Abstract
The genome of Lactobacillus salivarius UCC118 comprises a 1.83-Mb chromosome, a 242-kb megaplasmid (pMP118), and two smaller plasmids of 20 kb (pSF118-20) and 44 kb (pSF118-44). Annotation and bioinformatic analyses suggest that both of the smaller plasmids replicate by a theta replication mechanism. Furthermore, it appears that they are transmissible, although neither possesses a complete set of conjugation genes. Plasmid pSF118-20 encodes a toxin-antitoxin system composed of pemI and pemK homologs, and this plasmid could be cured when PemI was produced in trans. The minimal replicon of pSF118-20 was determined by deletion analysis. Shuttle vector derivatives of pSF118-20 were generated that included the replication region (pLS203) and the replication region plus mobilization genes (pLS208). The plasmid pLS203 was stably maintained without selection in Lactobacillus plantarum, Lactobacillus fermentum, and the pSF118-20-cured derivative strain of L. salivarius UCC118 (strain LS201). Cloning in pLS203 of genes encoding luciferase and green fluorescent protein, and expression from a constitutive L. salivarius promoter, demonstrated the utility of this vector for the expression of heterologous genes in Lactobacillus. This study thus expands the knowledge base and vector repertoire of probiotic lactobacilli.
Lactic acid bacteria and in particular members of the genus Lactobacillus are the most common microbes used as probiotics. They may beneficially affect the host upon ingestion by a variety of potential or proven mechanisms (13, 41). Similarly to bifidobacteria, lactobacilli are normal inhabitants of human and animal intestines. Among the more than 100 species of the genus Lactobacillus that have been identified, those that are used as probiotics include L. acidophilus, L. brevis, L. bulgaricus, L. casei, L. cellobiosus, L. crispatus, L. curvatus, L. delbrueckii, L. fermentum, L. gasseri, L. johnsonii, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus, and L. salivarius (32). Genome sequence availability considerably facilitates the identification of probiotic characteristics of these bacteria and the prediction of their behaviors in the human gastrointestinal (GI) tract. To date, 11 Lactobacillus genome sequences have been published, and at least 11 additional sequencing projects are in progress (6). This information has dramatically improved our understanding of the metabolic processes and genetics of these microorganisms, as well as their potential roles in health promotion of their hosts. However, for the targeted analysis of genes that contribute to probiotic characteristics, the development of molecular tools for these lactobacilli is of paramount importance.
Plasmids, autonomously replicating extrachromosomal genetic elements, are widely present in the genus Lactobacillus. About 38% of the species in this genus contain plasmids (60). Endogenous plasmids from Lactobacillus are of interest because of the traits they confer upon the host. For example, these plasmids may harbor genes encoding resistance to antibiotics (39, 54) and metal ions (55), genes encoding bacteriocins (30, 44), gene clusters for conjugation (55), genes involved in adherence and biotin metabolism (10), and genes encoding toxin-antitoxin (TA) proteins for plasmid maintenance (53). In addition to encoding such interesting traits, endogenous plasmids are the most commonly used systems to construct genetic tools especially for gene cloning and gene expression purposes (7, 52) due to their ability to replicate in the original hosts. Cryptic plasmids from L. delbrueckii (35), L. casei (2), L. plantarum (47), L. fermentum (1, 48), L. reuteri (40), L. helveticus (61), L. curvatus (33), and L. pentosus (49) have been adapted as Escherichia coli and Lactobacillus cloning and expression vectors (48). For the sequenced probiotic strain L. salivarius UCC118, there are limited genetic tools available. Previous studies in our laboratory showed that among the plasmids tested (pAMβ1 [4], pE194 [22], pCI305 [26], pLC2 [59], pUB110 [43], pSH71 [9], and pWV01 [46]), only plasmids containing pSH71 or pWV01 replication origins were successfully introduced (57). Therefore, there was significant incentive to adapt additional replicons to allow the development of gene expression vectors, promoter probe plasmids, and expression monitoring and gene mutagenesis systems for detecting and analyzing biologically relevant characteristics of probiotic lactobacilli. Here we describe the annotation of two endogenous plasmids from L. salivarius UCC118 and the adaptation of one of these plasmids for the purposes of cloning and expression in L. salivarius and other lactobacilli. The analysis of these two endogenous plasmids from strain UCC118 reveals their potential as genetic tools for probiotic lactobacilli.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study and their relevant features are listed in Table 1. The L. salivarius strains used in this study are listed in Table 2. E. coli Top10 was used as an intermediate host for the pCI341 and pEM constructs and was grown in Luria-Bertani broth (20) with aeration at 37°C. L. salivarius, L. plantarum, and L. fermentum were grown under microaerobic conditions (5% CO2) in de Man-Rogosa-Sharpe (MRS) medium (Oxoid Ltd., United Kingdom) at 37°C, except when simultaneously detecting bioluminescence and growth.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| L. lactis | ||
| MG1363 | Plasmid-free derivative of L. lactis subsp. cremoris NCDO712 | 16 |
| L. salivarius | ||
| UCC118 | Ileocecal isolate from a human adult | 12 |
| LS201 | pSF118-20-free derivative of strain UCC118 | This work |
| L. plantarum | ||
| NCIMB8826 | Isolated from human saliva, it is identical to the sequenced strain L. plantarum WCFS1, which is a single-colony isolate of strain NCIMB8826 | NCIMB |
| L. fermentum | ||
| DSM20055 | Isolated from saliva | DSM |
| Plasmids | ||
| pNZ8048 | Cmr, NcoI site has been used for translational fusions, nisin-induced gene expression vector | 34 |
| pVE6007 | Cmr, temperature sensitive, derivative of pWV01, lactococcal cloning vector | 42 |
| pCI341 | Cmr, replication probe vector | 25 |
| pLS201 | Cmr, pNZ8048 containing gene pemI and its own promoter region amplified from pSF118-20 | This work |
| pEM | Emr, pBluescript II SK(−) derivative in which amp was replaced by erm from pE194 | Unpublished resultsb |
| pLS202 | Cmr, pCI341 containing LSL_1963-LSL_1968 | 14 |
| pLS203 | Emr, pEM containing LSL_1963-LSL_1967 | This work |
| pLS204 | Cmr, pCI341 containing LSL_1965-LSL_1967 | 14 |
| pLS205 | Cmr, pCI341 containing LSL_1965-LSL_1966 | 14 |
| pLS206 | Cmr, pCI341 containing LSL_1965 and its 613-bp downstream region | 14 |
| pLS207 | Cmr, pCI341 containing LSL_1965 | 14 |
| pLS208 | Emr, pEM containing LSL_1960-LSL_1967 | This work |
| pLS209 | Emr; a derivative of pLS203 produced by PCR with primers FF122 and FF125, which has ClaI, NcoI, and HindIII sites in multiple cloning sites of pLS203 | This work |
| pLS210 | Emr, a derivative of pLS203 for expressing luxABCDE under the promoter of cysK (LSL_1718) | This work |
| pLS211 | Emr, a derivative of pLS209 for expressing pemI and pemK (LSL_1984 and LSL_1985, respectively) under the native promoter of these loci | This work |
| pLS212 | Cmr, a derivative of pVE6007 for expressing pemI and pemK (LSL_1984 and LSL_1985, respectively) under the native promoter of these loci | This work |
| pLS213 | Emr, a promoter probe vector derived from pLS203 containing gfp+ from pZEP08 | This work |
| pLS214 | Emr, a derivative of pLS213 for the production of GFP under the promoter of cysK (LSL_1718) | This work |
| pFT1 | Ampr, pUC19 containing the luxABCDE operon | Unpublished resultsc |
| pZEP08 | Cmr, Kmr, a pBR322 derivative containing gfp+ | 23 |
Strr, streptomycin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Ampr, ampicillin resistant; Kmr, kanamycin resistant.
Contributed by Mary O'Connell Motherway, UCC.
Contributed by Ian Monk, UCC.
TABLE 2.
L. salivarius strains used for detecting the presence of repA20- or repA44-related plasmids as described by Li et al. (38)
| Straina | Origin |
|---|---|
| UCC118 | Human ileal-cecal region |
| AH4231 | Human ileum-cecum |
| UCC119 | Chicken intestine |
| DSM20492 | Human saliva |
| DSM20554 | Human saliva, type strain |
| DSM20555 | Human saliva, type strain |
| NCIMB8816 | Italian human saliva |
| NCIMB8817 | Turkey feces |
| NCIMB8818 | St. Ivel cheese |
| NCIMB702343 | Not available |
| CCUG27530B | Human abdomen, abscess |
| CCUG38008 | Human gall, 73-year-old man |
| CCUG43299 | Human blood |
| CCUG45735 | Human blood |
| CCUG47825 | Human blood, 55-year-old woman |
| CCUG44481 | Bird |
| CCUG47171 | Human tooth plaque |
| CCUG47826 | Human blood, 55-year-old woman |
| JCM1040 | Human intestine |
| JCM1042 | Human intestine |
| JCM1045 | Human intestine |
| JCM1046 | Swine intestine |
| JCM1230 | Chicken intestine |
| 01M14315b | Human gallbladder pus |
| LMG14476 | Cat with myocarditis |
| LMG14477 | Parakeet with sepsis |
| L21c | Not available |
AH, Alimentary Health Culture Collection, Cork, Ireland; UCC, Department of Microbiology, University College Cork, Cork, Ireland; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Germany; NCIMB, National Collections of Industrial Food and Marine Bacteria, United Kingdom; CCUG, Culture Collection University Göteborg, Sweden; JCM, Japan Collection of Microorganisms, Japan; LMG, Laboratorium voor Microbiologie, University Gent, Belgium.
Contributed by Kwok-Yung Yuen, Hong Kong, China.
Contributed by Gerald W. Tannock, New Zealand.
Lactococcus lactis MG1363 was used as a cloning host for pNZ8048-based constructs. It was grown at 30°C in M17 broth supplemented with 0.5% glucose. When necessary, erythromycin (Em) was supplemented to a final concentration of 5 μg/ml for L. salivarius, L. plantarum, L. fermentum, and L. lactis and 300 μg/ml for E. coli Top10; chloramphenicol (Cm) was supplemented to a final concentration of 5 μg/ml for L. salivarius, L. plantarum, L. fermentum, and L. lactis. Ampicillin was supplemented at 100 μg/ml for E. coli.
Sequence analysis and annotation of pSF118-20 and pSF118-44.
The two endogenous plasmids pSF118-20 and pSF118-44 had previously been sequenced by an ordered library approach (14), and the annotated sequences were deposited in GenBank under accession numbers AF488831 and AF488832. The plasmid annotations were not reanalyzed or discussed when the genome sequence was determined (5). We revised the annotation of pSF118-20 and pSF118-44 and the updated GenBank annotations correspond to the original accession numbers AF488831 and AF488832. The revised annotation was performed essentially as for the genome sequence (5) using the ERGO platform of Integrated Genomics (Chicago, IL).
DNA manipulation.
Primers used for PCR were purchased from MWG Biotech (Ebersberg, Germany) and are listed in Table S1 in the supplemental material. An Expand long template kit (Roche, Mannheim, Germany) was used for the amplification of a 7-kb region of pSF118-20. Otherwise, Pwo polymerase (Roche, Mannheim, Germany) was used for the PCR amplifications. Restriction enzymes, T4 DNA ligase, and PCR purification kits were purchased from Roche (Mannheim, Germany) and were used as specified by the manufacturers. Ligation products were desalted by ethanol precipitation using pellet paint (Novagen, United Kingdom) prior to electrotransformation.
The genomic DNA of L. salivarius was isolated as previously described (15) with some modifications. Eighteen-hour stationary-phase cells of L. salivarius were harvested by centrifugation. The pelleted cells were washed once with 30 mM Tris-HCl buffer containing 3 mM MgCl2 and 25% sucrose (pH 8.0) and stored overnight at −20°C. Cells were thawed and treated with 10 mg/ml lysozyme at 37°C for 1.5 h and 2 mg/ml proteinase K at 55°C for 1 h before lysis. The DNA was further purified by a phenol-chloroform extraction protocol (51). For transformation, the preparation of electrocompetent cells of E. coli was performed as previously described by Sambrook et al. (51). The transformation of L. lactis was performed as described by Holo and Nes (27). L. salivarius was transformed as previously described (56). The procedure for the transformation of L. fermentum was the same as that for L. salivarius except for an incubation step for 1.5 h in MRS medium at 37°C immediately following electroporation. Plasmid DNA (up to 200 ng in 5 μl) was transformed by electroporation into E. coli Top10 at 2.5 kV, 25 μF capacitance, and 200 Ω resistance; L. lactis at 2.0 kV, 25 μF capacitance, and 200 Ω resistance; or Lactobacillus strains at 1.5 kV, 25 μF capacitance, and 400 Ω resistance.
Southern blot analysis followed a standard protocol (51). Amplicons used as Southern blot probes were generated by PCR using appropriate primers (see Table S1 in the supplemental material). For rehybridization, the previously hybridized membrane was stripped by washing once with distilled water, three times with 0.2 M NaOH containing 0.1% sodium dodecyl sulfate at 37°C for 30 min, and once with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min. The stripped membrane was then stored in 2× SSC before reuse.
Transcriptional analysis of target genes.
End point reverse transcription-PCR (RT-PCR) was employed to test if target genes were expressed in vitro. RNA was isolated from stationary-growth-phase cells using an RNA-easy kit (Ambion, Cambridgeshire, United Kingdom). Random primers were purchased from MWG Biotech, Germany. Improm-II reverse transcriptase (Promega, Madison, WI) was used to generate cDNA for further analysis.
Analysis of pSF118-20 replication.
For analysis of the replication mechanism of pSF118-20, we studied pLS203, an E. coli-Lactobacillus shuttle vector containing the replication origin of pSF118-20. pVE6007, a plasmid derivative of pWV01, which replicates via the rolling-cycle mechanism, was used as a positive control for single-stranded DNA (ssDNA) detection. L. salivarius LS201, harboring either pLS203 or pVE6007, was grown in MRS medium at 37°C until an optical density at 600 nm of 0.8 to 1.0 was reached, followed by treatment with 100 μg/ml rifampin and 100 μg/ml Em for 1 h at 37°C prior to harvest, to allow for the accumulation of ssDNA intermediates as described by Leenhouts et al. (36). Cell pellets from both strains were frozen and thawed, and then cell lysates were prepared by sodium perchlorate and chloroform extraction (37). For nuclease S1 treatment, cell lysates were treated with 10 units/μl nuclease S1 for 45 min at 37°C. The whole cell lysates of L. salivarius LS201 (pLS203 or pVE6007), either treated or not treated with nuclease S1, were electrophoresed on a 0.8% agarose gel. Both denatured and nondenatured gels were blotted to membrane (nitrocellulose) and hybridized with probes generated by PCR (for pLS203) or plasmid digestion (for pVE6007). Southern blot analysis was performed as described above in “DNA manipulation.”
Pulsed-field gel electrophoresis plug preparation, nuclease S1 treatment, and electrophoresis.
The preparation of agarose gel plugs of high-molecular-weight DNA for pulsed-field gel electrophoresis (PFGE), cell treatment, and electrophoresis was performed following the protocol described by Li et al. (38). A CHEF-DR II pulsed-field system (Bio-Rad Laboratories) was used to resolve DNA fragments at 6 V/cm in 0.5× Tris-borate-EDTA running buffer at 14°C for 20 h. A time setting of 3 to 50 s was employed for the linear ramped pulse.
Defining the minimal stable replicon of pSF118-20.
In order to determine the minimal stable replication region of pSF118-20, a series of deletion constructs were made by cloning PCR fragments, which were amplified by primers SF03 to SF09 into the replication probe vector pCI341. These constructs were then introduced into L. plantarum NCIMB8826 and L. lactis MG1363 by electroporation, and their replication abilities were tested by checking the cultures for growth on MRS agar plates containing Cm or Em. The segregational stabilities of the constructs were investigated in lactic acid bacteria by passaging them in the absence of antibiotic selection at the optimal growth temperature for 100 generations.
Construction of pLS203 and pLS208.
A stable replication region from pSF118-20 that functioned in both lactobacilli and lactococci was amplified by primers FF033 and FF034. The 4-kb amplicon was then cloned into the XhoI and PstI sites of the E. coli cloning vector pEM. The construct was then electroporated into E. coli Top10, yielding pLS203. Similarly, a 7-kb region, including the putative mobilization locus, the stable replication region of pSF118-20, and the loci between them, was amplified with primers FF009 and FF033 and then cloned into the XhoI and PstI sites of pEM, yielding pLS208. Plasmids pLS203 and pLS208 were introduced into L. lactis MG1363, L. plantarum NCIMB8826, L. salivarius LS201, and L. fermentum DSM20055 to analyze segregational stability and to perform mating experiments.
Curing of pSF118-20.
A putative promoter (TTGCCA-N13-TATAAT) was noted 202 bp upstream from the pemI (LSL_1984) start codon. A fragment including the pemI gene and this putative promoter was amplified by PCR (primers FF001 and FF004) and cloned into the NcoI and SpeI sites of pNZ8048. The ligation mixture was used to transform L. lactis by electroporation, resulting in Cmr transformants harboring pLS201. pLS201 was then transformed into L. salivarius UCC118. An overnight culture of UCC118(pLS201) was either inoculated into fresh MRS-Cm broth and grown at 30°C, 37°C, 42°C, 44°C, and 46°C for 30 generations or subcultured in fresh MRS-Cm broth containing novobiocin (0.2 to 10 μg/ml) at 37°C for 72 h. The corresponding cultures were plated on MRS agar plates containing Cm and screened for derivatives of L. salivarius UCC118 lacking pSF118-20 by colony PCR. Colonies confirmed to lack pSF118-20 were then grown at an elevated temperature without antibiotic selection to cure pLS201.
Construction of pVE6007 and pLS203 derivatives expressing the pSF118-20 pemI and pemK genes in L. salivarius.
For cloning purposes, pLS203 was modified at the multiple cloning sites. An amplicon generated by primers FF122 and FF125 using pLS203 as a template was digested with PstI, self-ligated, and transformed into E. coli, resulting in pLS209. The pemI and pemK genes (LSL_1984 and LSL_1985, respectively) and their promoter were amplified as a single expressing fragment and cloned into pLS209 and pVE6007. The resulting construct, pLS211, was then transformed into L. lactis MG1363, L. salivarius LS201, L. plantarum NCIMB8826, and L. fermentum DSM20055, and pLS212 was transformed into L. salivarius LS201 to investigate the segregational stabilities of those constructs in the absence of antibiotic selection.
Conjugation and species identification of transconjugants.
A filter mating method (18) was used to perform conjugation. The donor and recipient cells were grown in nonselective medium to log phase of growth and were mixed at ratios of 1:1, 1:5, and 1:10 (donor cells:recipient cells). Cells were collected by filtering through a sterile 0.45-μm-pore-size membrane (MF-Millipore membrane filter, HAWP 02500; Millipore, Dublin, Ireland). Membranes bearing cells were placed on nonselective MRS agar plates and incubated at 37°C (5% CO2) for 24 h. The bacteria were then washed from the membranes with 1 ml of one-quarter-strength Ringer's solution (Oxoid, United Kingdom). The mating mixtures were plated on MRS agar plates containing Cm and Em and incubated at 37°C (5% CO2) for 4 days. Individual control cultures of recipient and donor strains were treated using the same procedure and plated on MRS agar plates containing both Em and Cm to determine the number of spontaneous antibiotic-resistant mutants.
API 50 CH strips and CHL medium (bioMérieux) were used to detect the carbohydrate fermenting profile of transconjugants. Freshly grown overnight cultures of the respective strains were harvested and resuspended in sterile water to achieve a cell density of 1010 CFU/ml. An aliquot of the cell suspension (200 μl) was inoculated into 10 ml API 50 CHL medium; 120 μl of this suspension was inoculated into API 50 CH strips that were then overlaid with paraffin to maintain anaerobic conditions. Incubation was carried out at 37°C for 48 h.
Expression of lux and gfp in lactobacilli.
The backbone of pLS203 was used to construct plasmids for expressing heterologous genes in L. salivarius and other Lactobacillus species. The luxABCDE loci (50) and the gfp+ gene from Aequoria victoria (23) were chosen for expression in Lactobacillus. A native promoter (cysKp) of L. salivarius UCC118 was chosen as a constitutive promoter because it was ranked, by global transcriptional analysis, among the top 3% of highly expressed genes during exponential and early stationary growth phase (M. W. Mangan and P. W. O'Toole, unpublished data). The promoter fragment was amplified by primers FF128 and FF129. cysKp was then cloned into the SalI and SwaI sites of pFT1 (a derivative of pUC19 containing luxABCDE), resulting in pFT2. Subsequently, a SpeI-PstI fragment of pFT2, containing cysKp transcriptionally fused to luxABCDE, was subcloned into SpeI-PstI-digested pLS203. Em-resistant and ampicillin-sensitive colonies were screened for luminescence to select the desired construct pLS210. To detect bioluminescence in lactic acid bacteria, overnight cultures of L. plantarum NCIMB8826(pLS210), L. salivarius LS201(pLS210), and L. fermentum DSM20055(pLS210) were diluted 1/100 in fresh MRS-Em broth, transferred into 96-well plates, and incubated in a Xenogen IVIS 100 system (Xenogen, Alameda, CA) at 37°C. The levels of bioluminescence were determined in continuous imaging mode with 5-min exposure at high resolution.
A recombinant pLS203 plasmid for producing green fluorescent protein (GFP+) was constructed by cloning a promoterless gfp+ PCR product amplified from pZEP08 (Table 1), using primers FF179 and FF180, into the SmaI and PstI sites of pLS203, resulting in a promoter probe vector, pLS213. This was followed by subcloning of the cysKp amplicon described above into pLS213, yielding pLS214. To detect fluorescence, L. salivarius LS201(pLS214) was grown in MRS broth at 37°C till stationary phase. Cells were then harvested and washed with phosphate-buffered saline (PBS). Cell suspensions in PBS were examined using an epifluorescence microscope (Olympus BX-51; Olympus Co., Japan) equipped with a fluorescein isothiocyanate filter. The Olympus UPlan FI 100 X/1.30 Oil Iris objective lens was used. Images were captured with a DP70 camera (Olympus Co., Japan) with Olympus DP-Soft software version 3.2.
Growth rates of Lactobacillus strains with different constructs were monitored by using a Bioscreen C analyzer (Oy Growth Curves AB Ltd., Helsinki, Finland) in 100-well microtiter plates (Labsystems, Finland) at 37°C.
Challenge conditions and UV resistance measurement.
Stationary phase cells of L. salivarius UCC118 and LS201 were harvested by centrifugation. Cell pellets were washed once with PBS, resuspended in PBS, and incubated at 37°C for 24 h. Control cells were resuspended in fresh MRS broth at 37°C for 24 h. Both starved cells and control cells were harvested and washed with PBS before challenging with MRS medium containing 0.1% porcine bile. Samples were taken at time zero and 5, 10, and 30 min after challenging with 0.1% bile and plated for viable cell counting. To investigate the resistance of L. salivarius strains UCC118 and LS201 to UV light, overnight cultures of these two strains in MRS broth were harvested by centrifugation. Cell pellets were washed once with PBS buffer and then resuspended in PBS. A total of 105 CFU of both strains was dispensed into wells of a 96-well plate and irradiated with UV light for 0 to 60 s using a portable Ultra-Violet lamp (Hanovia, Slough, England) at a distance of 11 cm.
RESULTS
Annotation of L. salivarius UCC118 plasmids pSF118-20 and pSF118-44.
The primary annotations of genes located on pSF118-20 and pSF118-44 are provided in Table 3, with detailed annotations available in Table S2 and S3 in the supplemental material. Overall, 48% and 41% of the open reading frames on pSF118-20 and pSF118-44, respectively, are of unknown function.
TABLE 3.
Primary gene annotations for pSF118-20 and pSF118-44 of L. salivarius UCC118
| pSF118-20
|
pSF118-44
|
||
|---|---|---|---|
| Locus tag | Annotationa | Locus tag | Annotation |
| LSL_1960 | Putative nickase, TraA-like | LSL_1987 | Nicotinate phosphoribosyltransferase (EC 2.4.2.11) |
| LSL_1961 | Conserved hypothetical protein | LSL_1988 | Phosphohydrolase (MutT/nudix family protein) |
| LSL_1962 | Conserved hypothetical protein | LSL_1989 | Transcriptional regulator, TetR family |
| LSL_1963 | Conserved hypothetical protein | LSL_1990 | Pseudogene |
| LSL_1964 | Hypothetical protein | LSL_1991 | Hypothetical protein |
| LSL_1965 | RepA | LSL_1992 | Hypothetical protein |
| LSL_1966 | Hypothetical protein | LSL_1993 | Pseudogene |
| LSL_1967 | Plasmid partition protein | LSL_1994 | Antitoxin of TA stability system |
| LSL_1968 | Resolvase | LSL_1995 | Toxin of TA stability system |
| LSL_1969 | Pyridine nucleotide-disulfide oxidoreductase family protein | LSL_1996 | Toxin |
| LSL_1970 | Hypothetical protein | LSL_1997 | Putative antitoxin |
| LSL_1971 | Hypothetical protein | LSL_1998 | Plasmid partition protein |
| LSL_1972 | Hypothetical membrane-spanning protein | LSL_1999 | Hypothetical protein |
| LSL_1973 | General stress protein, Gls24 family | LSL_2000 | Replication initiator protein |
| LSL_1974 | Hypothetical membrane-spanning protein | LSL_2001 | Conserved hypothetical protein |
| LSL_1975 | Hypothetical cytosolic protein | LSL_2002 | Conserved hypothetical protein |
| LSL_1976 | Conserved hypothetical protein | LSL_2003 | Conserved hypothetical protein |
| LSL_1977 | Hypothetical protein | LSL_2004 | Conserved hypothetical protein |
| LSL_1978 | Transposase ISLasa5b, IS3 family | LSL_2005 | Nickase |
| LSL_1979 | Putative UV-resistance-like protein | LSL_2006 | Hypothetical protein |
| LSL_1980 | Hypothetical protein | LSL_2007 | Transcriptional regulator, MarR family |
| LSL_1981 | Hypothetical protein | LSL_2008 | Quinone oxidoreductase |
| LSL_1982 | Hypothetical protein | LSL_2009 | Hypothetical protein |
| LSL_1983 | Hypothetical protein | LSL_2010 | Transposase ISLasa18a, IS256 family |
| LSL_1984 | PemI-like protein | LSL_2011 | ABC transporter, ATP-binding protein |
| LSL_1985 | PemK protein | LSL_2012 | Putative ABC transporter, integral membrane protein |
| LSL_1986 | Transposase ISLasa17c, IS256 family | LSL_2013 | Putative transcriptional regulator |
| LSL_2014 | Conserved hypothetical protein | ||
| LSL_2015-LSL_2016 | Pseudogene | ||
| LSL_2017 | Hypothetical protein | ||
| LSL_2018 | Hypothetical protein | ||
| LSL_2019 | Hypothetical protein | ||
| LSL_2020 | Pyridine nucleotide-disulfide oxidoreductase family protein | ||
| LSL_2020b | Pseudogene | ||
| LSL_2021 | Hypothetical membrane-spanning protein | ||
| LSL_2022 | Cobalt transport protein CbiQ | ||
| LSL_2023 | Cobalt transport protein CbiQ | ||
| LSL_2024 | Cobalt transport ATP-binding protein CbiO | ||
| LSL_2025 | Hypothetical protein | ||
| LSL_2026 | Glycine betaine transport system permease protein/glycine betaine-binding protein | ||
| LSL_2027 | Glycine betaine transport ATP-binding protein | ||
| LSL_2028 | Glutathione reductase | ||
| LSL_2029 | Resolvase | ||
| LSL_2030 | Hypothetical protein | ||
| LSL_2031 | Hypothetical protein | ||
| LSL_2032 | DNA-damage-inducible protein J | ||
| LSL_2033 | Hypothetical protein | ||
| LSL_2034 | Hypothetical protein | ||
| LSL_2035 | Conserved hypothetical protein | ||
| LSL_2036 | Hypothetical protein | ||
| LSL_2037 | Hypothetical protein | ||
| LSL_2038 | Transcriptional regulator, PadR family | ||
BLAST top hit as of June 2007.
The replication regions from both plasmids are predicted to encode a replication initiator protein (repA), a plasmid partitioning protein (parA), and several conserved hypothetical proteins. The RepA proteins from pSF118-20 and pSF118-44 (LSL_1965 [repA20] and LSL_2000 [repA44], respectively) are 71% identical to each other and are 72% similar to the Rep protein of lactococcal plasmid pCI2000, which is predicted to replicate via a theta replication mechanism (31).
Bacterial plasmid TA systems encode both toxin and antitoxin molecules that control plasmid maintenance (17). Two putative TA system (24) gene pairs were present in pSF118-44 (LSL_1994 and LSL_1995 and LSL_1996 and LSL_1997) while we annotated one such system (LSL_1984 and LSL_1985) in pSF118-20. The TA systems in pSF118-44 are similar to those of the relB and relE family (21), while the single TA system in pSF118-20 (LSL_1984 and LSL_1985) encodes proteins showing 99% and 96% identity to those encoded by pemI and pemK from p256 (53). pemI and pemK are the type II TA system in which the antitoxin is a protein and the toxin, PemK, is an endoribonuclease, which cleaves cellular mRNAs and blocks protein synthesis (62).
Several stress-resistance-related proteins (those related to general, UV resistance, heavy metal, and hyperosmotic stress) appear to be encoded by pSF118-20 and pSF118-44. LSL_1973 is similar to the gene encoding the stress-inducible and starvation-inducible Gls24 family protein from Enterococcus faecalis, which maintains the growth rate of cells, resistance to bile salts, and chain length in starved cells (19). The product encoded by LSL_1979 is similar to a protein from Pediococcus pentosaceus, which has been defined as DNA repair nucleotidyltransferase. pSF118-44 encodes a glycine-betaine uptake system (LSL_2026 and LSL_2027) which contributed to resistance to high salt concentrations when expressed in L. lactis (14). Presumed ABC-type multidrug transporter systems (LSL_2011 and LSL_2012), cobalt transporter systems (LSL_2022 to LSL_2024), and a gene encoding mercuric reductase (LSL_2020) are also carried by pSF118-44.
Among the sequenced lactobacillus genomes, glutathione reductase genes are found in L. plantarum, L. johnsonii, L. acidophilus, and L. sakei but not in L. delbrueckii. LSL_2028 from pSF118-44 is the first plasmid-encoded glutathione reductase gene reported for Lactobacillus, and it is the only gene in L. salivarius UCC118 that encodes this enzyme. It has been shown that glutathione reductase contributes to oxygen tolerance in L. sanfranciscensis (28). LSL_2028 may also contribute to microaerophilic growth condition tolerance for the catalase-negative strain L. salivarius UCC118.
Replication analysis of pSF118-20.
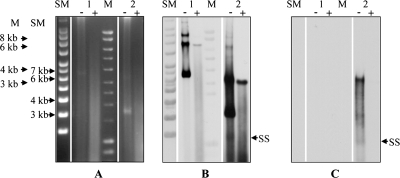
Annotation of pSF118-20 and pSF118-44 predicted that they would replicate via a theta replication mechanism. Since it was our intention to adapt pSF118-20 for vector construction, Southern blot analysis (Fig. 1) was employed to investigate the replication intermediates of pLS203, a shuttle vector containing the replication origin of pSF118-20 (see below). ssDNA intermediates indicative of rolling-circle replication were detected for pVE6007 (Fig. 1B and C), a plasmid containing the pWV01 replication origin which replicates through a rolling-cycle mechanism (36). However, no ssDNA intermediates were detected during the replication of pLS203 (Fig. 1B and C), which indicates that pSF118-20 replicates via a theta replication mechanism.
FIG. 1.
Analysis of the replication mechanism of pSF118-20. (A) Cell lysates of L. salivarius LS201 strains harboring pLS203 (an E. coli-Lactobacillus shuttle vector containing the replication origin of pSF118-20) or pVE6007 (a rolling-circle replication plasmid with pWV01 origin) with or without nuclease S1 treatment were electrophoresed on a 0.8% agarose gel. The PCR product of repA20 (LSL_1965) and NcoI-digested pVE6007 were used as probes to hybridize against blots prepared from either a denatured gel (B) or a nondenatured gel (C). Lane 1, pLS203; lane 2, pVE6007; lane SM, supercoiled DNA ladder (Sigma); lane M, linear DNA ladder (Bioline); −, untreated DNA sample; +, nuclease S1-treated DNA sample; SS, single-stranded DNA intermediates (indicated by arrows in the nuclease S1-treated DNA sample lane). The background smear in panel B represents degraded plasmid DNA.
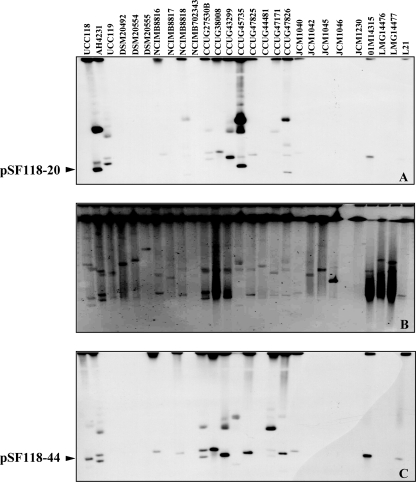
Distribution of related plasmids with theta-type replicons in L. salivarius strains.
The detection of pSF118-20 repA (repA20)- and pSF118-44 repA (repA44)-related plasmids in 27 strains of L. salivarius was performed by a combination of PFGE and Southern hybridization. Nuclease S1-treated genomic DNA samples of L. salivarius strains were resolved by PFGE (Fig. 2B). Probes based on repA20 and repA44 were generated by PCR and hybridized to membrane blotted with genomic DNA separated by PFGE. Cross-hybridization appeared as shown in Fig. 2A and C, as repA20 and repA44 are 73% identical in nucleotide sequences. Hybridization signals in Fig. 2A and C which do not correspond to linear plasmids in the PFGE are due to the hybridization of probes to another form of the plasmid that had not been linearized completely by nuclease S1. These are not the megaplasmids demonstrated by Li et al. (38), as different hybridization patterns were seen when the same membrane was probed with a fragment based on the megaplasmid (data not shown). Therefore, 52% of the tested 27 L. salivarius strains harbor plasmids ranging from 10 to 70 kb, which are all repA20- or repA44-related plasmids. Most of the plasmids from the L. salivarius strains are both repA20 and repA44 related due to their high sequence-relatedness identity, except for the small plasmids from NCIMB8818 (10 kb) and CCUG47826 (15 kb) that are repA20 related and the plasmids from CCUG47171 that are repA44 related.
FIG. 2.
Plasmid profiles of pSF118-20- and pSF118-44-related replication regions in 27 L. salivarius strains. Southern hybridization of nuclease S1-treated genomic DNA of 27 L. salivarius strains with the pSF118-20 repA probe (A) and the pSF118-44 repA probe (C). (B) PFGE of nuclease S1-treated genomic DNA of 27 L. salivarius strains.
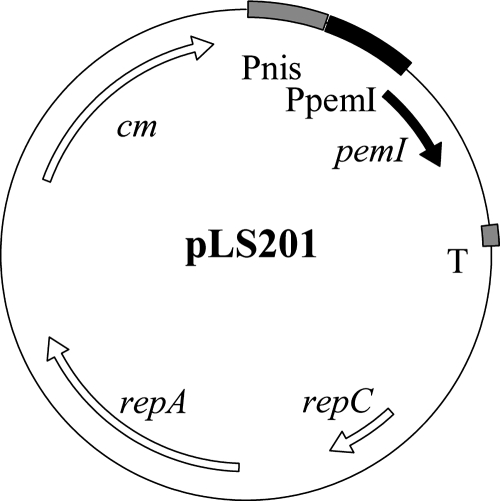
Curing of pSF118-20.
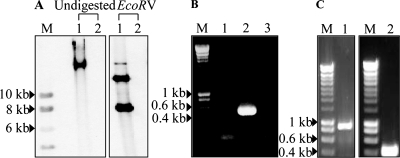
LSL_1984 (pemI) and LSL_1985 (pemK) in pSF118-20 were annotated as toxin and antitoxin plasmid addiction loci (29) which would provide segregational stability to pSF118-20 in a given host. This would explain why previous attempts to cure this plasmid were not successful (14). Presumably the relatively stable toxin kills the host once it loses pSF118-20 and its associated ability to make antitoxin. Therefore, construct pLS201 (Fig. 3) which produces antitoxin in trans was constructed (see Materials and Methods) and introduced into L. salivarius UCC118. The resulting transformants were grown in MRS medium containing novobiocin or passaged for 30 generations at different temperatures. Ninety-six colonies from the culture either treated with novobiocin or grown at different temperatures were picked and screened for the loss of pSF118-20 by colony PCR. One of them appeared to lack pSF118-20 (Fig. 4A and B), which was from the culture grown at 44°C. This derivative of UCC118 lacking pSF118-20 was designated as strain LS201. To confirm the loss of pSF118-20 in L. salivarius LS201, genomic DNA of L. salivarius UCC118 and LS201 was analyzed by Southern hybridization, using a PCR product amplified by primers FF005 and FF006 based on pSF118-20 as the probe. Figure 4 confirms the loss of pSF118-20 in strain LS201. Loss of the antitoxin-producing plasmid pLS201 from strain LS201 was subsequently obtained when the culture was passaged for 30 generations at 44°C in the absence of selection for pLS201. The other endogenous plasmids of UCC118, pSF118-44 and pMP118, were still present in strain LS201 as confirmed by PCR using primer pairs based on those plasmids (Fig. 4C).
FIG. 3.
Physical and genetic map of pLS201 (a derivative of pNZ8048 expressing pemI, LSL_1984). The region labeled pemIp contains a putative promoter (TTGCCA-N13-TATAAT) 202 nucleotides upstream of the pemI gene.
FIG. 4.
Confirmation of the curing of pSF118-20 from L. salivarius UCC118. (A) Southern hybridization analysis of L. salivarius UCC118 (lane 1) and its cured derivative LS201 (lane 2). Genomic DNA was either undigested or digested with EcoRV, and then blots were hybridized with a labeled 540-bp PCR amplicon from pSF118-20 as a probe. EcoRV cuts pSF118-20 into two fragments of 7.6 kb and 12.8 kb. Lane M, labeled DNA marker. DNA size markers are indicated. (B) PCR confirmation of the absence of pLS201 in strain LS201. Primers based on LSL_1984 (pemI) were used for PCR amplification. Lane M, DNA size markers; lane 1, negative control; lane 2, L. salivarius UCC118; lane 3, a derivative of L. salivarius UCC118 lacking pSF118-20 (strain LS201). (C) Retention of pSF118-44 (lane 1) and pMP118 (lane 2) by L. salivarius LS201. Lane M, Hyperladder I. Primer pairs YL007-YL008 and YL011-YL012 were used to confirm the retention of pSF118-44 (an 899-bp product should be produced) and pMP118 (a 410-bp product should be produced), respectively.
Properties of the cured derivative strain LS201.
The annotation of pSF118-20 suggested a number of functions conferred by this plasmid. We therefore compared L. salivarius UCC118 with the pSF118-20 cured strain LS201 for a number of properties. No differences in the morphologies of strains UCC118 and LS201 were found under standard culture conditions. Following challenge with 0.1% bile, the survival rates of 24-h-starved cells of UCC118 were 55-, 25-, and 7-fold higher than those of the unstarved control cells at 5, 10, and 30 min postchallenge, respectively. For strain LS201, the survival rate was the same for starved and unstarved cells at all time points. This may be related to the presence of LSL_1973 on pSF118-20, which encodes a general-stress protein belonging to the Gls24 family, which can be induced at the onset of starvation (19). The resistance to UV light of strains UCC118 and LS201 was also investigated, but no differences were detected (data not shown) despite the presence of LSL_1979 on pSF118-20.
Functional analysis of the TA system from pSF118-20.
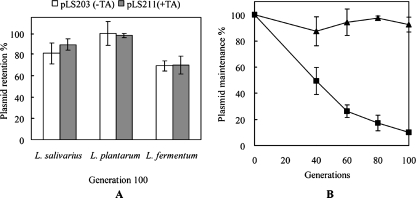
To confirm if the TA locus from pSF118-20 was involved in plasmid maintenance, LSL_1984 and LSL_1985 were cloned as an expression unit (with their presumptive native promoter) into plasmids pLS203 (see below; a plasmid containing the pSF118-20 replicon that is stable in L. salivarius) and pVE6007 (a cloning vector used in L. salivarius UCC118). The segregational stabilities of pLS203 and pLS211 (the pLS203 derivative carrying the TA system) were compared in L. lactis MG1363, L. salivarius LS201, L. plantarum NCIMB8826, and L. fermentum DSM20055 in the absence of antibiotic selection at the optimal growth temperature. The segregational stabilities of pVE6007 and pLS212 (a pVE6007 derivative with a TA system) were investigated in L. salivarius LS201. Transcriptional analysis (by RT-PCR) showed that the TA loci were not transcribed in L. lactis MG1363 (data not shown). Interestingly, the presence of the TA system did not increase the segregational stability of the relatively stable plasmid pLS203 (Fig. 5A) in Lactobacillus species, but it dramatically increased the stability of the unstable plasmid pVE6007 in the L. salivarius UCC118 derivative cured of pSF118-20 (strain LS201; Fig. 5B).
FIG. 5.
Function of the TA system on pSF118-20. (A) Contribution of the TA system to pLS203 maintenance in L. salivarius LS201, L. plantarum NCIMB8826, and L. fermentum DSM20055. Plasmid retention after 100 generations of growth for each strain was compared with that of generation 1. (B) Segregational stability of pVE6007 (▪) and pLS212 (▴; a derivative of pVE6007 containing the TA system from pSF118-20) in L. salivarius LS201 in the absence of antibiotic selection. Plasmid maintenance was measured as the percentage of chloramphenicol-resistant colonies. The data shown represent the mean values of the results from three independent experiments.
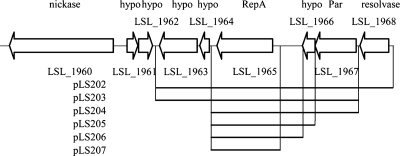
Identification of a minimal stable replicon from pSF118-20.
A series of deletion constructs was made to identify a stable replicon from pSF118-20 (Fig. 6). As shown in Table 4, the pCI341 derivative containing only the repA gene (LSL_1965) of pSF118-20 (i.e., pLS207) could replicate in Lactobacillus plantarum, while more genes were required for this plasmid to replicate in Lactococcus lactis. The presence of genes flanking repA increased the segregational stabilities of the constructs in lactic acid bacteria in the absence of antibiotic selection. However, the putative resolvase (LSL_1968) did not contribute to plasmid stability and could be deleted from the replicon. Therefore, a minimal stable replication region including LSL_1963 to LSL_1967 from pSF118-20 (i.e., pLS203) was identified as the minimal stable replicon for lactic acid bacteria.
FIG. 6.
Identification of a minimal stable replication region from pSF118-20. A series of deletion constructs from pSF118-20 was cloned into the replication probe vector pCI341, yielding pLS202 and pLS204 to pLS206. LSL_1963 to LSL_1967 were cloned into pEM, resulting in pLS203. hypo, hypothetical protein.
TABLE 4.
Segregational stabilities of constructs containing variously sized fragments of the replication region of pSF118-20 in lactic acid bacteria in the absence of antibiotic selection
| Strain | Generation | % Stability at optimal growth temp for constructa:
|
|||||
|---|---|---|---|---|---|---|---|
| pLS202 | pLS203 | pLS204 | pLS205 | pLS206 | pLS207 | ||
| L. lactis MG1363 | 50 | 46 | 3 | 33 | NR | NR | NR |
| 100 | 28 | 1 | 9 | NR | NR | NR | |
| L. plantarum NCIMB8826 | 50 | 100 | 100 | 81 | 72 | 70 | 64 |
| 100 | 100 | 98 | 62 | 57 | 53 | 49 | |
| L. salivarius LS201 | 50 | NT | 86 | NT | NT | NT | NT |
| 100 | NT | 81 | NT | NT | NT | NT | |
| L. fermentum DSM20055 | 50 | NT | 94 | NT | NT | NT | NT |
| 100 | NT | 72 | NT | NT | NT | NT | |
NR, no replication; NT, not tested.
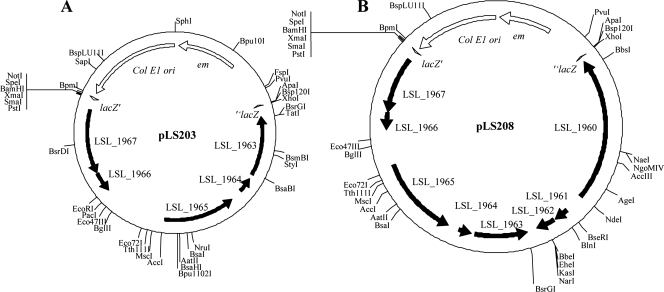
Construction of pLS203 and pLS208.
Based on the minimal stable replicon identified above, two shuttle vectors, pLS203 and pLS208 (Fig. 7A and B), were constructed, which contained either the stable replication region of pSF118-20 (LSL_1963 to LSL_1967) or a region containing both the putative mobilization region and the stable replication region of pSF118-20 (LSL_1960 to LSL_1967), respectively (see Materials and Methods). These plasmids were constructed in a shuttle format to facilitate cloning and manipulation. The direct introduction of pLS203 and pLS208 from E. coli into L. salivarius LS201 resulted in very few transformants. However, pLS203 and pLS208 derived from L. plantarum NCIMB8826 were successfully electroporated into L. salivarius LS201.
FIG. 7.
Construction of two shuttle vectors. (A) pLS203, E. coli pEM vector containing the 4-kb replication region of pSF118-20; (B) pLS208, E. coli pEM vector containing the 7-kb putative mobilization locus and replication region of pSF118-20.
Mobilization of pLS208 between Lactobacillus strains and species.
pLS208 contains both the replication region of pSF118-20 and a putative mobilization locus. Matings between L. salivarius LS201(pLS208) and L. fermentum DSM20055(pNZ8048), L. plantarum NCIMB8826(pLS208) and L. fermentum DSM20055(pNZ8048), and L. plantarum NCIMB8826(pLS208) and L. salivarius JCM1045(pNZ8048) were tested for the mobilization of pLS208 into the recipient cells. L. plantarum NCIMB8826 harbors a conjugative plasmid, pWCFS103 (55), which might conceivably have facilitated the transfer of pLS208. For the donor L. salivarius LS201(pLS208), putative loci related to conjugation are present in pMP118 according to the annotation of the genome of L. salivarius UCC118 (5). Thus, pMP118 might also have been capable of mobilizing pLS208 from L. salivarius LS201 into a recipient.
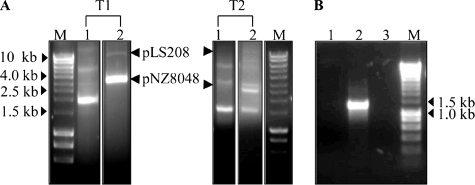
Transconjugants from filter mating experiments were selected on plates containing both Cm and Em. Whereas the transfer of pLS208 from L. salivarius LS201(pLS208) to L. fermentum DSM20055 did not occur, pLS208 was successfully mobilized from L. plantarum NCIMB8826 to L. salivarius JCM1045 and L. fermentum DSM20055 (Fig. 8A). The frequencies of conjugation between L. plantarum NCIMB8826(pLS208) and L. fermentum DSM20055(pNZ8048) and between L. plantarum NCIMB8826(pLS208) and L. salivarius JCM1045(pNZ8048) were 5 × 10−4 and 1.3 × 10−4 transconjugants per donor, respectively. The genotypes and phenotypes of transconjugants were confirmed by plasmid screening for the presence of pLS208, API 50 CH profiling (see Materials and Methods), and sequencing the 16S rRNA gene amplicon of transconjugants (data not shown). PCR was performed to investigate the transfer of pWCFS103 from the donor to the recipient. As shown in Fig. 8B, pWCFS103 was not transferred into the transconjugant.
FIG. 8.
Characterization of pLS208 transconjugants. (A) Plasmid profiles of two representative transconjugants obtained from mating between L. plantarum NCIMB8826(pLS208) and either L. fermentum DSM20055 (pNZ8048) (T1) or L. salivarius JCM1045(pNZ8048) (T2). Lane M, DNA size markers; lane 1, undigested samples; lane 2, PstI-restricted plasmids. (B) Absence of pWCFS103 in representative L. fermentum transconjugant. PCR was performed with primers based on pWCFS103 (FF071, FF072B). Lane 1, template was total genomic DNA of L. fermentum transconjugant; lane 2, template was total genomic DNA of L. plantarum NCIMB8826; lane 3, negative control, distilled water; lane M, Hyperladder I (Bioline).
Production and detection of bioluminescence and GFP in lactobacilli.
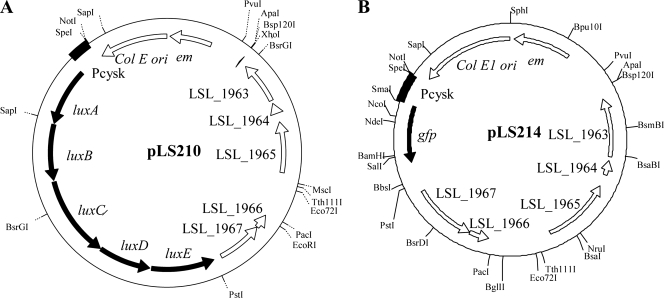
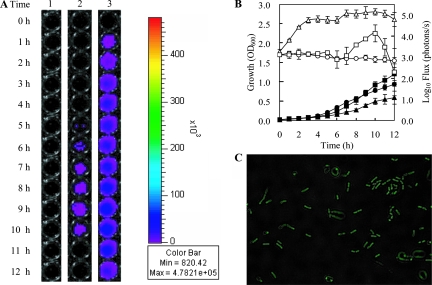
The recombinant plasmid pLS203 was considered potentially suitable for use as a cloning and expression vector for lactobacilli because of its high stability in this species even in the absence of selection (Table 4). For probiotic bacteria like L. salivarius UCC118, how and where it colonizes the GI tract are still largely unknown. The generation of lux- or GFP-tagged bacteria would significantly improve our ability to study the dynamics and microecology of L. salivarius in the GI tract of humans or model animals. We therefore attempted to express both the luciferase marker (lux) and gfp in Lactobacillus strains using pLS203 as a vector. The strategy of expressing lux has been used by colleagues to tag pathogens (3) and other commensal bacteria (45). A synthetic luxABCDE operon encoding luciferase (luxAB) and a fatty acid reductase complex (luxCDE) was obtained from pFT1 (a vector with the backbone of pUC19 containing the luxABCDE operon; I. Monk, unpublished data). The lux cassette was translationally fused to constitutive promoter cysK (LSL_1718) and cloned into pLS203, resulting in pLS210 (Fig. 9A). Bioluminescence was measured during the growth of Lactobacillus strains harboring pLS210. L. fermentum DSM20055(pLS210) showed significant bioluminescence production during a 12-h period of growth (Fig. 10). A low level of bioluminescence was detected from the culture of L. plantarum NCIMB8826(pLS210) for 4 h. No visible bioluminescence could be detected from the culture of L. salivarius LS201(pLS210), even though the lux cassette was proven to be transcribed by end point RT-PCR analysis (data not shown). The highest bioluminescence was detected at late log phase in L. fermentum DSM20055(pLS210).
FIG. 9.
Physical and genetic maps of luciferase-expressing construct pLS210 (A) and gfp+-expressing construct pLS214 (B), both derived from pLS203.
FIG. 10.
Growth of L. salivarius LS201(pLS210), L. plantarum NCIMB8826(pLS210), and L. fermentum DSM20055(pLS210) and the concomitant detection of bioluminescence (A, B). (A) Lane 1, L. salivarius LS201(pLS210); lane 2, L. plantarum NCIMB8826(pLS210); lane 3, L. fermentum DSM20055(pLS210); 0 h, time of inoculation. (B) Closed symbols, growth of Lactobacillus; open symbols, strength of bioluminescence; circle, L. salivarius LS201(pLS210); square, L. plantarum NCIMB8826(pLS210); triangle, L. fermentum DSM20055(pLS210). The error bars indicate the standard deviations of the results from three individual experiments. (C) Expression of gfp+ in L. salivarius LS201 by pLS214. Fluorescence was detected by fluorescence microscopy as described in Materials and Methods.
For the production of GFP in L. salivarius, we constructed a pLS203 derivative designated pLS214 (Fig. 9B), which was designed to express the gfp+ gene from the L. salivarius cysK promoter. Fluorescence was readily detected from stationary-phase cells of L. salivarius LS201(pLS214) (Fig. 10C).
DISCUSSION
The annotation of two endogenous plasmids (pSF118-20 and pSF118-44) from the sequenced probiotic strain L. salivarius UCC118, and the replication analysis of the former, suggest that both plasmids replicate via a theta replication mechanism. In this study, more than half of the 27 L. salivarius strains tested from various origins were shown by PFGE and Southern blotting to harbor plasmids related to repA20 of pSF118-20 or repA44 of pSF118-44. Due to the high stability expected for plasmids which contain theta-type replicons in the appropriate host, these repA20- and repA44-related plasmids are good candidates for developing suitable genetic tools for Lactobacillus strains to investigate their probiotic characteristics. Therefore, a gene cloning and gene expression vector, pLS203, and a mobilizable cloning vector, pLS208, for probiotic lactobacilli were constructed based on the plasmid pSF118-20.
pLS203 replicated in both L. lactis and Lactobacillus strains. Electrotransformation of pLS203 into other Lactobacillus species could be performed to examine the broader replication host range, if required. Segregational stability analysis showed that pLS203 is a stable plasmid in Lactobacillus strains in the absence of antibiotic selection. Novel cloning and gene expression vectors can now be developed based on pLS203 for L. salivarius based upon the high stability in the parental strain. A constitutive gene expression system based on pLS203 was tested for producing bioluminescence and GFP in Lactobacillus. As shown in Fig. 10, the production and intensity of bioluminescence were both species dependent and growth phase dependent in lactobacilli. In the light-emitting reaction, the expression of luxABCDE provides one of three required substrates (a long-chain fatty aldehyde), plus the luciferase enzyme. In the presence of oxygen, the bacterium needs to continuously provide reduced flavin mononucleotide (FMNH2) to support the reaction. Therefore, the reaction is highly dependent on FMNH2 (58). The high production level of luminescence we observed (from exponential-growth-phase cells of L. fermentum DSM20055) may be due to the accumulation of large amounts of NADH and FMNH2 during the log-growth phase. In luminescent bacteria, FMNH2 can be continuously produced from free FMN, catalyzed by NAD(P)H-flavin oxidoreductase (EC 1.6.8.1). However, no gene encoding NAD(P)H:FMN oxidoreductase is present in the genome of L. salivarius UCC118, and L. plantarum WCFS1 encodes two putative NADH-dependent flavin oxidoreductases (data obtained from ERGO, Integrated Genomics, Chicago, IL). The differences in the intensities of the luminescence in L. salivarius, L. plantarum, and L. fermentum could therefore be due to the different reducing powers (NADH, FMNH2) of the cytoplasmic environments in the respective species. L. plantarum NCIMB8826 and L. fermentum DSM20055 were both isolated from saliva, while strain UCC118 was isolated from the GI tract. Though isolation sites for lactobacilli must be treated with caution, this indicates that L. plantarum NCIMB8826 and L. fermentum DSM20055 may have different biological oxidation capabilities than strain UCC118. The production of GFP in L. salivarius confirms that the native cysKp is active in the endogenous host, and pLS203 derivatives harboring this promoter are suitable for expressing heterologous genes. In the case of GFP, this reporter offers attractive prospects for tracking the interaction of this probiotic commensal species with epithelial cells and lymphocytes, potentially in animal models.
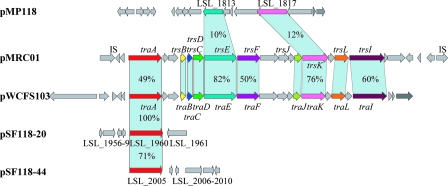
The conjugation between L. plantarum(pLS208) and either L. fermentum(pNZ8048) or L. salivarius JCM1045 (pNZ8048) showed that pSF118-20 is transmissible with the help of a conjugative plasmid within and between Lactobacillus species. Thus, transferring and expressing large DNA clusters in probiotic lactobacilli can be accomplished through bacterial conjugation. L. salivarius JCM1045 is plasmid free except for the megaplasmid (Fig. 2) and is bacteriocin nonproducing (38). Failure of the mobilization of pLS208 from L. plantarum NCIMB8826 to L. salivarius LS201 may be due to the bactericidal effect on the donor of bacteriocin produced by the L. salivarius recipient or the incompatibility of two plasmids containing a similarly functioning traA gene (Fig. 11; see below). The unsuccessful conjugation of pLS208 from L. salivarius LS201 to L. fermentum strongly suggests that the megaplasmid pMP118 is not capable of mobilizing pLS208 and that there are no other conjugation-related genes on the chromosome of L. salivarius LS201 that could help the transfer of pSF118-20. A comparison of genes for conjugal gene transfer in UCC118 (Fig. 11) with two loci of experimentally confirmed transfer capability illustrates that the whole process of conjugation may require more functional genes than are present on pSF118-20, pSF118-44, pMP118, or all acting in concert.
FIG. 11.
Comparison of the conjugal transfer (tra) regions in selected plasmids. pMP118, the megaplasmid from L. salivarius UCC118 (5); pMRC01, the conjugative plasmid from L. lactis DPC3147 (11); pWCFS103, the conjugative plasmid from L. plantarum NCIMB8826 (55); pSF118-20 and pSF118-44, plasmids from L. salivarius UCC118 (5). Percentages of identity in the translated nucleotide sequences of each gene are indicated. The identities of trsB, trsC, trsD, trsJ, and trsL from pMRCO1 and pWCFS103 are 72%, 73%, 80%, 48%, and 44%, respectively.
The curing of pSF118-44 was attempted by growing L. salivarius LS201 at an elevated temperature in order to make a L. salivarius strain harboring only the megaplasmid, but this has been so far unsuccessful. The failure to cure pSF118-44 may be because there are two putative TA systems in this plasmid. Moreover, pSF118-44 harbors a gene cluster (LSL_2026 to LSL_2027) that encodes an osmoprotection uptake system (Opu). Both combined, or either of the TA systems and opu, may make it more difficult to cure pSF118-44 under regular growth conditions. Therefore, a new strategy for curing pSF118- 44 will be required.
TA systems present on pSF118-20 and pSF118-44 are the second-reported plasmid-encoded TA loci in lactobacilli, while the first was from L. plantarum plasmid p256 (53). Bacterial TA systems were initially identified in plasmids (17) and are presumed to maintain the stability of the plasmid in corresponding hosts (24). The employment of standard approaches to cure pSF118-20 from L. salivarius UCC118 was unsuccessful. Curing pSF118-20 by producing antitoxin in trans indicated that the TA system may improve the stability of the plasmid in the host through either killing plasmid-free segregants or by inhibiting cell division (17, 29). The TA system from pSF118-20 contributed to the increased segregational stability of pVE6007 in L. salivarius LS201 (Fig. 5B). This system does not work by killing plasmid-free segregants, as the survival rates of viable cells of strains harboring pVE6007 and pLS212 grown at an elevated temperature were the same. However, the plasmid-encoded TA system from pSF118-20 did not increase the segregational stability of a relatively stable plasmid in the lactobacilli tested (Fig. 5A). We noted that the genes repA20 (LSL_1965) and repA44 (LSL_2000) are 71% identical. It could be that, as suggested (8) for a single resident stable plasmid, the TA systems may act to mediate the exclusion of competing plasmids. Once this hypothesis has been confirmed, the TA system can be applied to cure a plasmid by constructing a compatible plasmid containing the TA system from pSF118-20. Moreover, the TA locus can be used to construct novel gene deletion, disruption, and expression vectors as accomplished for Lactobacillus (63). In L. salivarius, this opens the possibility for the confirmation of a second functioning pemI (LSL_1984) and pemK (LSL_1985) system for exploitation in other lactobacilli.
Supplementary Material
Acknowledgments
This research was supported by Science Foundation Ireland through a Centre for Science, Engineering and Technology award to the Alimentary Pharmabiotic Centre, UCC.
We thank Mary O'Connell Motherway, Ian Monk, and Lothar Steidler for sharing unpublished information. We acknowledge Michael W. Mangan for valuable discussions on the manuscript.
Footnotes
Published ahead of print on 4 April 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aleshin, V. V., E. V. Semenova, V. G. Doroshenko, Y. V. Jomantas, B. V. Tarakanov, and V. A. Livshits. 1999. The broad host range plasmid pLF1311 from Lactobacillus fermentum VKM1311. FEMS Microbiol. Lett. 178:47-53. [DOI] [PubMed] [Google Scholar]
- 2.An, H. Y., and T. Miyamoto. 2006. Cloning and sequencing of plasmid pLC494 isolated from human intestinal Lactobacillus casei: construction of an Escherichia coli-Lactobacillus shuttle vector. Plasmid 55:128-134. [DOI] [PubMed] [Google Scholar]
- 3.Bron, P. A., I. R. Monk, S. C. Corr, C. Hill, and C. G. Gahan. 2006. Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl. Environ. Microbiol. 72:2876-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruand, C., S. D. Ehrlich, and L. Janniere. 1991. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAM beta 1. EMBO J. 10:2171-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claesson, M. J., D. van Sinderen, and P. W. O'Toole. 2007. The genus Lactobacillus-a genomic basis for understanding its diversity. FEMS Microbiol. Lett. 269:22-28. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N. 1993. Bacterial plasmids: their extraordinary contribution to molecular genetics. Gene 135:67-76. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David, S., G. Simons, and W. M. De Vos. 1989. Plasmid transformation by electroporation of Leuconostoc paramesenteroides and its use in molecular cloning. Appl. Environ. Microbiol. 55:1483-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmond, C., R. P. Ross, G. Fitzgerald, and C. Stanton. 2005. Sequence analysis of the plasmid genome of the probiotic strain Lactobacillus paracasei NFBC338 which includes the plasmids pCD01 and pCD02. Plasmid 54:160-175. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 12.Dunne, C., L. Murphy, S. Flynn, L. O'Mahony, S. O'Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly, B. Kiely, E. M. Quigley, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie van Leeuwenhoek 76:279-292. [PubMed] [Google Scholar]
- 13.Fayol-Messaoudi, D., C. N. Berger, M. H. Coconnier-Polter, V. Lievin-Le Moal, and A. L. Servin. 2005. pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 71:6008-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn, S. 2001. Molecular characterisation of bacteriocin producing genes and plasmid encoded functions of the probiotic strain Lactobacillus salivarius subsp. salivarius UCC118. Ph.D. thesis. University College Cork, Cork, Ireland.
- 15.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 18.Gevers, D., G. Huys, and J. Swings. 2003. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol. Lett. 225:125-130. [DOI] [PubMed] [Google Scholar]
- 19.Giard, J.-C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosalbes, M. J., C. D. Esteban, J. L. Galán, and G. Pérez-Martínez. 2000. Integrative food-grade expression system based on the lactose regulon of Lactobacillus casei. Appl. Environ. Microbiol. 66:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 22.Gryczan, T. J., J. Hahn, S. Contente, and D. Dubnau. 1982. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J. Bacteriol. 152:722-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hautefort, I., M. J. Proenca, and J. C. D. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, F., C. Daly, and G. F. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes, F., P. Vos, G. F. Fitzgerald, W. M. de Vos, and C. Daly. 1991. Molecular organization of the minimal replicon of novel, narrow-host-range, lactococcal plasmid pCI305. Plasmid 25:16-26. [DOI] [PubMed] [Google Scholar]
- 27.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansch, A., M. Korakli, R. F. Vogel, and M. G. Ganzle. 2007. Glutathione reductase from Lactobacillus sanfranciscensis DSM20451T: contribution to oxygen tolerance and thiol exchange reactions in wheat sourdoughs. Appl. Environ. Microbiol. 73:4469-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 30.Kanatani, K., and M. Oshimura. 1994. Plasmid-associated bacteriocin production by a Lactobacillus plantarum strain. Biosci. Biotechnol. Biochem. 58:2084-2086. [DOI] [PubMed] [Google Scholar]
- 31.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein, G., A. Pack, C. Bonaparte, and G. Reuter. 1998. Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 41:103-125. [DOI] [PubMed] [Google Scholar]
- 33.Klein, J. R., C. Ulrich, and R. Plapp. 1993. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid 30:14-29. [DOI] [PubMed] [Google Scholar]
- 34.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 35.Lee, J.-H., J. S. Halgerson, J.-H. Kim, and D. J. O'Sullivan. 2007. Comparative sequence analysis of plasmids from Lactobacillus delbrueckii and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 73:4417-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leenhouts, K. J., B. Tolner, S. Bron, J. Kok, G. Venema, and J. F. M. L. Seegers. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55-66. [DOI] [PubMed] [Google Scholar]
- 37.Lessard, P., X. O'Brien, D. Currie, and A. Sinskey. 2004. pB264, a small, mobilizable, temperature sensitive plasmid from Rhodococcus. BMC Microbiol. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Y., C. Canchaya, F. Fang, E. Raftis, K. A. Ryan, J.-P. van Pijkeren, D. van Sinderen, and P. W. O'Toole. 2007. Distribution of megaplasmids in Lactobacillus salivarius and other lactobacilli. J. Bacteriol. 189:6128-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, C. F., Z. F. Fung, C. L. Wu, and T. C. Chung. 1996. Molecular characterization of a plasmid-borne (pTC82) chloramphenicol resistance determinant (cat-TC) from Lactobacillus reuteri G4. Plasmid 36:116-124. [DOI] [PubMed] [Google Scholar]
- 40.Lin, C. F., J. L. Ho, and T. C. Chung. 2001. Characterization of the replication region of the Lactobacillus reuteri plasmid pTC82 potentially used in the construction of cloning vector. Biosci. Biotechnol. Biochem. 65:1495-1503. [DOI] [PubMed] [Google Scholar]
- 41.Ljungh, A., and T. Wadstrom. 2006. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 7:73-89. [PubMed] [Google Scholar]
- 42.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenzie, T., T. Hoshino, T. Tanaka, and N. Sueoka. 1986. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid 15:93-103. [DOI] [PubMed] [Google Scholar]
- 44.Miller, K. W., P. Ray, T. Steinmetz, T. Hanekamp, and B. Ray. 2005. Gene organization and sequences of pediocin AcH/PA-1 production operons in Pediococcus and Lactobacillus plasmids. Lett. Appl. Microbiol. 40:56-62. [DOI] [PubMed] [Google Scholar]
- 45.Oozeer, R., J. P. Furet, N. Goupil-Feuillerat, J. Anba, J. Mengaud, and G. Corthier. 2005. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 71:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otto, R., W. M. De Vos, and J. Gavrieli. 1982. Plasmid DNA in Streptococcus cremoris Wg2: influence of pH on selection in chemostats of a variant lacking a protease plasmid. Appl. Environ. Microbiol. 43:1272-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, W. J., K. H. Lee, J. M. Lee, H. J. Lee, J. H. Kim, J. H. Lee, H. C. Chang, and D. K. Chung. 2004. Characterization of pC7 from Lactobacillus paraplantarum C7 derived from Kimchi and development of lactic acid bacteria-Escherichia coli shuttle vector. Plasmid 52:84-88. [DOI] [PubMed] [Google Scholar]
- 48.Pavlova, S. I., A. O. Kilic, L. Topisirovic, N. Miladinov, C. Hatzos, and L. Tao. 2002. Characterization of a cryptic plasmid from Lactobacillus fermentum KC5b and its use for constructing a stable Lactobacillus cloning vector. Plasmid 47:182-192. [DOI] [PubMed] [Google Scholar]
- 49.Posno, M., R. J. Leer, N. van Luijk, M. J. van Giezen, P. T. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 52.Shareck, J., Y. Choi, B. Lee, and C. B. Miguez. 2004. Cloning vectors based on cryptic plasmids isolated from lactic acid bacteria: their characteristics and potential applications in biotechnology. Crit. Rev. Biotechnol. 24:155-208. [DOI] [PubMed] [Google Scholar]
- 53.Sorvig, E., M. Skaugen, K. Naterstad, V. G. Eijsink, and L. Axelsson. 2005. Plasmid p256 from Lactobacillus plantarum represents a new type of replicon in lactic acid bacteria, and contains a toxin-antitoxin-like plasmid maintenance system. Microbiology 151:421-431. [DOI] [PubMed] [Google Scholar]
- 54.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 55.van Kranenburg, R., N. Golic, R. Bongers, R. J. Leer, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Functional analysis of three plasmids from Lactobacillus plantarum. Appl. Environ. Microbiol. 71:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Pijkeren, J.-P., C. Canchaya, K. A. Ryan, Y. Li, M. J. Claesson, B. Sheil, L. Steidler, L. O'Mahony, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 72:4143-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Pijkeren, J. P. 2007. Functional characterization of surface components in Lactobacillus salivarius UCC118. Ph.D. thesis. University College Cork, Cork, Ireland.
- 58.Vetrova, E. V., N. S. Kudryasheva, and V. A. Kratasyuk. 2007. Redox compounds influence on the NAD(P)H:FMN-oxidoreductase-luciferase bioluminescent system. Photochem. Photobiol. Sci. 6:35-40. [DOI] [PubMed] [Google Scholar]
- 59.Vogel, R. F., M. Lohmann, A. N. Weller, M. Hugas, and W. P. Hammes. 1991. Structural similarity and distribution of small cryptic plasmids of Lactobacillus curvatus and L. sake. FEMS Microbiol. Lett. 68:183-190. [DOI] [PubMed] [Google Scholar]
- 60.Wang, T. T., and B. H. Lee. 1997. Plasmids in Lactobacillus. Crit. Rev. Biotechnol. 17:227-272. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto, N., and T. Takano. 1996. Isolation and characterization of a plasmid from Lactobacillus helveticus CP53. Biosci. Biotechnol. Biochem. 60:2069-2070. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, X. Z., X. Yan, Z. L. Cui, Q. Hong, and S. P. Li. 2006. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 34:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.