Abstract
Viruses are believed to be significant pathogens for phytoplankton. Usually, they infect a single algal species, and often their infection is highly strain specific. However, the detailed molecular background of the strain specificity and its ecological significance have not been sufficiently understood. Here, we investigated the temporal changes in viral RNA accumulation and virus-induced cell lysis using a bloom-forming dinoflagellate Heterocapsa circularisquama and its single-stranded RNA virus, HcRNAV. We observed at least three host response patterns to virus inoculation: sensitive, resistant, and delayed lysis. In the sensitive response, the host cell culture was permissive for viral RNA replication and apparent cell lysis was observed; in contrast, resistant cell culture was nonpermissive for viral RNA replication and not lysed. In the delayed-lysis response, although viral RNA replication occurred, virus-induced cell lysis was faint and remarkably delayed. In addition, the number of infectious virus particles released to the culture supernatant at 12 days postinoculation was comparable to that of the sensitive strain. By further analysis, a few strains were characterized as variants of the delayed-lysis strain. These observations indicate that the response of H. circularisquama to HcRNAV infection is highly diverse.
Viruses are enormously abundant in seawater and are believed to be significant pathogens of the planktonic primary producers (phytoplankton) (27). Most are DNA viruses, including cyanophages that infect cyanobacteria (19) and phycodnaviruses that infect eukaryotic algae (10). Recent investigations show that previously unknown RNA viruses (RNAV) also infect marine unicellular algae (1, 5, 16, 23, 24, 26, 32). HaRNAV (16, 28) and RsRNAV (23, 26) are positive-sense single-stranded RNA viruses that infect the toxic-bloom-forming microalga Heterosigma akashiwo (Raphidophyceae) and the bloom-forming diatom Rhizosolenia setigera (Bacillariophyceae), respectively. Although the genomes of HaRNAV and RsRNAV have some characteristics similar to those of viruses in the proposed order Picornavirales, phylogenetic analysis based on the deduced amino acid sequence of the RNA-dependent RNA polymerase domain suggests that these two viruses have an independent lineage that evolved from a deep internal branch in Picornavirales (8, 26). HcRNAV belongs to a previously unrecognized positive-sense single-stranded RNA virus group and infects the bloom-forming bivalve-killing dinoflagellate Heterocapsa circularisquama (Dinophyceae) (26, 32). MpRV is a double-stranded RNA virus in the family Reoviridae that infects the bloom-forming cosmopolitan photosynthetic protist Micromonas pusilla (Prasinophyceae) (1, 5). Observations of virus-induced cell lysis indicate that these RNA viruses infect a single algal species and that their infection is highly strain specific (5, 23, 28, 32). To estimate the ecological impact of these viruses on the population dynamics of phytoplankton, a detailed understanding of the strain specificity is essential.
In a previous study, 107 clonal HcRNAV strains were isolated from nine different locations in western Japan during 2000 to 2001 and independently inoculated into 53 clonal H. circularisquama strains (collected during 1994 to 2001 from eight locations in western Japan) (32). Observation of virus-induced cell lysis demonstrated HcRNAV and H. circularisquama strains were categorized into two (CY and UA) and three (A, B, and C) distinct types, respectively (32). Type B host strains exhibited apparent cell lysis (80 to 90% decline in cell number) when inoculated with type CY HcRNAV, but not with type UA HcRNAV. In contrast, type C host strains exhibited apparent cell lysis when inoculated with type UA HcRNAV, but not with type CY HcRNAV. However, type A host strains did not exhibit apparent cell lysis when inoculated with any of the 107 clonal HcRNAV strains tested (32).
In this study, to gain further insight into the host strain specificity of HcRNAV, temporal changes in host cell number and intracellular viral RNA accumulation of representative host-virus combinations were analyzed. Here, we describe the high diversity of host response patterns to virus infection and discuss its ecological implication for the population dynamics of phytoplankton.
MATERIALS AND METHODS
Hosts and viruses.
The origins of the virus isolates (HcRNAV34 and HcRNAV109) and Heterocapsa circularisquama isolates (HU9433-P, HCLG-1, and MZ2) used in this study were previously reported (Table 1) (32); originally, they were all clonal. Eight strains of H. circularisquama (05HC01 to 05HC08) were isolated from Ago Bay (Mie Prefecture, Japan) in 2005. The samples for host isolation were sent to the laboratory without fixation at 4°C, and isolation of the H. circularisquama strains was conducted within 30 h of collection. Cell cultures were grown in modified SWM3 medium enriched with 2 nM Na2SeO3 (7, 13, 14) and incubated under a 12-h light (12-h dark cycle); the light (130 to 150 μmol photons m−2 s−1) was provided by cool white fluorescent illumination at 20°C. Until use, the clonal host cultures (i.e., those originated from a single cell) were maintained under the laboratory conditions mentioned above in the absence of HcRNAV. From 2001 to the present, host cell cultures HCLG-1, HU9433-P, and MZ2 have exhibited the same response to virus infection at the light-microscopic level.
TABLE 1.
Relationships of strains of Heterocapsa circularisquama and HcRNAV used in this study
| Host strain | Cell lysis induced bya:
|
|
|---|---|---|
| HcRNAV34 | HcRNAV109 | |
| HCLG-1 | − | + |
| HU9433-P | + | − |
| MZ2 | − | − |
+, virus-host combination in which apparent cell lysis (80 to 90% decline in cell number) was observed. −, virus-host combination in which apparent cell lysis was not observed.
To assess the variety among cells in a clonal host culture, we conducted an additional experiment comparing the virus sensitivity of recloned cultures originated from H. circularisquama HU9433-P (type C strain sensitive to HcRNAV34) and MZ2 and 05HC08 (delayed-lysis strains; see below). These recloned cultures were incubated for 18 days in 48-well culture plates; then, 15 randomly selected cultures (from each strain) were maintained in 50 ml medium for 1 to 3 weeks. Virus susceptibility tests were conducted as described below.
Because HcRNAV infection is strain specific (i.e., each HcRNAV isolate has a type-specific host range) (32), HcRNAV34 and HcRNAV109 were maintained using their suitable hosts, HU9433-P and HCLG-1, respectively (Table 1). Each virus stock was inoculated into a fresh culture of its suitable H. circularisquama strain and incubated until lysis. The lysate was filtered through a 0.2-μm-pore-size polycarbonate membrane filter (Whatman, Middlesex, United Kingdom) to remove host cell debris. The viral titer of the filtrate was estimated by means of the extinction dilution method as described previously (32). An exponentially growing culture of H. circularisquama was inoculated with the virus lysate at a multiplicity of infection of 10 and incubated under the conditions mentioned above.
Host cell counts.
The H. circularisquama cell count was performed immediately after sampling without fixation using an Epics XL-MCL flow cytometer (Beckman Coulter, Inc., Fullerton, CA) equipped with a 488-nm air-cooled argon ion laser (15 mW) and standard filters. Fluorescent microspheres (FlowCount, 10 μm; Beckman Coulter, Inc.) were added as an internal reference. Readings were collected for 1 min in the logarithmic mode and analyzed using EXPO32ADC software (Beckman Coulter, Inc.). Chlorophyll autofluorescence, total count, and side scatter were recorded. A gate was created around the areas in which intact H. circularisquama cells were observed in the chlorophyll autofluorescence versus the side scatter histogram.
Chlorophyll fluorescence intensities were analyzed to estimate the influence of virus infection using a Turner Designs fluorometer (model 10-005R) equipped with a 436-nm excitation filter and a >650-nm emission filter.
Northern blot analysis.
An aliquot of a cell suspension (1.5 ml) was removed from the culture and centrifuged (13,000 × g for 3 min), and the pellets were stored at −80°C until RNA extraction. Frozen cells were homogenized with plastic pestles in 100 μl RNase-free water and 40 μl RNA extraction buffer (15). The aqueous phase was extracted using phenol prior to ethanol precipitation. Northern blot analysis of the purified RNA was performed as previously described (9). Viral negative-stranded RNA-specific digoxigenin-labeled RNA probes were transcribed from the previously constructed plasmid, pBSSK+MCP (22). RNA signals were detected using a luminescence image analyzer (LAS-3000 mini; Fuji Photo Film, Tokyo, Japan).
Virus titer.
The virus titer was estimated as described below. The lysate that was filtrated through a 0.2-μm-pore-size polycarbonate membrane filter was diluted with SWM3 medium in a series of 10−1 to 10−10 dilutions, and aliquots of each dilution were added to 8 wells in a 96-well microtiter plate containing exponentially growing H. circularisquama HU9433-P culture. The cultures in the micotiter plate were incubated and checked daily for 14 days for cell lysis (29). The dilution of the culture wells in which cell lysis occurred was scored, and the most probable number of infectious viral units was determined using the BASIC program (25).
RESULTS AND DISCUSSION
Characterization of the host responses to HcRNAV inoculation.
Algal host strains MZ2, HCLG-1, and HU9433-P were used as representatives of H. circularisquama types A, B, and C, respectively (Table 1) (32). Representative virus strains were HcRNAV109 (type CY) and HcRNAV34 (type UA) (Table 1) (32). Full nucleotide sequences of HcRNAV109 and HcRNAV34 were determined; they are ∼97% identical to each other at the nucleotide sequence level (24).
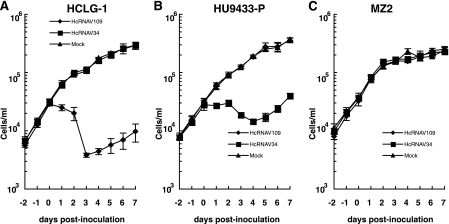
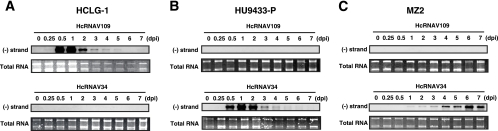
Following inoculation of HcRNAV109 into HCLG-1, a decline in host cell number was observed between 1 to 3 days postinoculation (dpi) (Fig. 1A). A similar decline in cell number was also observed following inoculation of HcRNAV34 into HU9433-P (Fig. 1B). After 3 to 4 dpi, regrowth of H. circularisquama cells was observed, even in the presence of infectious viruses in the cultures (Fig. 1A and B). To further investigate this phenomenon, total RNA was extracted from these cells and the accumulation of genome-length negative-stranded RNA was measured (Fig. 2). An increase in the viral negative-stranded RNA accumulation was not observed in the regrowing cells, indicating that viral replication did not occur at this stage (3 to 7 dpi) (Fig. 2A and B). These data apparently show that the cells are changing upon infection; the survivor cells can grow even under the condition of being exposed to HcRNAV attacks. Although the mechanism supporting these phenomena has not been elucidated, similar phenomena were observed in bacterial cultures inoculated with a bacteriophage (18) and in algal cultures inoculated with a DNA virus (34). Just recently, we found that viral RNA replication does not occur in the survivor cells physically transfected with viral RNA (Y. Tomaru, H. Mizumoto, and K. Nagasaki, unpublished data); these data strongly support the hypothesis of rapid cell change (i.e., the dominance of resistance-expressing cells) upon infection.
FIG. 1.
Change in the abundance of Heterocapsa circularisquama cells. Algal cell numbers were measured using flow cytometry. Algal host strains HCLG-1 (A), HU9433-P (B), and MZ2 (C) were inoculated with HcRNAV109 and HcRNAV34 at day 0. As a mock control, sterilized SWM3 medium was added to a cell culture. The assays were performed at least three times. The averages (symbols) and standard deviations (error bars) calculated from three experiments are shown in the graphs.
FIG. 2.
Accumulation of negative-stranded RNA of HcRNAV in Heterocapsa circularisquama strains HCLG-1 (A), HU9433-P (B), and MZ2 (C) inoculated with HcRNAV34 or HcRNAV109. Total RNA was extracted from H. circularisquama cells, separated by gel electrophoresis, and blotted onto membranes. The membranes were then probed with strand-specific digoxigenin-labeled RNA probes (22). Total RNA stained with SYBR gold (Molecular Probes, OR) is also shown. The assays were performed at least twice.
On the other hand, the growth of H. circularisquama cells was not affected in the HU9433-P culture (type C) inoculated with HcRNAV109 (type CY) or in the HCLG-1 culture (type B) inoculated with HcRNAV34 (type UA) (Fig. 1A and B).
When virus-induced cell lysis was observed (Fig. 1A and B, HCLG-1 inoculated with HcRNAV109 and HU9433-P inoculated with HcRNAV34), accumulation of viral negative-stranded RNA was clearly observed at 12 h postinoculation (0.5 dpi), peaked at 1 dpi, and decreased at 2 dpi (Fig. 2A and B). Based on this observation, the host response patterns to HcRNAV inoculation were defined as sensitive. In contrast, when no apparent virus-induced cell lysis was observed (Fig. 1A and B, HCLG-1 inoculated with HcRNAV34 and HU9433-P inoculated with HcRNAV109), no accumulation of viral negative-stranded RNA was detected throughout this experimental period (until 7 dpi) (Fig. 2A and B). The patterns of host response to HcRNAV inoculation were defined as resistant.
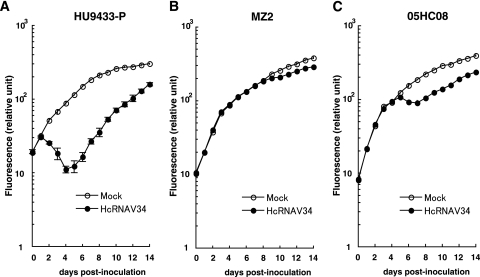
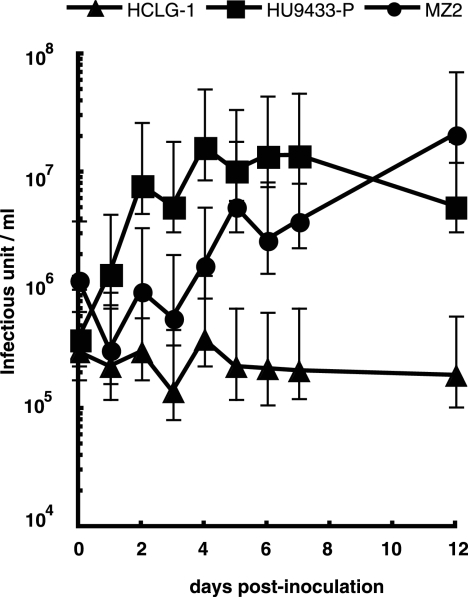
When H. circularisquama strain MZ2 was inoculated with HcRNAV109, no decline in cell number or accumulation of viral negative-stranded RNA was observed (Fig. 1C and 2C); this is consistent with the resistant response mentioned above. On the other hand, when MZ2 was inoculated with HcRNAV34, there was also no apparent decline in cell number until 7 dpi (Fig. 1C); nevertheless, as shown in Fig. 2C, accumulation of viral negative-stranded RNA was observed from 1 dpi (very faint band) and peaked at 6 to 7 dpi. To further investigate the effect of the inoculation of HcRNAV34 into the MZ2 culture, chlorophyll fluorescence intensities were extendedly measured until 14 dpi (Fig. 3B). Compared with mock-inoculated MZ2 culture (control), the chlorophyll fluorescence intensity of the MZ2 culture inoculated with HcRNAV34 showed a slight growth inhibition after 8 dpi (Fig. 3B). In contrast to the HU9433-P culture (Fig. 3A), the chlorophyll fluorescence intensity of the HCLG-1 culture was not affected by inoculation with HcRNAV34 throughout the experimental period (until 14 dpi) (data not shown). These observations indicate that strain MZ2 is permissive for intracellular replication of HcRNAV34. To determine whether progeny virions were released from the MZ2 cells after inoculation with its suitable virus strain, the virus titer (most probable number) in the culture supernatant was measured (Fig. 4). The results showed that the number of infectious HcRNAV34 particles released into the MZ2 culture supernatant at 12 dpi was comparable to that of the sensitive host strain (HU9433-P inoculated with HcRNAV34), although the increase in the viral titer was delayed (Fig. 4). In contrast, no increase in the virus titer in the culture supernatant was observed in HCLG-1 inoculated with HcRNAV34 (Fig. 4). These observations indicate that strain MZ2 supports the reproduction of HcRNAV34. Therefore, the response observed in the MZ2 culture was concluded to differ from both the sensitive and resistant responses and was termed “delayed lysis.”
FIG. 3.
Change in chlorophyll fluorescence intensity (relative units) of MZ2 cultures inoculated with or without HcRNAV34. Chlorophyll fluorescence intensity was measured using a chlorophyll fluorometer. The averages (symbols) and standard deviations (error bars) calculated from four experiments are shown in the graph.
FIG. 4.
Changes in virus titer in culture supernatants of Heterocapsa circularisquama. Strains HCLG-1, HU9433-P, and MZ2 were inoculated with equivalent infectious units of HcRNAV34. Error bars indicate 95% confidence intervals. The assays were performed twice, and similar results were obtained.
Variations in the delayed-lysis response.
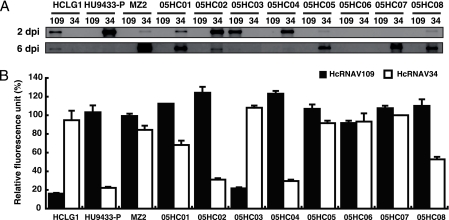
An additional eight H. circularisquama strains (05HC01 to 05HC08) were newly isolated from Ago Bay, Japan, in 2005 and inoculated with virus strains HcRNAV109 and HcRNAV34, and then viral negative-stranded RNA accumulation and the effect of virus inoculation were examined (Fig. 5). Of the eight isolates tested, two were sensitive to HcRNAV34 (05HC02 and 05HC04; similar to HU9433-P); one was sensitive to HcRNAV109 (05HC03; similar to HCLG-1); four (05HC01, -05, -07, and -08) were defined as delayed-lysis type because of the dense accumulation of viral negative-stranded RNA at 6 dpi, which is similar to MZ2; and one was resistant to both HcRNAV34 and HcRNAV109 (05HC06), the virus sensitivity of which is of much interest (Fig. 5).
FIG. 5.
Viral negative-stranded RNA accumulation at 2 and 6 dpi (A) and relative chlorophyll fluorescence intensity at 6 dpi (B) in newly isolated Heterocapsa circularisquama strains (05HC01 to 05HC08). Chlorophyll fluorescence intensity was analyzed using a chlorophyll fluorometer. Relative chlorophyll intensity at 6 dpi was expressed as the percentage of chlorophyll intensity compared to that of the mock culture. The averages plus standard deviations (error bars) calculated from three experiments are shown in the graph.
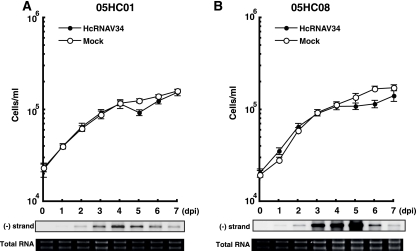
Because strains 05HC05 and 05HC07 showed similar patterns to MZ2 in both viral negative-stranded RNA accumulation (Fig. 5A) and the relative fluorescence unit (Fig. 5B), no further analysis of them was conducted in this study. Although strains 05HC01 and 05HC08 had viral RNA accumulation patterns similar to the delayed-lysis strains (MZ2, 05HC05, and 05HC07) (Fig. 5A), the decreases in chlorophyll fluorescence intensity were more moderate (Fig. 5B). Hence, in order to determine how different the responses of 05HC01 and 05HC08 were from MZ2, the temporal change in host cell number and viral negative-stranded RNA accumulation of the first two strains were measured in time course experiments (Fig. 6) and compared to MZ2 (Fig. 1C and 2C). Compared to strain MZ2 inoculated with HcRNAV34 (Fig. 2C), viral negative-stranded RNA accumulation of 05HC01 and 05HC08 peaked 1 to 2 days earlier (4 to 5 dpi) (Fig. 6). In addition, in the cases of 05HC01 and 05HC08, a faint decrease in cell abundance (compared to control cultures) was observed at 5 to 6 dpi (Fig. 6), which was not detected in MZ2 (Fig. 1C). Comparison of the chlorophyll fluorescence intensities among the HU9433-P, MZ2, and 05HC08 cultures inoculated with HcRNAV34 supported this observation (Fig. 3). Based on these observations, strains 05HC01 and 05HC08 were considered to be variants of the delayed-lysis strain, showing the high diversity of host response to HcRNAV infection.
FIG. 6.
Changes in cell abundance and accumulation patterns of negative-stranded RNA of HcRNAV in Heterocapsa circularisquama strains 05HC01 (A) and 05HC08 (B). For others, refer to Fig. 1 and 3.
Homogeneity of the host cell cultures.
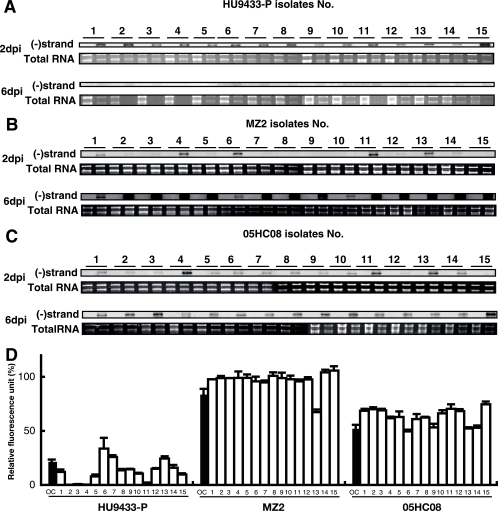
It may be possible that delayed-lysis strains are a “mixture” of genetically heterogeneous cells (containing both resistant cells and sensitive cells), although they were originally clonal. If this is true, both sensitive isolates (showing responses similar to those of HU9433-P inoculated with HcRNAV34) and resistant isolates (showing responses similar to those of HCLG-1 inoculated with HcRNAV34) could have been obtained from the cultures of MZ2 and 05HC08. To verify this possibility, we compared the virus sensitivity of recloned cultures originated from H. circularisquama HU9433-P (type C strain sensitive to HcRNAV34) and MZ2 and 05HC08 (delayed-lysis strains; see above). As shown in Fig. 7, accumulation of viral negative-stranded RNA was detected in all recloned cultures at 2 and/or 6 dpi (Fig. 7A to C). The results also indicated that (i) apparent differences in chlorophyll fluorescence intensities at 6 dpi among the three isolate groups exist (Fig. 7D), (ii) accumulation of negative-sense RNA occurred more quickly in HU9433-P-originated isolates than in those originated from MZ2 and 05HC08 (Fig. 7A to C), and (iii) a little variety was observed in MZ2-originated clonal isolates concerning negative-sense RNA accumulation rates (Fig. 7B), but levels of chlorophyll fluorescence intensity at 6 dpi were almost similar (Fig. 7D). This observation supported our indication that the host cultures are all clonal and that host responses to HcRNAV infection are diverse among host cultures. Alternatively, it may also be possible that the variation is due to preincubation for about a month prior to the test. Based on this observation, however, the delayed-lysis strains (MZ2 and 05HC08) were shown not to be a “mixture” of resistant clones and sensitive clones.
FIG. 7.
Susceptibility to HcRNAV34 of Heterocapsa circularisquama cell cultures recloned from original strains (HU9433-P [a sensitive host strain] and MZ2 and 05HC08 [delayed-lysis host strains]). Viral negative-stranded [(−)strand] RNA accumulation at 2 and 6 dpi in 15 cultures (lanes 1 to 15) recloned from original strains HU9433-P (A), MZ2 (B), and 05HC08 (C). For others, refer to Fig. 2. (D) Relative chlorophyll fluorescence intensity of recloned cell cultures (bars 1 to 15) and each original culture (OC) at 6 dpi. Chlorophyll fluorescence intensity was analyzed using a chlorophyll fluorometer. Relative chlorophyll intensity was expressed as the percentage of chlorophyll intensity relative to those of the corresponding host cultures without virus inoculation. The averages plus standard deviations (error bars) of four experiments are shown in this graph.
Implication.
In a previous study conducted by Tomaru et al. (32), the type A H. circularisquama strains did not exhibit apparent cell lysis when inoculated with any of the 107 clonal HcRNAV strains. Based solely on this result, type A strains cannot be categorized as a host for HcRNAV. However, a detailed analysis of viral RNA accumulation clearly showed that MZ2 (a representative of H. circularisquama type A) supports replication of HcRNAV34 (Fig. 2C and 4). Characterization of the delayed-lysis response will be of value in gaining not only molecular-level insights into the strain specificity of single-stranded RNA viruses infecting microalgae but also insights into the ecological interaction between algal hosts and their viruses in the marine environment as discussed below.
A general hypothesis is that viruses have important roles in determining the dynamics of algal blooms (3, 4, 12, 20, 30, 31, 33). One of the strongest lines of evidence for this is that a transient increase in algal abundance is closely followed by a peak in abundance of infectious viruses in the water column and sediment (31, 33). This hypothesis is also supported by cell culture studies that show a decline in algal host abundance associated with an increase in extracellular infectious virus abundance (2, 6, 32). Here, it is noticeable that although the culture of delayed-lysis H. circularisquama strain MZ2 did not show quick and severe HcRNAV-induced cell lysis (Fig. 1C and 2C), progeny viruses were produced at a productivity level comparable to that of the sensitive host strains (Fig. 4). If delayed-lysis host strains have an advantage under conditions favoring bloom formation, it is possible that the viral impact on bloom dynamics has been overestimated. However, because most of the H. circularisquama strains isolated from natural blooms were sensitive to HcRNAV (32), this possibility is unlikely. Results of our recent survey were also inconsistent with this possibility, as we have observed that 85% of H. circularisquama strains (62 out of 73 clonal strains isolated from Ago Bay, Japan, in 2006) showed apparent virus-induced cell lysis when inoculated with either HcRNAV109 or HcRNAV34 (H. Mizumoto, Y. Tomaru, and K. Nagasaki, unpublished data). Both HcRNAV type CY and type UA were isolated from the water samples, but viruses lytic to strain MZ2 were not discovered from samples from Ago Bay in 2006 (H. Mizumoto, Y. Tomaru, and K. Nagasaki, unpublished data). Then, why were the delayed-lysis strains isolated at very low frequencies from natural waters, even in the presence of viruses causing cell lysis of the sensitive host strains? One possible explanation is that they may be placed at a physiological disadvantage in the ambient conditions favoring bloom formation, as has been proposed for the relationship between bacteria and bacteriophages (11, 17, 21). Further surveys determining the proportion of hosts exhibiting various responses to HcRNAV will be essential for the interpretation of the above results.
In this study, the patterns of viral negative-stranded RNA accumulation apparently differed between sensitive and delayed-lysis host strains (Fig. 2 and 5). Based on these observations, it is possible that the efficiency of proliferation (including viral RNA replication and/or viral entry into the host cell) defines the host's response to HcRNAV infection, where early and quick virus proliferation induces intensive cell lysis, while late and slow virus proliferation results in weak and late cell lysis. In addition, as discussed above, the host response to viral infection may be affected by the physiological activity of the host strains. To understand in more detail the ecological implication of HcRNAV for the population dynamics of H. circularisquama, analysis of the biological relevance of the efficiency of HcRNAV replication, host response, and physiological activity in H. circularisquama strains will be required.
Acknowledgments
We are grateful to J. L. Van Etten (University of Nebraska) for his critical reading of this paper and to M. Yamaguchi and S. Nagai (National Research Institute of Fisheries and Environment of Inland Sea, Fisheries Research Agency, Japan) for their helpful advice. We also thank I. Imai (Kyoto University, Japan) for providing the HU9433-P culture and N. Hata (Mie Prefectural Science and Technology Promotion Center, Japan) for providing water and sediment samples.
This work was partially supported by funding from the Fisheries Agency of Japan; the Ministry of Agriculture, Forestry, and Fisheries of Japan; and the Ministry of Education, Science, Sports, and Culture Grant-in-Aid for JSPS Fellows.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Attoui, H., F. M. Jaafar, M. Belhouchet, P. de Micco, X. de Lamballerie, and C. P. Brussaard. 2006. Micromonas pusilla reovirus: a new member of the family Reoviridae assigned to a novel proposed genus (Mimoreovirus). J. Gen. Virol. 87:1375-1383. [DOI] [PubMed] [Google Scholar]
- 2.Baudoux, A. C., and C. P. Brussaard. 2005. Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology 341:80-90. [DOI] [PubMed] [Google Scholar]
- 3.Bratbak, G., J. K. Egge, and M. Heldal. 1993. Viral mortality of marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93:39-48. [Google Scholar]
- 4.Brussaard, C. P., R. S. Kempers, A. J. Kop, R. Riegman, and M. Heldal. 1996. Virus-like particles in a summer bloom of Emiliania huxleyi in the North Sea. Aquat. Microb. Ecol. 10:105-113. [Google Scholar]
- 5.Brussaard, C. P., A. A. Noordeloos, R. A. Sandaa, M. Heldal, and G. Bratbak. 2004. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319:280-291. [DOI] [PubMed] [Google Scholar]
- 6.Brussaard, C. P. D. 2004. Viral control of phytoplankton populations: a review. J. Eukaryot. Microbiol. 51:125-138. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L. C. M., T. Edelstein, and J. McLachlan. 1969. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 8.Culley, A. I., A. S. Lang, and C. A. Suttle. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795-1798. [DOI] [PubMed] [Google Scholar]
- 9.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 10.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 12.Hennes, K. P., and M. Simon. 1995. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl. Environ. Microbiol. 61:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai, I., S. Itakura, Y. Matsuyama, and M. Yamaguchi. 1996. Selenium requirement for growth of a novel red tide flagellate Chattonella verruculosa (Raphidophyceae) in culture. Fish. Sci. 62:834-835. [Google Scholar]
- 14.Itoh, K., and I. Imai. 1987. Raphidophyceae, p. 122-130. In Japan Fisheries Resource Conservation Association (ed.), A guide for studies of red tide organisms. Shuwa, Tokyo, Japan.
- 15.Kroner, P., D. Richards, P. Traynor, and P. Ahlquist. 1989. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 63:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 17.Lenski, R. E. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T14. Evolution 42:425-432. [DOI] [PubMed] [Google Scholar]
- 18.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann, N. H. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17-34. [DOI] [PubMed] [Google Scholar]
- 20.Martínez, J. M., D. C. Schroeder, A. Larsen, G. Bratbak, and W. H. Wilson. 2007. Molecular dynamics of Emiliania huxleyi and cooccurring viruses during two separate mesocosm studies. Appl. Environ. Microbiol. 73:554-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middelboe, M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114-124. [DOI] [PubMed] [Google Scholar]
- 22.Mizumoto, H., Y. Tomaru, Y. Takao, Y. Shirai, and K. Nagasaki. 2007. Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection. J. Virol. 81:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagasaki, K., Y. Tomaru, N. Katanozaka, Y. Shirai, K. Nishida, S. Itakura, and M. Yamaguchi. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagasaki, K., Y. Shirai, Y. Takao, H. Mizumoto, K. Nishida, and Y. Tomaru. 2005. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 71:8888-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishihara, T., N. Kurano, and S. Shinoda. 1986. Calculation of most probable number for enumeration of bacteria on a micro-computer. Jpn. J. Toxicol. Environ. Health 32:226-228. [Google Scholar]
- 26.Shirai, Y., Y. Takao, H. Mizumoto, Y. Tomaru, D. Honda, and K. Nagasaki. 2006. Genomic and phylogenetic analysis of a single-stranded RNA virus infecting Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Assoc. UK 86:475-483. [Google Scholar]
- 27.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 28.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 29.Tarutani, K., K. Nagasaki, and M. Yamaguchi. 2006. Virus adsorption process determines virus susceptibility in Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 42:209-213. [Google Scholar]
- 30.Tarutani, K., K. Nagasaki, and M. Yamaguchi. 2000. Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo. Appl. Environ. Microbiol. 66:4916-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomaru, Y., N. Hata, T. Masuda, M. Tsuji, K. Igata, Y. Masuda, T. Yamatogi, M. Sakaguchi, and K. Nagasaki. 2007. Ecological dynamics of bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 9:1376-1383. [DOI] [PubMed] [Google Scholar]
- 32.Tomaru, Y., N. Katanozaka, K. Nishida, Y. Shirai, K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34:207-218. [Google Scholar]
- 33.Tomaru, Y., K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Quantitative and qualitative impacts of viral infection on a Heterosigma akashiwo (Raphidophyceae) bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34:227-238. [Google Scholar]
- 34.Zingone, A., F. Natale, E. Biffali, M. Borra, G. Forlani, and D. Sarno. 2006. Diversity in morphology, infectivity, molecular characteristics and induced host resistance between two viruses infecting Micromonas pusilla. Aquat. Microb. Ecol. 45:1-14. [Google Scholar]