 (V. parahaemolyticus RIMD 2210633) | x | 1E−06 | 68 | ZP_797946 | |
| 129 | ET023968 | 344 | Unknown COG | x | |||||
| 130 | ET023969 | 366 | 906 | Ferrous iron efflux protein F (V. cholerae O1 bv. eltor strain N16961) | x | 6E−44 | 76 | ZP_232318 | |
| 135 | ET023970 | 219 | 1,038 | iSSod13, transposase (V. vulnificus YJ016) | x | 1E−28 | 94 | ZP_934531 | |
| 154 | ET023971 | 485 | 2,058 | ATPase involved in DNA repair-like protein (Shewanella frigidimarina NCIMB 400) | x | 3E−41 | 70 | YP_750769 | |
| 166 | ET023972 | 375 | Unknown COG | x | |||||
| 173 | ET023973 | 365 | Unknown COG | x | |||||
| 176 | ET023974 | 347 | 3,840 | Hypothetical protein CburD_01002029 (Coxiella burnetii Dugway 7E9-12) | x | 3E−12 | 45 | ZP_01298115 | |
| 196 | ET023975 | 389 | 1,539 | Phage integrase (Thiomicrospira crunogena XCL-2) | x | 1E−17 | 44 | YP_390599 | |
| 197 | ET023976 | 372 | 1,038 | iSSod13, transposase (V. vulnificus YJ016) | x | 7E−67 | 95 | ZP_934531 | |
| 198 | ET023977 | 336 | 8,811 | RTX protein or autotransporter adhesin (V. vulnificus CMCP6) | x | 2E−072 | 42 | ZP_761533 | |
| 205 | ET023978 | 450 | 894 | Glutamyl-tRNA synthetase (Chromobacterium violaceum ATCC 12472) | x | 5E−31 | 50 | ZP_903103 | |
| 214 | ET023979 | 355 | 5,874 | Conserved hypothetical protein, putative DNA helicase (Desulfovibrio vulgaris subsp. vulgaris DP4) | x | 7E−11 | 48 | ZP_01458409 | |
| 216 | ET023980 | 480 | 2,034 | Chain A, chondroitinase Ac Lyase (Flavobacterium heparinum) | x | 7E−09 | 24 | ICB8_A | |
| 227 | ET023981 | 414 | 498 | GCN5-related N-acetyltransferase (Psychromonas ingrahamii 37) | 7E−21 | 57 | ZP_01350703 | ||
| 732 | Hypothetical protein P3TCK_08758 (Photobacterium profundum 3TCK) | 1E−05 | 89 | P3TCK_08758 | |||||
| 260 | ET023982 | 432 | 1,071 | Transposase, IS4 (Shewanella baltica OS195) | x | 8E−17 | 73 | ZP_01432822 | |
| 269 | ET023983 | 403 | Unknown COG | x | |||||
| 273 | ET023984 | 367 | 801 | ISPsy9, transposase OrfB (Alphaproteobacterium HTCC2255) | x | 2E−33 | 68 | ZP_01448232 | |
| 278 | ET023985 | 369 | 951 | Peptide ABC transporter, permease protein (Brucella abortus bv. 1 strain 9-941) | x | 2E−24 | 45 | YP_223694 | |
| 289 | ET023986 | 449 | 1,947 | Putative epimerase/dehydratase (V. parahaemolyticus RIMD 2210633) | x | 4E−77 | 96 | ZP_796614 | |
| 293 | ET023987 | 447 | Unknown COG | x | |||||
| 318 | ET023988 | 506 | 1,194 | Putative ABC transporter (Actinobacillus actinomycetemcomitans) | x | 3E−22 | 33 | BAA82537 | |
| 320 | ET023989 | 281 | 1,080 | Binding-protein-dependent transport system inner membrane component (Psychromonas ingrahamii 37) | x | 5E−14 | 62 | ZP_01350492 | |
| 342 | ET023990 | 383 | 510 | Hypothetical protein PBPRB0091 (P. profundum SS9) | x | 3E−40 | 62 | YP_131764 | |
| 348 | ET023991 | 348 | 1,530 | Deoxyguanosinetriphosphate triphosphohydrolase (P. profundum SS9) | x | 7E−32 | 60 | YP_130718 | |
| 351 | ET023992 | 365 | 801 | ISPsy9, transposase OrfB (Alphaproteobacterium HTCC2255) | x | 2E−20 | 61 | ZP_01449847 | |
| 368 | ET023993 | 517 | 1,227 | 2-Oxoisovalerate dehydrogenase alpha subunit (Oceanicaulis alexandrii HTCC2633) | x | 6E−36 | 54 | ZP_00953146 | |
| 376 | ET023994 | 436 | 10,947 | Alpha-aminoadipyl-l-cysteinyl-d-valine synthetase (Amycolatopsis lactamdurans) | x | 6E−20 | 40 | CAA40561 | |
| 384 | ET023995 | 319 | 1,164 | Nucleotide sugar dehydrogenase (V. vulnificus) | x | 6E−42 | 96 | AAO32664 | |
| 417 | ET023996 | 333 | 1,143 | Hypothetical protein VP1567 (V. parahaemolyticus RIMD 2210633) | x | 2E−07 | 37 | ZP_797946 | |
| 424 | ET023997 | 323 | Unknown COG | x | |||||
| 430 | ET023998 | 372 | Unknown COG | x | |||||
| 431 | ET023999 | 227 | 1,155 | Putative acyl-CoA dehydrogenase (Oceanospirillum sp. strain MED92) | x | 4E−25 | 72 | ZP_01165327 | |
| 439 | ET024000 | 421 | 1,056 | Putative ATP-binding ABC transporter (Rhizobium leguminosarum bv. viciae 3841) | x | 1E−30 | 47 | CAK10505 | |
| 458 | ET024001 | 252 | 8,361 | McyA (Microcystis aeruginosa) | x | 1E−14 | 54 | BAA83992 | |
| 461 | ET024002 | 284 | 2,343 | Organic solvent tolerance protein (V. parahaemolyticus RIMD 2210633) | x | 9E−37 | 72 | BAC58602 | |
| 476 | ET024003 | 439 | 6,348 | Putative nonribosomal peptide synthetase (Erwinia carotovora subsp. atroseptica SCRI1043) | x | 3E−22 | 44 | YP_048600 | |
| 486 | ET024004 | 481 | 1,023 | Capsule biosynthesis protein CapA (Bacteroides thetaiotaomicron VPI-5482) | x | 4E−14 | 50 | ZP_810259 | |
| 490 | ET024005 | 444 | Unknown COG | x | |||||
| 505 | ET024006 | 468 | 2,022 | Methyl-accepting chemotaxis protein (Colwellia psychrerythraea 34H) | x | 3E−23 | 54 | YP_270583 | |
| 507 | ET024008 | 384 | Unknown COG | x | |||||
| 526 | ET024009 | 482 | 1,035 | PTS system N-acetylgalactosamine- specific IID component (Symbiobacterium thermophilum IAM 14863) | x | 3E−30 | 53 | YP_075076 | |
| 541 | ET024010 | 422 | 5,835 | Unknown (Pseudomonas syringae pv. syringae) | x | 8E−33 | 51 | AAK83337 | |
| 553 | ET024011 | 369 | 2,157 | Probable toxin transporter (Pseudomonas aeruginosa PAO1) | x | 4E−38 | 66 | AAG07530 | |
| 563 | ET024012 | 374 | 1,434 | Tn7-like transposition protein C (Shewanella baltica OS155) | x | 3E−34 | 60 | ZP_00584340 | |
| 566 | ET024013 | 404 | 942 | Hypothetical protein OS145_02860 (Idiomarina baltica OS145) | x | 4E−13 | 52 | ZP_01041994 | |
| 595 | ET024014 | 395 | 2,145 | Type III restriction enzyme, res subunit (Shewanella frigidimarina NCIMB 400) | x | 3E−32 | 65 | YP_750771 | |
| 617 | ET024015 | 191 | Unknown COG | x | |||||
| 618 | ET024016 | 230 | 888 | 3-Hydroxyisobutyrate dehydrogenase (Marinomonas sp. strain MED121) | x | 6E−17 | 60 | ZP_01075946 | |
| 622 | ET024017 | 390 | 1,124 | Hypothetical protein V12B01_04853 (V. splendidus 12B01) | x | 7E−32 | 79 | ZP_00990364 |
Virulence gene candidates are in boldface. COG, cluster of orthologous groups; CoA, coenzyme A.
Identification and genetic organization of the pSFn1 plasmid.
Within the subgroup of SSH fragments detected only in HP isolates, three clones (clones 16, 68, and 155) contained a partial ORF with high similarity to two genes (Z2Z3 and Z8) found in one of the plasmids evidenced in V. shilonii (Z. Pancer, A. Kushmaro, A. Toren, E. Ron, Y. Loya, and E. Rosenberg, unpublished data).
Previous experiments using the protocol described by Kado and Liu (15) failed to demonstrate the presence of a plasmid in V. nigripulchritudo isolates. Since the detection of fragments Z2Z3 and Z8 suggested the presence of a plasmid in SFn1, three additional extraction protocols were tested; only the Qiafilter Plasmid Midi kit (Qiagen) allowed purification of a replicon from this isolate.
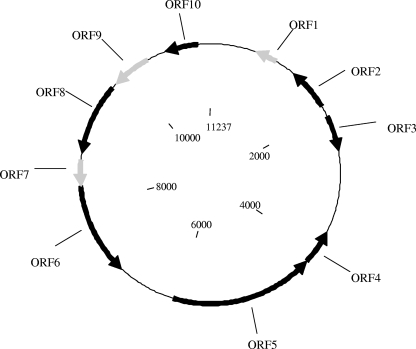
The complete sequence of the replicon named pSFn1 (11,237 bp) was obtained, and the putative ORFs were identified using GeneMark software. A graphical representation of the 10 predicted ORFs appears in Fig. 2. Their relationships to their homologues in databases are detailed in Table 2. Five ORFs showed significant similarity to known genes coding for a putative partitioning protein (ORF2), a putative phage tail protein (ORF4), a phage head-tail tape measure protein (ORF5), an S49 family serine peptidase (ORF6), and an activator of the ProP osmoprotectant transporter (ORF10). Two ORFs corresponded to conserved hypothetical proteins (ORF3 and ORF8), and three are unknown (ORF1, ORF7, and ORF9).
FIG. 2.
ORF map of the 11,237-bp plasmid pSFn1. The orientations of the putative ORFs are indicated by the orientations of the arrows; black arrows, ORFs with significant sequence similarities to the BlastX algorithm on GenBank; gray arrows, putative ORFs for which no significant similarity was found.
Among the 13 DNA fragments that were demonstrated to be present in all HP isolates, 10 were localized in the plasmid pSFn1.
Correlation between plasmid and virulence.
The successful plasmid extraction procedure was conducted with a larger panel of isolates. A single plasmid was evidenced in four of four additional HP isolates (SFn27, SFn135, POn19, and POn3). In one of eight MP (AQn1) and one of seven NP (AgMn1) isolates, one or more plasmid(s) were also purified.
Restriction fragment length polymorphism analysis of the plasmids was performed using the EcoRI or XhoI restriction enzyme and demonstrated that these four HP isolates harbor a plasmid identical or very similar to pSFn1, with three EcoRI restriction fragments (1.1, 3.4, and 6.6 kbp) and one XhoI-linearized plasmid of 11.2 kbp. In isolates AQn1 and AgMn1, the EcoRI and XhoI plasmid restriction profiles were found to be clearly distinct (data not shown). Furthermore, double digestions suggested a single larger plasmid or the existence of several plasmids. The results were confirmed by Southern blotting using SSH fragment 16 as a probe. An EcoRI-digested fragment of 3.4 kbp (in pSFn1) was evidenced in all tested HP isolates.
Comparison between pSFn1 and the plasmid pAK1 of V. shilonii.
The same plasmid extraction procedure was used successfully to purify a plasmid from V. shilonii strain AK1. In agreement with Rosenberg et al. (unpublished), more than one plasmid was obtained. Among several primers designed on the basis of the pSFn1 sequence, primers 9F and 9R, localized between ORF5 and ORF6, were successfully used to amplify by inverse PCR a fragment of 13.4 kbp, which was further sequenced to provide the complete sequence of one of the AK1 plasmids, pAK1. As for pSFn1, the putative ORFs were identified using GeneMark software.
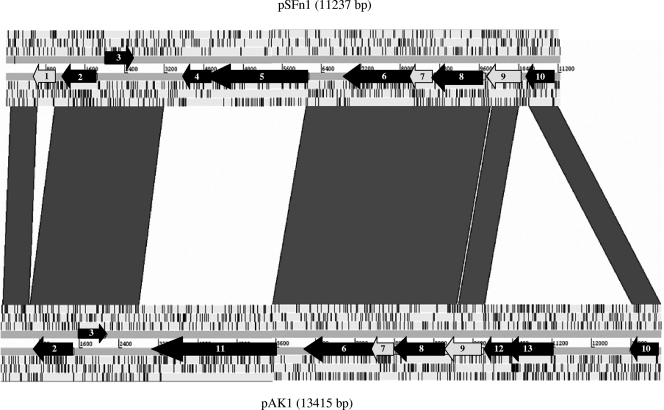
A DNA-DNA comparison between plasmids showed that 71.8% of pSFn1 was shared with pAK1, with 93% nucleotide identity for the sequences (Fig. 3). Synteny analysis revealed that five regions were significantly similar between pSFn1 and pAK1.
FIG. 3.
Linear comparison of pSFn1 and pAK1 plasmids. The ORFs of the two strands are indicated by gray arrows when no significant BLAST matches were obtained and by black arrows when significant BLAST matches were obtained. ORF2 encodes an ATPase involved in a partitioning protein; ORF4 and ORF5 encode phage tail tape measure protein TP901; ORF6 encodes an S49 family serine peptidase; ORF10 encodes an activator of a ProP osmoprotectant transporter; ORF11 encodes a putative tail length determinant; ORF3,ORF 8, ORF12, and ORF13 encode conserved hypothetical proteins; and ORF1, ORF7, and ORF9 encode unknown hypothetical proteins. The gray lines between the plasmids represent DNA-DNA similarities (BlastN matches between the two sequences with scores of >500).
DISCUSSION
Compared to human bacterial pathogens, little is known concerning Vibrio pathogenesis in marine invertebrates. The genetic diversity of Vibrio, as well as the complexity of virulence mechanisms, causes difficulties in diagnosing vibriosis. Among the approaches that can be proposed to investigate genetic markers of pathogenicity, whole-genome sequencing appears to be the most informative, as it has significantly improved our understanding of the physiology and pathogenicity of many microbes and has provided insights into the mechanisms and history of genome evolution (5). The genomes of four Vibrio species have already been sequenced: V. cholerae (12), V. parahaemolyticus (23), V. vulnificus (3), and V. fischeri (29). This makes comparative genomics an attractive approach to investigate the basis of virulence in Vibrio. However, in spite of recent progress in high-density sequencing methods, this approach is still laborious and expensive, and as a consequence, it is restricted to a limited number of strains. Furthermore, if whole-genome sequencing allows us to rapidly and extensively hypothesize virulence mechanisms based on putative virulence determinants deduced from known functions of heterologous or orthologous known sequences, the functional demonstration of their involvement in virulence still relies on mutagenesis and complementation of the candidate genes.
Subtractive hybridization methods are techniques designed to identify genomic regions that are present in one genome but absent from another (32). The application of such a method has led to the identification of genomic islands (26), mobile genetic elements (31), and plasmids (18). In comparisons of virulent and nonvirulent strains, such regions could correspond to virulence genes or regulators. Therefore, this relatively simple and cheap technique is attractive for investigating genomic variation and identifying virulence factors.
In a former study, a collection of V. nigripulchritudo isolates was studied in order to gain a better understanding of the epidemiology of the pathogen in New Caledonia (7, 8). Bacteria phenotypically identified as V. nigripulchritudo were isolated from shrimps suffering summer syndrome or from other contexts and over a wide geographic area (Table 1). Molecular typing using two different techniques, AP-PCR and MLST, were congruent and permitted the definition of a cluster that included all summer syndrome isolates from diseased animals from the two affected farms, whatever their dates of isolation. Together with these isolates, a few environmental isolates from the affected farms (sediment or pond water) suggested that they might be environmentally transmitted.
By experimental infection, the isolates of this cluster were demonstrated to be MP to HP. These data led us to hypothesize that the summer syndrome is attributable to a single pathogenic clone surviving from one year to the next in the shrimp farm environment and then redeveloping inside the grow-out system at the next farming cycle.
The correlation between the virulence phenotype and taxonomic markers suggests that virulence genes, at least in part, are carried by one of the two chromosomes. However, because genotyping studies do not allow us to distinguish HP isolates from MP isolates, more recent genetic events can be suspected to be the origin of HP isolate emergence inside this cluster. Such a recent evolution often implies mobile elements that can be tracked by the SSH approach.
In the present study, the SSH approach comparing an HP to an NP isolate allowed us to identify 13 fragments specific to the HP isolates. Among these fragments, 10 corresponded to putative ORFs harbored by a plasmid, pSFn1, evidenced only in the HP isolates and showing high similarity to a 13.5-kbp plasmid described in V. shilonii (Rosenberg et al., unpublished). This Vibrio was also putatively identified, together with V. nigripulchritudo, in corals along the coasts of Florida (27), suggesting that V. nigripulchritudo and V. shilonii can coexist in the same ecological niche.
Our hypothesis is that coral or shrimp, as well as other marine invertebrates, with the millions of resident bacteria that are concentrated in their different compartments as a niche, could be a suitable place for horizontal gene transfer. The exchange could impact different adaptive functions, leading to the capacity to colonize different ecological niches and ultimately the emergence of a specific clone. Therefore coral, shrimp, or other invertebrates could be the origins of plasmid transfer.
V. shilonii has been associated with coral-bleaching events in Oculina patagonica in the Mediterranean Sea. Many data concerning the temperature-regulated mechanism of infection, virulence mechanisms, and pathogen transmission have been obtained experimentally with the strain AK1 (28). However, no data concerning the epidemiological survey in situ are available. As a consequence, the absence of results concerning the identification of plasmids within a collection of V. shilonii prevents a discussion of the role of this plasmid in the virulence of V. shilonii.
Here, the presence of the plasmid pSFn1 has been clearly correlated with the HP status of V. nigripulchritudo isolates, suggesting that this element played a role in the emergence of HP.
One hypothesis is that the plasmid harbors one or more genes involved in bacterial virulence and could be considered a plasmid linked to virulence. However, because no ORFs annotated in the plasmid can be clearly assigned to a pathogenicity factor, genetic approaches should be developed to investigate the role of the plasmid in virulence: pSFn1 curing from an HP isolate, pSFn1 transferring to NP/MP isolates, and ORF deletion require experimental developments that are currently in progress.
Furthermore, the identification of three SSH fragments, HP specific and absent from the plasmid, suggests that several virulence determinants are chromosomally localized. Further knockout strategies should target genomic virulence markers.
Hybridization analysis using 68 SSH-derived fragments appears to be more discriminating than MLST or AP-PCR because it allows the distinction of HP from MP isolates. Our results could lead to the development of relevant tools for the diagnosis of HP isolates, for instance, a plasmid-specific PCR, thereby avoiding the need to characterize virulence by experimental infection. These operational tools will allow evaluation of the impact of this vibriosis on shrimp aquaculture in New Caledonia.
Acknowledgments
We acknowledge Dominique Ansquer for technical help, Mohammed Zouine for plasmid comparison analysis, and Collin Tinsley for critical reading of the manuscript.
This study was carried out with financial support from the South and North Provinces and the Government of New Caledonia, the Institut Français de Recherche pour l'Exploitation de la Mer (IFREMER), the Institut Pasteur (CNRS URA2171), and the Institut de Génomique Marine (contrat Ministère de la Recherche no. 0425).
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachere, E. 2000. Shrimp immunity and disease control. Aquaculture 191:3-11. [Google Scholar]
- 3.Chen, C., K. Wu, Y. Chang, C. Chang, H. Tsai, T. Liao, Y. Liu, H. Chen, A. B. Shen, J. Li, T. Su, C. Shao, C. Lee, L. Hor, and S. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11:505-510. [DOI] [PubMed] [Google Scholar]
- 5.Fraser-Liggett, C. M. 2005. Insights on biology and evolution from microbial genome sequencing. Genome Res. 151:1603-1610. [DOI] [PubMed] [Google Scholar]
- 6.Goarant, C., J. Herlin, D. Ansquer, R. Brizard, and A. L. Marteau. 2004. Vibrio penaeicida and Syndrome 93 in New Caledonian prawn farming: review and perspectives, p. 203-209. In C. Goarant, A. Herbland, Y. Harache, and C. Mugnier (ed.), Styli 2003, trente ans de crevetticulture en Nouvelle-Calédonie, Ifremer ed., Actes Colloq. Editions Ifremer, Plouzané, France. (In French with English abstract.)
- 7.Goarant, C., D. Ansquer, J. Herlin, D. Domalain, F. Imbert, and S. De Decker. 2006. “Summer syndrome” in Litopenaeus stylirostris in New Caledonia: pathology and epidemiology of the etiological agent, Vibrio nigripulchritudo. Aquaculture 253:105-113. [Google Scholar]
- 8.Goarant, C., Y. Reynaud, D. Ansquer, S. de Decker, D. Saulnier, and F. Le Roux. 2006. Molecular epidemiology of Vibrio nigripulchritudo, a pathogen of cultured penaeid shrimp (Litopenaeus stylirostris) in New Caledonia. Syst. Appl. Microbiol. 29:570-580. [DOI] [PubMed] [Google Scholar]
- 9.Goarant, C., Y. Reynaud, D. Ansquer, S. de Decker, and F. Merien. 2007. Sequence polymorphism-based identification and quantification of Vibrio nigripulchritudo at the species and subspecies level targeting an emerging pathogen for cultured shrimp in New Caledonia. J. Microbiol. Methods 70:30-38. [DOI] [PubMed] [Google Scholar]
- 10.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 11.Harache, Y., and A. Herbland. 2004. Le programme DESANS (Défi Santé Stylirostris): une démarche comparable au Défi MOREST appliquée à la filière crevette Calédonienne, p. 31-38. In C. Goarant, A. Herbland, Y. Harache, and C. Mugnier (ed.), Styli 2003, trente ans de crevetticulture en Nouvelle-Calédonie, Ifremer ed., Actes Colloq. Editions Ifremer, Plouzané, France. (In French with English abstract.)
- 12.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez, G., and J. Olmos. 2004. Molecular identification of pathogenic and nonpathogenic strains of Vibrio harveyi using PCR and RAPD. Appl. Microbiol. Biotechnol. 63:722-727. [DOI] [PubMed] [Google Scholar]
- 14.Juíz-Río, S., C. R. Osorio, V. de Lorenzo, and M. L. Lemos. 2005. Subtractive hybridization reveals a high genetic diversity in the fish pathogen Photobacterium damselae subsp. piscicida: evidence of a SXT-like element. Microbiology 151:2659-2669. [DOI] [PubMed] [Google Scholar]
- 15.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kautsky, N., P. Ronnback, M. Tedengren, and M. Troell. 2000. Ecosystem perspectives on management of disease in shrimp pond farming. Aquaculture 191:145-161. [Google Scholar]
- 17.Le, T. X., Y. Munekage, and S. Kato. 2005. Antibiotic resistance in bacteria from shrimp farming in mangrove areas. Sci. Total Environ. 349:95-105. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C., C. Amaro, E. Sanjuán, and L. Hor. 2005. Identification of DNA sequences specific for Vibrio vulnificus biotype 2 strains by suppression subtractive hybridization. Appl. Environ. Microbiol. 71:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Roux, F., M. Gay, C. Lambert, J. L. Nicolas, M. Gouy, and F. Berthe. 2004. Phylogenetic study and identification of Vibrio splendidus-related strains based on gyrB gene sequences. Dis. Aquat. Org. 58:143-150. [DOI] [PubMed] [Google Scholar]
- 20.Lightner, D. V. 1988. Vibrio disease of penaeid shrimp, p. 42-47. In C. J. Sindermann and D. V. Lightner (ed.), Disease diagnosis and control in North American marine aquaculture. Elsevier, Amsterdam, The Netherlands.
- 21.Lightner, D. V., and D. Lewis. 1975. A septicemic bacterial disease syndrome of penaeid shrimp. Mar. Fish. Rev. 37:25-28. [Google Scholar]
- 22.Lightner, D. V., and R. M. Redman. 1998. Shrimp diseases and current diagnostic methods. Aquaculture 164:201-220. [Google Scholar]
- 23.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of Vibrio cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 24.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 25.Rañoa, D. R. E., and C. T. Hedreyda. 2005. Sequence analysis of partial toxR gene from Philippine Vibrio isolates and design of toxR-targeted primers for detection. J. Gen. Appl. Microbiol. 51:343-351. [DOI] [PubMed] [Google Scholar]
- 26.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie, K. B. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322:1-14. [Google Scholar]
- 28.Rosenberg, E., and L. Falkovitz. 2004. The Vibrio shiloi/Oculina patagonica model system of coral bleaching. Annu. Rev. Microbiol. 58:143-159. [DOI] [PubMed] [Google Scholar]
- 29.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 31.Sawada, K., S. Kokeguchi, H. Hongyo, S. Sawada, M. Miyamoto, H. Maeda, F. Nishimura, S. Takashiba, and Y. Murayama. 1999. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect. Immun. 67:5621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winstanley, C. 2002. Spot the difference: applications of subtractive hybridisation to the study of bacterial pathogens. J. Med. Microbiol. 51:459-467. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y. L., C. T. Ong, and K. Y. Leung. 2000. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology 146:999-1009. [DOI] [PubMed] [Google Scholar]