Abstract
World requirements for fossil energy are expected to grow by more than 50% within the next 25 years, despite advances in alternative technologies. Since conventional production methods retrieve only about one-third of the oil in place, either large new fields or innovative strategies for recovering energy resources from existing fields are needed to meet the burgeoning demand. The anaerobic biodegradation of n-alkanes to methane gas has now been documented in a few studies, and it was speculated that this process might be useful for recovering energy from existing petroleum reservoirs. We found that residual oil entrained in a marginal sandstone reservoir core could be converted to methane, a key component of natural gas, by an oil-degrading methanogenic consortium. Methane production required inoculation, and rates ranged from 0.15 to 0.40 μmol/day/g core (or 11 to 31 μmol/day/g oil), with yields of up to 3 mmol CH4/g residual oil. Concomitant alterations in the hydrocarbon profile of the oil-bearing core revealed that alkanes were preferentially metabolized. The consortium was found to produce comparable amounts of methane in the absence or presence of sulfate as an alternate electron acceptor. Cloning and sequencing exercises revealed that the inoculum comprised sulfate-reducing, syntrophic, and fermentative bacteria acting in concert with aceticlastic and hydrogenotrophic methanogens. Collectively, the cells generated methane from a variety of petroliferous rocks. Such microbe-based methane production holds promise for producing a clean-burning and efficient form of energy from underutilized hydrocarbon-bearing resources.
The recognition that the earth's petroleum supplies are finite and dwindling has sparked the development of many non-fossil-fuel-based energy alternatives. However, even optimistic projections suggest that such energy sources will comprise less than 10% of world requirements through 2030 (10). In fact, the demand for oil is expected to increase given world population growth, the accompanying human dependence on fossil fuels for power, the existing energy infrastructure, and the use of petroleum components for manufacturing feedstocks. Currently, oil recovery techniques can extract only up to 40% of existing resources, leaving the remainder stranded in mature fields (38). Developing new technologies to recover even a fraction of such a large energy pool is of great interest to help nations reduce reliance on foreign imports and increase the value of domestic reserves (www.fossil.energy.gov/programs/oilgas/marginalwells/index.html). Here, we investigated whether it is possible to convert at least a fraction of trapped oil in marginal fields into methane gas as an alternate energy source by using a hydrocarbon-degrading methanogenic consortium.
The ability of anaerobic microorganisms to biodegrade a variety of hydrocarbons has now been well documented and widely reviewed (e.g., see reference 41). Most work has been done in the interest of environmental restoration in response to anthropogenic fuel releases, but geological evidence suggests that anaerobic hydrocarbon biodegradation has occurred for millennia. For example, many of the earth's petroleum reserves have been altered to various extents, depending on such factors as nutrient and water availability, temperature, and the requisite microorganisms, presumably in the absence of oxygen (18). Indeed, Aitken et al. (1) detected anaerobic metabolites characteristic of polycyclic aromatic hydrocarbon metabolism in 52 of 77 oil samples from across the globe. The detection of numerous anaerobes, including methanogens, associated with petroleum reservoir fluids also supports this contention (26, 28). In fact, gasses of biological origin, including methane, are believed to be primary by-products of anaerobic oil decomposition in petroliferous deposits (18, 29), where oil quality has diminished due to the preferential consumption of “light” or shorter-chain hydrocarbons. n-Alkanes, a major fraction of most crude oils, have recently been found to be biodegradable under methanogenic conditions, both as pure substrates (3, 43) and in oily mixtures (23, 32, 37), and could be a substantial source of methane in biodegraded oil resources. Indeed, Jones et al. (23) recently combined isotopic fractionation measurements and the modeling of CH4 and CO2 in variously biodegraded oilfields with observations of methanogenic oil biodegradation in laboratory incubations to link biodegraded oil patterns in reservoirs with anaerobic biodegradation processes. In many cases, though, the biogenic methane that accumulated in petroliferous deposits was likely produced long ago; thus, methane production from these resources on a human time scale may not be substantial or economically viable without stimulation or inoculation (13). Following the first report of methanogenic alkane decay (43), it was speculated that oil entrained in marginal fields could be recovered as natural gas following biodegradation by microbes (30), but no report evaluating such a prospect exists. Here, we found that oil residing in marginal reservoir samples can be converted into methane by using a methanogenic microbial consortium as an inoculant.
MATERIALS AND METHODS
Culture incubations.
The oil-utilizing methanogenic consortium was enriched from gas-condensate-contaminated subsurface sediments. The microbial population was previously shown to utilize a suite of hydrocarbons, including crude oil, under either sulfate-reducing or methanogenic conditions (8, 14, 15, 31, 37), and had been exposed to residual oil core material (34) prior to the start of the experiments described here. The consortium was maintained by repeated transfers (10%, vol/vol) into reduced, sulfate-free, bicarbonate/CO2-buffered, mineral salts freshwater medium containing resazurin, trace metals, and vitamins (14). In some cases, NaCl was added to incubations up to a 2% final concentration from a sterile anoxic stock solution to determine the salinity tolerance of the freshwater methanogenic culture. Crushed residual-oil-bearing core material was routinely used as the substrate for the consortium. The residual oil sandstone core used for most experiments was obtained from a depth of approximately 200 m from a mature oilfield undergoing secondary recovery (waterflooding) in Nowata County, OK. The sandstone sample had a residual oil saturation of approximately 30 to 40% and contained about 0.013 g of oil per gram of core. Crude oil from this oilfield (e.g., formation oil) was also separately acquired and used in some experiments. The core was stored in the laboratory at room temperature but was placed in an anaerobic glove bag (containing up to 10% H2 in N2) for at least 1 week prior to use in experiments. To prepare incubations, interior core portions were dissected with chisels and the resulting rock fragments were crushed to various extents with mortars and pestles before being dispensed into serum bottles. All implements that came in contact with the crushed core fragments were sterile, and the core processing was conducted inside an anaerobic glove bag. The amount of core material added to incubations varied depending on the experiment, but most were supplied with 5 to 20 g core material and 10 to 25 ml of medium. To obtain core material of different grain sizes, crushed core was shaken through appropriately sized sieves that had been autoclaved. Most incubations contained core material that had been crushed to a grain size of less than 1.18 mm. After we added sterile medium to incubation vessels and prior to inoculation, bottles were capped with butyl rubber stoppers, crimp sealed, and removed from the glove bag and the headspace of each was aseptically exchanged three times with 20% CO2 in N2 to remove traces of H2 and to provide a buffering capacity for the medium (35, 40). All incubations were in the dark at room temperature (21 ± 2°C). Inoculated, substrate-unamended (e.g., core-free) controls were included for every experiment conducted to account for any background levels of methane production. Uninoculated core incubations were also included as experimental controls. In some cases, 2-bromoethanesulfonic acid (BESA) was added as an inhibitor of methanogenesis to a final concentration of 25 μmol/ml of culture fluid. Five additional archived marginal field core samples were obtained from the Oklahoma Geological Survey Core Library (Norman, OK) and were prepared for incubations as described above. These core samples were originally obtained from different depths and were characterized by different levels of salinity. Details and designations of these samples are as follows: core 2 (from Pittsburg County, OK), depth of 1,768 m, 120 mM Cl−; core 3 (from Latimer County, OK), depth of 3,871 m, 22 mM Cl−; core 4 (from Creek County, OK), depth of 876 m, 51 mM Cl−; core 5 (from Pittsburg County, OK), depth of 1,966 m, 30 mM Cl−; and core 6 (from Creek County, OK), depth of 960 m, 24 mM Cl−.
Chemical analyses.
Methane was monitored regularly by injecting 0.2 ml of an incubation headspace into a Varian 3300 gas chromatograph (GC) equipped with a flame ionization detector and a packed stainless steel column (Poropak Q with 80/100 mesh; Supelco, Bellefonte, PA). The injector and column temperatures were held at 100°C, and the detector was at 125°C. Acetate concentrations in the culture fluids were determined on the same model GC and flame ionization detector, but with a Carbopack B-DA (80/120 mesh)/4% Carbowax 20-M glass column (2 m by 2 mm inside diameter; Supelco) held isothermally at 155°C, with injector and detector temperatures at 200°C. To determine the loss of residual oil components in a time course experiment, multiple replicate incubations containing inoculated or uninoculated crushed core material were established (10 g core material [<1.18 mm] in 15 ml medium, prepared as described above) and duplicates were sacrificed at various time points by extraction with four aliquots of methylene chloride. Organic layers were pooled, dried over sodium sulfate, and concentrated under N2. A standard compound (C24D50; Isotec, Miamisburg, OH) was added to the cultures prior to organic extraction in order to quantify the alkane fraction of the residual oil extracts. Quantification was performed by summing the GC peak areas of alkanes ranging from C12 to C29 and dividing this sum by the peak area of the added C24D50 to obtain an n-alkane-to-standard ratio. For such residual oil analysis, an Agilent 6890 model GC, coupled with a 5973 model mass spectrometer, was used. One microliter of each extracted oil sample was injected at 270°C, and components were separated on an HP-5MS capillary column (30 m by 0.25 mm inside diameter, 0.25-μm film thickness; Agilent Technologies, Inc.) by initially holding the oven at 45°C for 5 min, increasing the temperature at a rate of 8°C/min to 270°C, and holding at this final temperature for 15 min. In order to prepare oil-free core material, methylene chloride was used to wash residual oil components from crushed core material until hydrocarbons could not be detected by GC, and the solvent was allowed to evaporate. Such oil-free core material was used in a set of experiments designed to pinpoint the substrates in the core material that were driving methanogenesis. In some cases, formation oil was aseptically added to the solvent-extracted, core-material-containing incubations through the stoppers using sterile N2-flushed syringes. Sulfate concentrations were measured using ion chromatography as described previously (8).
DNA analyses.
Two milliliters of culture fluid from the consortium was added to microfuge tubes containing sterile silica beads and centrifuged to pellet the cells, and the supernatant was removed (31). Commercially available DNA extraction kits were used to extract DNA from the cell pellets; each protocol combined mechanical (e.g., bead-beating) and chemical means to lyse the cells. Controls for sterility included processing one or two reagent controls to which no cells were added. Aliquots from reagent control tubes were used as the template DNA in the same set of PCRs used to create the clone libraries; however, no visible PCR products were formed. Following DNA extraction of the oil-degrading methanogenic consortium, three 16S rRNA gene libraries (two eubacterial and one archaeal) and three mcrA (methyl coenzyme M reductase) libraries were constructed. For the first eubacterial library (library 1) (clone series “E”), cells were collected on 26 May 2005 from two consortium samples (e.g., L2 and L4), extracted separately using the FastDNA spin kit for soil (Quantum Biotechnologies, Inc., Carlsbad, CA), and amplified separately. Nearly full-length 16S rRNA gene sequences (Escherichia coli positions 8 to 1492) were obtained from DNA purified from cells by amplification with primers targeting conserved regions and the cycling conditions described previously by Herrick et al. (19). Primers 27f and 907r (22) were used to create the PCR products for cloning eubacterial library 2 (clone series “lg1”) from the L2 and L4 consortium samples collected on 8 September 2005 using the same PCR cycling conditions as those used for library 1. After an initial bead-beating step identical to that used for the May samples, DNA extraction of the September 2005 samples followed the protocol for the QIAamp DNA stool mini kit (Qiagen, Valencia, CA). Primers ARC333 and 958r and the PCR amplification conditions detailed previously by Struchtemeyer et al. (33) were used to obtain archaeal 16S rRNA gene sequences from the same template DNA as that used for eubacterial library 1 and from the L2 resampled on 27 April 2007. DNA was extracted from the 2007 sample with the MO BIO PowerSoil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA), which has an initial bead-beating step. The mcrA libraries were created by using primers ME1 and ME2 (17) to amplify DNA extracted from the L2 sample collected on 26 May 2005 and resampled on 27 April 2007. Primers ME1 and ME2 were also used to amplify mcrA sequences after enrichment under conditions selective for methanogens utilizing H2/CO2. The PCR products were cloned into the TOPO-TA vector (Invitrogen, Carlsbad, CA). A preliminary screening of the eubacterial and archaeal 16S rRNA gene diversity was performed by randomly selecting a few white colonies from eubacterial library 1 (for L2, three colonies, and for L4, four colonies) and the archaeal 16S rRNA gene library (for L2, three colonies; for L4, four colonies; and for the L2-2007 sample, 11 colonies). Cloned DNA was directly amplified from transformed cells by using flanking M13 vector sequences and purified by spin filter centrifugation (Montage PCR purification units; Thermo Fisher Scientific). Sequencing was performed at the University of Oklahoma DNA Sequencing Facility on an ABI model 377 automated sequencer. The amplification primers and two internal primers (704f and 907r [22]) were employed to sequence eubacterial library 1. White colonies from eubacterial library 2 (for L2, 60 colonies, and for L4, 36 colonies) and the mcrA (for the L2-May 2005 sample, 12 colonies, and for the L2-April 2007 resample, 12 colonies) libraries were transferred with sterile toothpicks into 96-well microtiter plates containing 200 μl tryptone-yeast extract-glycerol broth with ampicillin (9), grown overnight at 37°C, and stored at −85°C until DNA isolation and sequencing could be performed at the Advanced Center for Genome Technology (Norman, OK), as described previously (9). For all libraries, initial phylogenetic assignments were made following BLASTN searches (Basic Local Alignment Search Tool [2]). Sequencher (Windows version 4.2; Gene Codes Corp., Ann Arbor, MI) was used to trim vector regions from the cloned sequences and to examine each cloned sequence for the presence of universally conserved regions (e.g., primer regions). Chimera-Check (Ribosomal Database Project [27]), Bellerophon (20), and Pintail (4) were used to check for putative chimeric sequences that were then removed from the data set. Many of the clones from eubacterial library 2 and the mcrA library had poor growth; the numbers of full-length, nonchimeric sequences for eubacterial library 2 were as follows: for L2, 33 sequences; for L4, 21 sequences; for the mcrA library May 2005 sample, 1 sequence; and for the April 2007 sample, 2 sequences. Sequences with greater than 97% similarity were grouped into operational taxonomic units (OTUs), and one sequence was chosen to represent each OTU used for phylogenetic tree construction. Phylogenetic placement of 16S sequences was further examined using the Classifier program (Ribosomal Database Project [27]). Sequences (from the BLASTN search) that most closely matched the sequences from the clones and sequences of selected outgroup strains were trimmed to a common region of approximately 800 bp, aligned using ClustalX (version 1.81) (36), and corrected manually as needed. An evolutionary distance tree was constructed using the neighbor-joining algorithm, and 1,000 bootstrap replicates were performed to estimate the support for each branch (11).
RESULTS
Methanogenic oil decay in residual oil core.
An anaerobic consortium originally enriched on crude oil was found to produce enhanced levels of methane when transferred into a medium containing crushed residual-oil-bearing core material relative to the levels in the controls. Initial rates of methanogenesis ranged from 0.27 to 0.42 μmol CH4/day/g of core. With repeated transfers, methane production from the residual-oil-bearing core incubations ensued at rates ranging from 0.15 to 0.25 μmol CH4/day/g crushed core material (Fig. 1, line 6). The presence of crushed core material in the incubations was important. When the inoculum was added to medium containing only the same amount of formation oil (e.g., ∼0.06 g of oil, the amount present in 5 g of core), methane production was significantly diminished (Fig. 1, line 3), suggesting that the core itself was providing a solid surface for the inoculum or perhaps some unidentified nutrient(s). No difference in methanogenesis was observed when the added core material was crushed to grain sizes ranging from <149 μm to >1.18 mm, suggesting that the culture could access the residual oil under these conditions (data not shown).
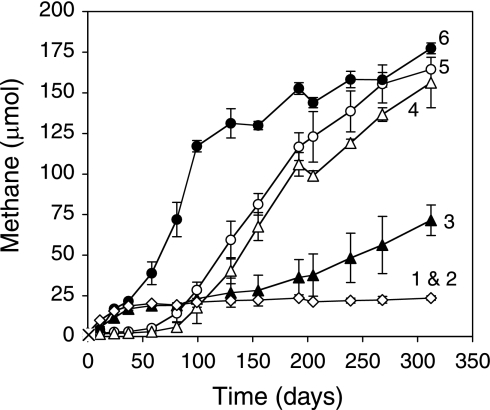
FIG. 1.
Methane production from inoculated, crushed residual-oil-bearing core material incubated under different conditions as follows: (lines 1 and 2 [overlapping data]) inoculum in medium only and inoculum in medium with solvent-extracted core, (line 3) inoculum in medium with formation oil (0.06 g), (line 4) inoculum in medium with solvent-extracted core plus formation oil (0.06 g) plus 1 mM sulfate, (line 5) inoculum in medium with solvent-extracted core plus formation oil (0.06 g), and (line 6) inoculum in medium with whole core (no solvent extraction). These incubations contained 5 g core material and 10 ml medium. Error bars represent 1 standard deviation of the mean of triplicate incubations.
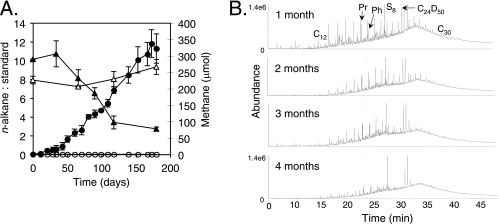
No methane was ever produced in uninoculated core-containing incubations, attesting to the absence of requisite endogenous core microflora and to the requirement for the hydrocarbon-degrading consortium. Further, only low levels of methane were formed in incubations containing the inoculum alone (Fig. 1, line 1), suggesting that hydrocarbons within the core material were biodegrading and being converted to methane. Three lines of evidence support this notion. First, when inoculated incubations were supplied with 5, 10, or 20 g of crushed core, the extent of methane production over a 230-day incubation was roughly proportional to the amount of oil-bearing core material added, with 204, 376, or 744 μmol CH4 produced, respectively (rates approximated 0.16 μmol CH4/day/g core, similar to the rate observed for Fig. 1, line 6). Given the residual oil saturation of 0.013 g oil per gram of the sandstone core, these results equated to methane yields of 3.14, 2.90, and 2.86 mmol CH4 per gram of residual oil. Second, when crushed core material was solvent extracted to remove residual oil and then inoculated, methane production mirrored that of the control incubations containing inoculum alone (Fig. 1, lines 1 and 2), showing that the hydrocarbon substrates were necessary for the observed levels of methanogenesis. When formation oil was added to solvent-extracted, oil-free core material and inoculated, comparable methane formation was observed relative to the incubations that received unextracted crushed core material (e.g., hydrocarbons were not removed) following a short lag period (Fig. 1, line 5). Third, over a 4-month incubation period, the n-alkane-to-standard ratio of alkanes ranging from C12 to C29 decreased from 10.1 to 3.4, showing that approximately two-thirds of the n-alkane fraction of the residual oil in the sandstone was removed relative to uninoculated controls as methane was concomitantly produced over time (Fig. 2A). Gas chromatographic evidence revealed that the n-alkane profile was progressively altered during 4 months of incubation relative to branched alkanes, such as pristane and phytane (Fig. 2B). Analysis of oil components after 6 months showed no further alkane depletion, although methane production continued (Fig. 2A). This result suggests that other residual oil fractions were also subject to biodegradation, as was previously shown when the microbial population was initially enriched on crude oil (37). When BESA was added to crushed core-containing incubations as an inhibitor of methanogenesis, methane was not produced, but acetate (4.89 ± 0.64 mM) accumulated. In contrast, acetate did not accumulate in any of the methane-producing incubations (data not shown).
FIG. 2.
(A) n-Alkane consumption and methane production in crushed residual-oil-bearing core incubations. Closed triangles, n-alkane-to-standard peak area ratio in inoculated residual oil cultures; open triangles, n-alkane-to-standard peak area ratio in uninoculated and sterile controls; closed circles, methane production in inoculated residual oil cultures; open circles, methane production in uninoculated residual oil incubations. (B) Total ion chromatograms showing the consumption of n-alkanes over 4 months when crushed residual-oil-bearing core material was inoculated with a methanogenic, hydrocarbon-degrading consortium. Pr, pristane; Ph, phytane; C24D50, extraction standard; S8, sulfur.
Methanogenesis in the presence of sulfate or NaCl.
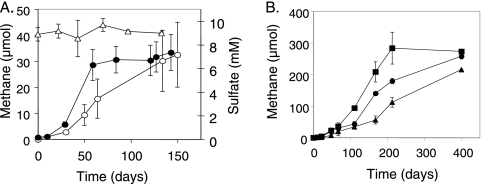
The sandstone core routinely yielded sulfate (<4 mM) to the medium, and we repeatedly observed that this alternate electron acceptor was simultaneously depleted as methane was produced. For example, in a time course experiment (Fig. 2A), approximately 4 mM sulfate was consumed in the inoculated incubations over a 4-month period as methanogenesis ensued (no sulfate was consumed in the uninoculated controls). In fact, we found in all of our incubations that neither the rate nor extent of methanogenesis was impeded by sulfate. For example, when solvent-extracted crushed core material was reamended with formation crude oil and 1 mM sulfate and inoculated, methane production paralleled that of the sulfate-unamended incubations (Fig. 1, compare lines 4 and 5). In a separate experiment, the onset of methanogenesis was slightly delayed when approximately 10 mM sulfate was exogenously provided to the incubations, but the extent of methane production after 130 days was equivalent under the two conditions and no sulfate was consumed during this time in the sulfate-amended incubations (Fig. 3A). In addition to its ability to carry out methanogenesis in the presence of sulfate, the consortium was also able to convert residual oil components to methane gas when the salinity levels were as high as 2%, despite its freshwater origin (Fig. 3B), suggesting its potential utility in more brackish reservoirs.
FIG. 3.
(A) Methane production from inoculated crushed residual-oil-bearing sandstone core incubations in the absence (closed circles) or presence (open circles) of exogenously added sulfate. Open triangles represent the sulfate concentrations in the sulfate-amended incubations. (B) Methane production from inoculated residual-oil-bearing sandstone core incubations at different salinity levels. Squares, no added NaCl; circles, 1% NaCl; triangles, 2% NaCl. Error bars represent 1 standard deviation of the mean of triplicate incubations.
Molecular analysis of the methanogenic consortium.
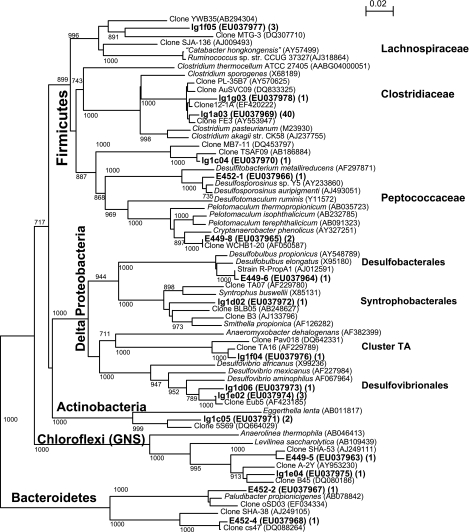
In the oil-degrading consortium, we recovered 16S rRNA gene sequences that affiliated with the sulfate-reducing bacteria (SRB) genera Desulfobulbus, Desulfosporosinus, and Desulfovibrio as well as with the syntrophic bacteria Desulfotomaculum subcluster Ih and Smithella (Fig. 4). Numerous fermentative bacteria affiliated with Chloroflexi (subphylum I), Clostridiales, and Bacteroidetes (Fig. 4) were also present. Sequences similar (>97%) to that of lg1a03 were obtained from both consortium samples in September 2005 (23 sequences from L2 and 17 from L4). All of the archaeal 16S rRNA gene clones sequenced (18 total, representative OTU E460-1, EU037979) showed 98 to 100% similarity to each other and to members of the obligately aceticlastic genus Methanosaeta. However, separate cultivation efforts directed toward H2/CO2-utilizing methanogens (data not shown) revealed that H2/CO2-utilizing methanogens must also have been present in the inoculum (L48-2, EU037980, 99% similar to the 16S rRNA gene of Methanobacterium subterraneum). In order to interrogate the consortium for the presence of such methanogens, we used primers specific for the mcrA gene that were previously reported to be unsuitable for amplification of Methanosaeta mcrA (ME1 and ME2 [25]). Our analysis of the 16S rRNA archaeal gene libraries indicated that Methanosaeta was dominant, and it was difficult (poor yield of amplification products) to obtain mcrA gene sequences using the ME1-ME2 primer pair. The mcrA gene sequences from the consortium were most similar to the mcrA gene from the hydrogenotrophic methanogen Methanoculleus marisnigri (CP000562, 92% similarity, one sequence), while another was most similar to the mcrA sequence from an uncultured clone (AF525519, 92% similarity), both in the Methanomicrobiales. The difficulty in amplifying mcrA from the consortium stood in stark contrast to the ease with which mcrA could be amplified after selection for H2/CO2-utilizing methanogens.
FIG. 4.
Phylogenetic relationships of eubacterial clones from the oil-utilizing methanogenic consortium with respect to related sequences. The tree is constructed from approximately 800 bp 16S rRNA gene sequence using the neighbor-joining algorithm. One thousand bootstrap replications were performed; only values greater than 750 are shown. The numbers in parentheses following the accession numbers indicates the total number of clones represented by the OTU.
Methanogenesis from other petroliferous cores.
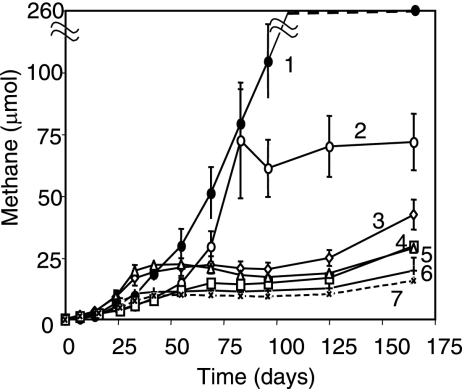
To determine whether the oil-degrading methanogenic inoculum might have broad utility, we tested it for the ability to generate methane from diverse petroliferous cores. Relative to uninoculated controls established for each core sample (no methane production [data not shown]) and incubations containing the inoculum alone, significantly enhanced levels of methane were measured for four of the five petroliferous samples (Fig. 5). Although changes in oil components were not monitored from these other core samples, this initial survey shows that the inoculum may be of more general utility to enhance methane production from a variety of mature petroliferous deposits.
FIG. 5.
Methane production from six different inoculated petroliferous rocks sampled from different formations in Oklahoma relative to an inoculum-only control (line 7). Line 1 represents the sandstone core obtained from Nowata County. Core samples 2 to 6 (lines 2 through 6) are described in the text in Material and Methods. Error bars represent 1 standard deviation of the mean of triplicate incubations.
DISCUSSION
We found that crude oil entrained in mature reservoir samples can be converted to methane gas by a hydrocarbon-degrading methanogenic consortium. Although the various core materials tested were provided with nutrients, such as nitrogen and phosphorus salts, CO2, trace elements, and vitamins, we never observed methane production unless the core samples received the hydrocarbon-utilizing inoculum. These results attest to the requirement for inoculation in an oil-to-methane energy recovery process in mature reservoirs. Clearly, we have examined only a minute fraction of marginal resources that could potentially be exploited for methane recovery, so it is certainly possible that different formations may respond to stimulatory strategies other than inoculation. To date, though, an indigenous anaerobic hydrocarbon-degrading consortium or isolate from a reservoir remains elusive (26). This report provides a “proof of concept” that hydrocarbons entrained in reservoir materials can be bioconverted to methane as an alternate energy source.
The methanogenic decomposition of organic matter requires syntrophic organisms able to convert complex substrates in a series of reactions to acetate and H2 and two types of methanogens to consume these intermediates. Hydrocarbon-degrading consortia are no exception. Indeed, in a study of methanogenic hexadecane metabolism, sequence analysis revealed that most bacterial clones from the consortium were closely related to the genus Syntrophus within the Deltaproteobacteria, whereas archaeal sequences affiliated with both aceticlastic (Methanosaeta) and hydrogenotrophic (Methanospirillum and Methanoculleus) methanogens (43). Similar organisms were also identified in a methanogenic toluene-degrading enrichment (12). Jones et al. (23) found that an oil-degrading consortium derived from river sediment was dominated by Syntrophus sp. and H2/CO2-using methanogens among several other unidentified eubacterial OTUs. The researchers proposed that methanogenic oil biodegradation in the consortium occurs mainly through syntrophic alkane conversion to acetate and H2, with subsequent syntrophic acetate oxidation to CO2, coupled with hydrogenotrophic methanogenesis. This hypothesis was supported by CO2 isotopic signature data, both measured and modeled, suggesting that CO2 reduction to methane predominates in biodegraded reservoirs (23). In our residual oil-degrading inoculum, the predominant archaeal sequences retrieved most closely affiliated with the genus Methanosaeta, which suggests that aceticlastic methanogenesis can also be an important pathway for methane production from oil. In a consistent fashion, when BESA was added to residual-oil-amended incubations to inhibit methanogenesis, acetate was found to accumulate relative to uninhibited incubations. Nevertheless, we also obtained sequences indicating the presence of Methanoculleus sp. in the consortium, albeit in low abundance, so H2/CO2-based methane production is also a relevant process, but possibly to a lesser extent. The finding of predominantly aceticlastic methanogens in our inoculum is consistent with the findings of in a separate investigation, wherein methanogenesis was the predominant fate of acetate even in the presence of sulfate, with 95% (180 out of 190) of the sequenced archaeal clones found to affiliate with members of the Methanosaetaceae (33). Our observation of significant methane production from residual oil in the presence of 10 mM sulfate agrees with that study (33) in that methanogenesis is the preferred electron-accepting process, even in the presence of alternate electron acceptors. It should be noted that the microbial populations in both studies were derived from the same gas-condensate-contaminated aquifer. The predominant or particular route of hydrocarbon metabolism to methane likely varies with the source of a given consortium (7, 28).
Fermentative bacteria such as the Clostridia present in the inoculum are currently not known to directly catalyze alkane transformations, but probably utilize various hydrocarbon intermediates to generate methanogenic precursors. However, it has recently been speculated that such traditional fermentative organisms may be able to oxidize hydrocarbons (24). In contrast, SRB have definitely been shown to utilize oil components directly (e.g., see references 6 and 41). Individually, SRB have a relatively narrow substrate range (5, 41), so a complex assemblage of organisms to metabolize the range of components in oil is consistent with our observations. Some members of the Desulfotomaculum cluster I organisms, typically known as gram-positive SRB, have lost the ability for sulfate respiration and opt for a syntrophic lifestyle in concert with methanogens (21). Based on its phylogenetic position, we can only speculate that the Cryptanaerobacter clone E449-8 (Fig. 4) plays a comparable role in our culture.
As has been found by others who have examined oil field environments (26, 28), many of our sequences show no close affiliation to 16S rRNA eubacterial gene sequences from named strains (e.g., lg1a03, which has 91.8% similarity to Clostridium akagii; lg1c04, which has 86% similarity to Cryptanaerobacter phenolicus; and lg1f05, which has 86.7% similarity to Ruminococcus sp. strain CCUG 37327). The repeated occurrence of 16S rRNA gene sequences from environmental clones similar to that of lg1f04 has been noted (42) and designated “cluster TA” in recognition of their distance from sequences of named strains (e.g., 82.2% 16S rRNA gene sequence similarity to that of Anaeromyxobacter dehalogenans). In addition to the low similarity of many of our 16S rRNA eubacterial gene sequences to those of named species, our clone libraries are too small for us to claim complete representation of the diversity. However, as an examination of diversity by cloning and sequencing entails certain biases (25), it will be important for future research to also use alternative approaches to determine the species composition and physiology in these consortia. Determining the exact function of the consortium members will be of great interest for designing and optimizing methane recovery efforts from marginal domestic reservoirs.
In the United States, energy recovery from marginal wells approximates 1 million barrels of oil per day, which equates to about 19% of domestic oil production (www.fossil.energy.gov/programs/oilgas/marginalwells/index.html). It may be possible to increase this percentage by converting a portion of the oil in marginal wells to methane. The extent of the effect can be extrapolated from our data to determine how much methane can theoretically be recovered from known domestic U.S. reservoirs (e.g., 375 billion barrels [38]). For example, if we take into account our measured rates of 0.1 to 0.4 μmol methane/day/g core, the average residual oil saturation of our model marginal core (0.013 g oil per gram core), and the density of the model formation oil (0.79 g/ml) and assume that 1% of residual oil supplies (e.g., 3.75 billion barrels) would be amenable to biological transformation, the resulting production could be 3 to 13 Bcf of CH4 per day or 1 to 5 Tcf of CH4 per year, a substantial fraction of current natural gas consumption in the United States, which is nearing 30 Tcf per year (10). While an oversimplification, these calculations provide some insight as to how an oil-to-methane bioconversion process can potentially recover economically valuable energy. As it will likely be necessary to utilize a suite of new methods and processes in combination to meet future world energy demands (16, 39), the microbial conversion of residual oil to natural gas could occupy a niche in energy recovery. Clearly, the nutritional and physiological limits (such as temperature and pressure limits) of the inoculum described here, its ability to migrate through formations, and the engineering design for such an energy recovery scheme will need to be determined.
Acknowledgments
This work received partial support from the International Petroleum Environmental Consortium under EPA grant number X83242801.
We thank M. Brackin of Arrow Oil & Gas, Inc., Norman, OK, for the petroliferous sandstone core and formation crude oil from Nowata County, OK, used in this study.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Aitken, C. M., D. M. Jones, and S. R. Larter. 2004. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 341:291-294. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, R. T., and D. R. Lovley. 2000. Hexadecane decay by methanogenesis. Nature 404:722-723. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidova, I. A., and J. M. Suflita. 2005. Enrichment and isolation of anaerobic hydrocarbon-degrading bacteria. Methods Enzymol. 397:17-34. [DOI] [PubMed] [Google Scholar]
- 6.Davidova, I. A., K. E. Duncan, O. K. Choi, and J. M. Suflita. 2006. Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane degrading sulfate reducing bacterium. Int. J. Syst. Evol. Microbiol. 56:2737-2742. [DOI] [PubMed] [Google Scholar]
- 7.Dolfing, J., S. R. Larter, and I. M. Head. 2007. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 1:1-11. [DOI] [PubMed] [Google Scholar]
- 8.Elshahed, M. S., L. M. Gieg, M. J. McInerney, and J. M. Suflita. 2001. Signature metabolites attesting to the in situ attenuation of alkylbenzenes in anaerobic environments. Environ. Sci. Technol. 35:682-689. [DOI] [PubMed] [Google Scholar]
- 9.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Energy Information Administration. 2007. Annual energy outlook. DOE/EIA-0383. U.S. Department of Energy, Washington, DC.
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:666-670. [DOI] [PubMed] [Google Scholar]
- 12.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein, M., R. P. DeBruyn, J. L. Weber, and J. B. Dodson. 2005. Buried hydrocarbons: a resource for biogenic methane generation. World Oil 226:61-67. [Google Scholar]
- 14.Gieg, L. M., R. V. Kolhatkar, M. J. McInerney, R. S. Tanner, S. H. Harris, Jr., K. L. Sublette, and J. M. Suflita. 1999. Evidence for intrinsic bioremediation in a gas condensate-contaminated aquifer. Environ. Sci. Technol. 33:2550-2560. [Google Scholar]
- 15.Gieg, L. M., and J. M. Suflita. 2002. Detection of anaerobic metabolites of saturated and aromatic hydrocarbons in petroleum-contaminated aquifers. Environ. Sci. Technol. 36:3755-3762. [DOI] [PubMed] [Google Scholar]
- 16.Giles, J. 2004. Every last drop. Nature 429:694-695. [DOI] [PubMed] [Google Scholar]
- 17.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Head, I. M., D. M. Jones, and S. R. Larter. 2003. Biological activity in the deep subsurface and the origin of heavy oil. Nature 426:344-352. [DOI] [PubMed] [Google Scholar]
- 19.Herrick, J. B., E. L. Madsen, C. A. Batt, and W. C. Ghiorse. 1993. Polymerase chain reaction amplification of naphthalene catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl. Environ. Microbiol. 59:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 21.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Loy, Y.-L. Qiu, P. Hugenholtz, N. Kimura, M. Wagner, A. Ohashi, and H. Harada. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. L. 1994. Similarity analysis of DNAs, p. 655-682. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 23.Jones, D. M., I. M. Head, N. D. Gray, J. J. Adams, A. K. Rowan, C. M. Aitken, B. Bennett, H. Huang, A. Brown, B. F. J. Bowler, T. Oldenburg, M. Erdmann, and S. R. Larter. 2008. Crude oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451:176-180. [DOI] [PubMed] [Google Scholar]
- 24.Kunapuli, U., T. Lueders, and R. Meckenstock. 2007. The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J. 1:643-653. [DOI] [PubMed] [Google Scholar]
- 25.Lueders, T., K.-J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analysis of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 26.Magot, M. 2005. Indigenous microbial communities in oil fields, p. 21-33. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, DC.
- 27.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nazina, T. N., N. M. Shestakova, A. A. Grigor'yan, E. M. Mikhailova, T. P Tourova, A. B. Poltaraus, C. Feng, F. Ni, and S. S. Belyaev. 2006. Phylogenetic diversity and activity of anaerobic microorganisms of high-temperature horizons of the Dagang oil field (China). Microbiology 75:55-65. (In Russian.) [PubMed] [Google Scholar]
- 29.Pallasser, R. J. 2000. Recognising biodegradation in gas/oil accumulations through the ∂13C composition of gas components. Org. Geochem. 31:1363-1373. [Google Scholar]
- 30.Parkes, J. 1999. Cracking anaerobic bacteria. Nature 401:217-218. [DOI] [PubMed] [Google Scholar]
- 31.Rios-Hernandez, L. A., L. M. Gieg, and J. M. Suflita. 2003. Biodegradation of an alicyclic hydrocarbon by a sulfate-reducing enrichment from a gas condensate-contaminated aquifer. Appl. Environ. Microbiol. 69:434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddique, T., P. M. Fedorak, and J. M. Foght. 2006. Biodegradation of short-chain n-alkanes in oil sands tailings under methanogenic conditions. Environ. Sci. Technol. 40:5459-5464. [DOI] [PubMed] [Google Scholar]
- 33.Struchtemeyer, C. G., M. S. Elshahed, K. E. Duncan, and M. J. McInerney. 2005. Evidence for aceticlastic methanogenesis in the presence of sulfate in a gas condensate-contaminated aquifer. Appl. Environ. Microbiol. 71:5348-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suflita, J. M., I. A. Davidova, L. M. Gieg, M. Nanny, and R. C. Prince. 2004. Anaerobic hydrocarbon biodegradation and the prospects for microbial enhanced energy production, p. 283-305. In R. Vazquez-Duhalt and R. Quintero-Ramirez (ed.), Petroleum biotechnology: developments and perspectives, vol. 151. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 35.Tanner, R. S. 2002. Cultivation of bacteria and fungi, p. 62-70. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, DC.
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmongin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Townsend, G. T., R. C. Prince, and J. M. Suflita. 2003. Anaerobic oxidation of crude oil hydrocarbons by the resident microorganisms of a contaminated anoxic aquifer. Environ. Sci. Technol. 37:5213-5218. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Department of Energy. 2006. Undeveloped domestic oil resources: the foundation for increasing oil production and a viable domestic oil industry. www.fossil.energy.gov/programs/oilgas/eor/Undeveloped_Domestic_Oil_Resources_Provi.html.
- 39.Whitesides, G. M., and G. W. Crabtree. 2007. Don't forget long-term fundamental research in energy. Nature 315:796-798. [DOI] [PubMed] [Google Scholar]
- 40.Widdel, F., and F. Bak. 1992. Gram-negative sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.) The prokaryotes, vol. 4. Springer, New York, NY. [Google Scholar]
- 41.Widdel, F., A. Boetius, and R. Rabus. 2006. Anaerobic biodegradation of hydrocarbons including methane, p. 1028-1049. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: ecophysiology and biochemistry, vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 42.Wu, J.-H., W.-T. Liu, I.-C. Tseng, and S.-S. Cheng. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373-382. [DOI] [PubMed] [Google Scholar]
- 43.Zengler, K., H. H. Richnow, R. Rossello-Mora, W. Michaelis, and F. Widdel. 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266-269. [DOI] [PubMed] [Google Scholar]