Abstract
A field study was conducted to determine the microbial community structures of streambed sediments across diverse geographic and climatic areas. Sediment samples were collected from three adjacent headwater forest streams within three biomes, eastern deciduous (Pennsylvania), southeastern coniferous (New Jersey), and tropical evergreen (Guanacaste, Costa Rica), to assess whether there is biome control of stream microbial community structure. Bacterial abundance, microbial biomass, and bacterial and microbial community structures were determined using classical, biochemical, and molecular methods. Microbial biomass, determined using phospholipid phosphate, was significantly greater in the southeastern coniferous biome, likely due to the smaller grain size, higher organic content, and lower levels of physical disturbance of these sediments. Microbial community structure was determined using phospholipid fatty acid (PLFA) profiles and bacterial community structure from terminal restriction fragment length polymorphism and edited (microeukaryotic PLFAs removed) PLFA profiles. Principal component analysis (PCA) was used to investigate patterns in total microbial community structure. The first principal component separated streams based on the importance of phototrophic microeukaryotes within the community, while the second separated southeastern coniferous streams from all others based on increased abundance of fungal PLFAs. PCA also indicated that within- and among-stream variations were small for tropical evergreen streams and large for southeastern coniferous streams. A similar analysis of bacterial community structure indicated that streams within biomes had similar community structures, while each biome possessed a unique streambed community, indicating strong within-biome control of stream bacterial community structure.
Recent interest in microbial biogeography has been sparked by the potential for biogeographic patterns to reveal the roles of evolutionary and ecological forces acting on microbial species (43), and rates of speciation, dispersal, and extinction are the three fundamental processes considered the likely candidates responsible for producing biogeographical patterns (29). Environmental heterogeneity and spatial distance also determine microbial community composition (44). For free-living microbes, especially prokaryotic heterotrophs, physical mixing counteracts the influence of spatial distance and the diversity of energy sources modifies environmental heterogeneity. For example, marine bacterioplankton communities, mixed by ocean currents (8) and exposed to ubiquitous algal exudates (1), exhibit global distributions for a few dominant microbial clades and relatively few operational taxonomic units (OTU) (10, 26, 39), although estimates of the underlying diversity are being revised (47, 57). In contrast, soils present barriers to mixing and support a vast spectrum of terrestrial vegetation, creating conditions sufficient for the development of distinct biogeographic patterns (29) and even endemism. Soils contain relatively high numbers of bacterial OTU and, unless water logged, few dominant species (10, 15, 16, 46, 54, 60). Planktonic microbial communities in freshwater rivers and lakes show an intermediate pattern (40); there are cosmopolitan or representative species (30, 41, 50, 61), yet there appear to be regional community structure patterns driven by biological, chemical, and physical differences among habitats (55, 59). Low-order streams, while serving as important links between terrestrial and larger aquatic systems, are the least studied (31). Gao et al. (25) examined several streams across the southeastern and midwestern United States and observed differences attributed to variations in chemical characteristics of the habitats but not to geographic location.
This study examined microbial communities from streambed sediments in low-order, forested streams within three biomes, geographic areas distinguished by climate and their predominant terrestrial vegetation. Forested headwater streams derive most of their organic energy from allochthonous sources, either directly from the adjacent terrestrial vegetation or as products of decaying vegetation modified by soil diagenesis. Our study was designed to investigate the general question of how similar are stream microbial communities within and among biomes and, more specifically, do heterotrophic bacteria within streambed communities exhibit biogeographic patterns at the biome level? Nine streams, three located in each of three biomes, were assayed for bacterial abundance, microbial biomass, and microbial and bacterial community structures using a combination of classical, biochemical, and molecular methods. Multivariate statistical analyses were performed to compare the patterns of community structure within and among biomes.
MATERIALS AND METHODS
Study sites and experimental design.
Nine streams were selected to represent three different biomes and physiographic provinces: eastern deciduous forest, Pennsylvania piedmont; southeastern coniferous forest, Pinelands, New Jersey coastal plain; and tropical evergreen forest, Cordillera de Guanacaste, Costa Rica. The streams were matched, as closely as possible, with respect to watershed size and discharge; streams within a biome were located within approximately 10 km of each other; and study sites included the following: for eastern deciduous forest, White Clay Creek (E1), Birch Run (E2), and West Creek (E3); for southeastern coniferous forest, McDonalds Branch (S1), Shinns Branch (S2), and Mount Misery (S3); and for tropical evergreen forest, Rio Tempisquito (T1), Rio Tempisquito Sur (T2), and Quebrada Katia (T3). Triplicate sediment samples were collected from each stream, all streams within a biome were sampled on the same day, and all biomes were sampled between May and June 2000 to avoid seasonal differences and when water temperatures in all streams were between 20 and 21°C.
Environmental variables.
Concomitant with benthic sampling, temperature was measured with a hand-held field thermometer and water was collected for analyses of dissolved organic carbon (DOC), anions, cations, pH, and conductivity. Samples for DOC analyses were collected in borosilicate glassware rendered organic C free by combustion (500°C for 6 h), and then filtered through precombusted glass fiber filters (Whatman GF/F) with a syringe and syringe-type filter holder (32). DOC concentration was determined by Pt-catalyzed persulfate oxidation with either an OI 700 or OI 1010 total organic carbon analyzer. Anions and cations were measured with a Dionex DX 500 ion chromatography system equipped with an ED40 electrochemical detector after passing each sample through a sterile 0.22-μm Pall Gelman HF Tuffryn acrodisc filter. Conductivity was determined with a YSI model 32 conductance meter, and pH was measured with a Fisher Scientific pH probe and meter.
Sampling of benthic microbial communities.
Sediments were delimited with a 2- by 10-cm (height by inside diameter) Plexiglas ring that was inserted 2 cm deep into the sediment. Plexiglas plates were slipped under and over the ring, effectively trapping the sediments and allowing them to be removed from the stream without disturbance. Sediments in the upper 2 mm were transferred with a methanol-washed spatula to a clean plastic weigh boat and thoroughly mixed. Sediments were subsampled for bacterial abundance, lipid analysis, and molecular analysis (approximately 0.5, 1.0, and 0.5 g [wet weight] of sediment, respectively), and each subsample was placed in separate 2-ml microcentrifuge tubes. This procedure, beginning with delimiting of the sediment, was repeated twice to yield three true replicate samples from each stream. Tubes receiving subsamples for molecular analysis contained 0.5 g of prebaked (180°C for at least 2 h) zirconium beads (0.1-mm diameter; Biospec). Subsamples were preserved (2.5% formaldehyde for bacterial abundance and freezing for lipid and molecular analyses) and shipped to the appropriate laboratory for further analysis. Several subsamples designated for molecular analysis were lost in shipping or analysis.
Epifluorescence microscopic counts.
Bacterial abundance was estimated by epifluorescence direct microscopic counts (28). Briefly, preserved sediments were treated with 0.1 mM sterile tetrasodium pyrophosphate (56) for 10 min, and cells were removed by sonication (75 W, 60 s). Suspensions were vortexed, and subsamples (1 ml) were transferred into sterile vials containing 1 ml of tetrasodium pyrophosphate, thoroughly vortexed, and sonicated (75 W, 30 s). Samples were mixed with sterile glycerol (30% final concentration) in 2.5% formaldehyde, vortexed, and spun at low speed (3 min, 25 × g) to pellet bacterium-free particles. After staining with propidium iodide, bacterial cells in the supernatant fluid were filtered onto a black 0.2-μm-pore-size Nuclepore filter and 20 randomly selected fields or 300 to 500 cells were enumerated (4). One filter was counted per sediment sample, with duplicate filters counted for 10% of the samples.
Phospholipid analysis.
Microbial biomass and community structure were determined using phospholipid analysis following the methods of Findlay (19). Briefly, lipids were extracted from frozen sediment samples by dichloromethane-methanol. The extraction mixture was partitioned into aqueous and organic fractions through the addition of dichloromethane and water, and the organic fraction containing the lipids was recovered. The organic fraction was subsampled for total microbial biomass, determined by phospholipid phosphate (PLP) (21). Phospholipid fatty acids (PLFAs) were recovered from the remaining lipid by differential elution from silicic acid columns (J. T. Baker) and were analyzed as their methyl esters (19). Fatty acid methyl esters were quantified by gas chromatography, with identification based on relative retention times, coelution with known standards, and mass spectrometry analysis. Using polyenoic fatty acids as indicators of microeukaryotes, total biomass was divided between prokaryotic and microeukaryotic organisms (20) and the results were expressed as percentages. To investigate bacterial community structure, PLFAs assigned a priori to the functional group microeukaryotes (19) and those known to be common to both bacteria and microeukaryotes were removed from the PLFA profiles (58), leaving only PLFAs of bacterial origin, and weight percentages were recalculated. Prokaryotic biomass was converted into bacterial abundance using a conversion factor of 100 nmol PLP = 4 × 109 cells (3).
DNA extraction.
Phosphate and MT buffers from the Fast DNA spin kit for soils (Qbiogene, Carlsbad, CA) were added to the sample tubes containing the frozen sediment and zirconium beads. The samples were shaken in a Bio101 FastPrep (Qbiogene, Carlsbad, CA) at speed 4.5 for 15 s (twice) and placed on ice for 1 min between mechanical disruptions. The samples were centrifuged at 4°C at 15,000 × g for 5 min. Supernatants were placed into new tubes, and samples were processed according to the manufacturer's protocol. Genomic DNA was eluted in 50 μl diethyl pyrocarbonate (Ambion, Austin, TX)-treated water and stored at −80°C until further analysis. To evaluate quality, the DNA was resolved on 0.8% (wt/vol) high-melt agarose gels with 1× TAE (Tris-acetate-EDTA) buffer and visualized by ethidium bromide staining (48).
tRFLP analysis.
For terminal restriction fragment length polymorphism (tRFLP) analysis, bacterial 16S rRNA gene sequences were amplified using primer S-D-Bact-1512-a-A-21 (35) and a second primer (S-D-Bact-008-a-S-17) 5′ end labeled with 6-carboxyfluorescein (35). Each 50-μl PCR mixture contained primers at 0.7 μmol liter−1, 1.0 μmol liter−1 Tris-HCl (pH 8.8), 5.0 μmol liter−1 KCl, and 0.15 μmol liter−1 MgCl2; 125 μmol liter−1 of each deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA); 25 μg of bovine serum albumin; and 2.5 U Taq DNA polymerase (Invitrogen, Carlsbad, CA). Each reaction mixture was incubated on a PTC-100 thermal cycler (MJ Research, Waltham, MA) using the following “touchdown” parameters: initial denaturation at 94°C for 1 min, followed by four cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 30 s. The annealing temperature for the reaction was subsequently reduced by 1°C per cycle until a 50°C annealing temperature was reached, cycled five more times at a 50°C annealing temperature, and terminated with a final extension at 72°C for 5 min. The PCR products were visualized on a 0.8% (wt/vol) high-melt agarose gel stained with ethidium bromide and quantified based on relative intensity using a quantitative DNA ladder (Invitrogen, Carlsbad, CA) and NIH image v 1.63 (http://rsb.info.nih.gov/nih-image).
PCR products, derived from environmental samples, were digested overnight in the dark at 37°C using 10 U of the restriction enzyme HaeIII, which has a 4-bp recognition site, in a standard restriction buffer (One-Phor-All Plus; Amersham-Pharmacia). Digested samples were ethanol precipitated (48), dried, and analyzed using an ABI 377 DNA sequencer (Applied Biosystems, Inc., Fremont, CA). At least 100 fmol of each digested sample was loaded onto a 4% polyacrylamide gel for fragment analysis in Gene Scan mode. The Genescan TAMRA 500 size standard (HaeIII) was added to each sample lane. ASCII files of electropherograms were analyzed using Dax analysis software (van Mierlo, Inc., Holland). Peaks representing tRFLP fragments were measured as integrated peak areas. Peaks less than 2 bp apart were binned and normalized as the relative percentage of total peak area. Peaks less than 1% of the total area or found in less than three replicate profiles were excluded from further analysis.
Statistical analysis.
Comparisons of water chemistry, microbial biomass, and bacterial abundance were made using nested analyses of variance (streams within and among biomes) (Systat 10 and SAS). Relationships among variables were investigated using linear regression (Microsoft Excel 2004 for Mac 11.2). Fatty acid profiles were subjected to principal component analysis (PCA) after log transformation [ln(x + 1)] of weight percent fatty acid data (SPSS, version 7.5, and Minitab, release 12.23); PLFA profiles were interpreted with a functional group approach (19).
Nonmetric multidimensional scaling, which does not require the assumption of linearity, was used to compare tRFLP fragment composition among streams (PC-ORD version 4) (38) following the calculation of Sorensen's distance (34). The raw data set consisted of 15 samples made up of 16 fragment lengths or OTU. To reduce skew, the square root of the relative percent area was arcsine transformed (52). For the ordination, the autopilot option was set to “slow and thorough.” Choosing the axis that minimizes stress and maximizes the interpretation of the data assessed the amount of variability explained in the data set. Stress, an indicator of goodness of fit, measured the inverse of the fit between the original data matrix and the reduced dimension ordination matrices. Stress values between 0 and 15 give a good approximation of the data in multivariable space with a low risk of drawing false inferences (38). A plot between the final stress and the number of iterations was used to assess the stability of the solutions. Monte Carlo tests identified the dimensions that gave solutions that were significantly different from those due to random chance. The proportion of the variance of the distances between the original matrix and the ordination space explained was described for each axis.
RESULTS
Water chemistry and sediment organic content.
Pinelands streams within the southeastern coniferous forest had low conductivity, pH, and concentrations of anions, cations, and nutrients, while the streambed sediments were enriched in organic matter (Table 1). Streams from the eastern deciduous forest had high values for these parameters, while tropical evergreen forest streams showed intermediate values. The exceptions to this pattern were pH, sodium, and sediment organic content, which were higher in the tropical evergreen streams than in the eastern deciduous forest streams. Annual temperature ranges for streams from the Pinelands and eastern deciduous forest were comparable (∼0 to 21°C), while the tropical evergreen streams had nearly constant temperatures (range, 19 to 22°C). Biome-level average DOC concentrations at baseflow were lowest in the tropical evergreen streams (0.7 mg C/liter), twofold higher in the deciduous forest streams (1.5 mg C/liter), and highest in the coniferous forest streams (8.6 mg C/liter).
TABLE 1.
Summary of chemical characterizations of stream water and sedimentsa
| Biome and stream | Conductance (μS/cm) | pH | Anion or cation content (mg/liter)
|
DOC (mg/liter) | Sediment organic content (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chloride | Sulfate | Nitrate-N | Calcium | Sodium | Potassium | Magnesium | |||||
| Southeastern coniferous | |||||||||||
| McDonalds Branch | 45 ± 9 B | 4.2 ± 0.2 A | 2.4 ± 1.2 | 3.3 ± 1.2 | 0.01 ± 0.01 | 0.2 ± 0.1 | 1.5 ± 0.5 | 0.2 ± 0.1 AB | 0.4 ± 0.1 | 6.6 ± 4.5 B | 26.4 ± 8.8 |
| Shinns Branch | 59 ± 7 A | 3.9 ± 0.1 B | 3.3 ± 1.5 | 4.3 ± 2.3 | 0.01 ± 0.01 | 0.1 ± 0.2 | 1.9 ± 0.6 | 0.4 ± 0.1 A | 0.3 ± 0.2 | 10.9 ± 4.2 A | 33.3 ± 5.5 |
| Mount Misery | 45 ± 11 B | 4.1 ± 0.2 AB | 2.5 ± 1.4 | 3.3 ± 1.9 | 0.01 ± 0.01 | 0.2 ± 0.3 | 1.5 ± 0.6 | 0.2 ± 0.1 B | 0.3 ± 0.1 | 8.4 ± 4.2 AB | 29.8 ± 1.9 |
| Eastern deciduous | |||||||||||
| White Clay Creek | 205 ± 16 A | 7.6 ± 0.6 | 12.2 ± 0.7 B | 21.4 ± 3.4 A | 4.60 ± 0.60 A | 21.2 ± 3.4 A | 5.4 ± 0.9 | 1.8 ± 0.4 A | 7.6 ± 1.0 A | 1.7 ± 0.4 A | 1.6 ± 1.0 |
| Birch Run | 168 ± 7 B | 7.3 ± 0.4 | 13.3 ± 7.1 A | 8.9 ± 4.2 A | 6.20 ± 3.00 A | 11.1 ± 1.7 B | 5.8 ± 1.5 | 1.3 ± 0.3 B | 6.8 ± 1.1 A | 1.1 ± 0.2 B | 2.3 ± 1.3 |
| West Creek | 143 ± 10 C | 7.5 ± 0.6 | 5.7 ± 0.5 C | 32.0 ± 4.9 B | 1.80 ± 0.50 B | 13.7 ± 0.9 B | 5.1 ± 0.6 | 2.3 ± 0.4 A | 4.7 ± 0.5 B | 1.8 ± 0.3 A | 2.5 ± 1.5 |
| Tropical evergreen | |||||||||||
| Rio Tempisquito | 84 ± 23 B | 8.3 ± 0.4 | 4.6 ± 0.6 B | 1.2 ± 0.3 C | 0.20 ± 0.05 A | 6.4 ± 1.7 B | 4.2 ± 0.7 B | 0.9 ± 0.2 A | 2.6 ± 0.5 | 1.0 ± 0.7 | 5.7 ± 0.7 A |
| Rio Tempisquito Sur | 135 ± 42 A | 8.0 ± 0.5 | 13.6 ± 5.0 A | 36.2 ± 11.2 A | 0.08 ± 0.03 B | 9.5 ± 2.8 A | 7.5 ± 1.8 A | 1.4 ± 0.2 A | 3.1 ± 0.9 | 0.6 ± 0.4 | 3.9 ± 0.4 B |
| Quebrada Katia | 65 ± 10 B | 8.5 ± 0.5 | 4.2 ± 0.7 B | 2.1 ± 0.4 B | 0.18 ± 0.05 A | 4.4 ± 1.2 B | 3.5 ± 0.6 B | 0.7 ± 0.1 B | 1.9 ± 0.4 | 0.6 ± 0.2 | 4.2 ± 0.8 B |
Data are presented as mean ± standard deviation of 10 to 13 monthly samples for water chemistry and two to six sampling dates for sediments. Values for all analytes were significantly different at the biome level, with the exception of sodium, for which values in the eastern deciduous and tropical evergreen biomes were greater than for the southeastern coniferous biome. Significant differences among streams within a biome are designated by different letters following the standard deviations.
Total microbial biomass and bacterial abundance.
Total microbial biomass was ∼10- to 160-fold greater in southeastern coniferous biome streams than in eastern deciduous and tropical evergreen streams (P < 0.001); there were no significant differences among streams within biomes (P = 0.22) (Table 2). The range of total microbial biomass for streams within the eastern deciduous biome overlapped the range observed from streams within the tropical evergreen biome. Prokaryotes comprised between 39 and 88% of total microbial biomass, and in seven of the nine streams, bacteria accounted for greater than 60% of total biomass.
TABLE 2.
Microbial biomass and bacterial abundance in sediments of nine streams
| Biome and stream | PLP (nmol g−1 dry wt)a | % Eukaryotic/ prokaryoticb | Bacterial abundance (g−1 dry wt)
|
|
|---|---|---|---|---|
| Calculatedc | Epifluorescence microscopy countsd | |||
| Southeastern coniferous | ||||
| McDonalds Branch | 2,475.78 ± 1003.42 | 39/61 | 9.90 × 1010 | 6.25 × 1010 ± 4.67 × 1010 |
| Shinns Branch | 3,339.24 ± 1743.16 | 24/76 | 1.34 × 1011 | 7.19 × 1010 ± 3.41 × 1010 |
| Mount Misery | 1,608.03 ± 576.78 | 16/84 | 6.43 × 1010 | 6.22 × 1010 ± 1.76 × 1010 |
| Eastern deciduous | ||||
| White Clay Creek | 36.09 ± 5.45 | 61/39 | 1.44 × 109 | 6.90 × 109 ± 2.35 × 109 |
| Birch Run | 164.44 ± 115.39 | 56/44 | 6.58 × 109 | 4.29 × 1010 ± 2.20 × 1010 |
| West Creek | 70.18 ± 45.86 | 26/74 | 2.81 × 109 | 1.64 × 1010 ± 1.32 × 1010 |
| Tropical evergreen | ||||
| Rio Tempsiquito | 48.78 ± 3.23 | 12/88 | 1.95 × 109 | 8.43 × 109 ± 1.36 × 109 |
| Rio Tempisquito Sur | 21.46 ± 7.84 | 18/82 | 8.58 × 108 | 6.57 × 109 ± 2.78 × 109 |
| Quebrada Katia | 35.37 ± 15.87 | 12/88 | 1.41 × 109 | 7.27 × 109 ± 2.62 × 109 |
Mean ± standard deviation (n = 3).
Percentage that microeukaryotes comprise of total microbial biomass, calculated from PLFA profiles (mean of three replicate samples).
Calculated from PLP × % prokaryotic (expressed as a decimal fraction) and the conversion factor given in reference 3.
Mean ± standard deviation (n = 3).
The relationships between microscopic and biochemical estimates of bacterial abundance differed among biomes. In southeastern coniferous streams, biochemical estimates of bacterial abundance were 2.4 times greater than those based on epifluorescence microscopic counts and showed little correlation between the estimates made on the same samples. In contrast, epifluorescence microscopic determinations of bacterial abundance were, on average, 7.3-fold greater than biochemical estimates of bacterial abundance for streams from the tropical evergreen and eastern deciduous biomes and were highly correlated (R2 = 0.78, n = 18).
Microbial community structure.
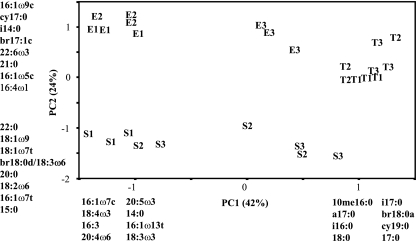
PCA of PLFA profiles based on a data set of 48 PLFAs separated the streams within the three biomes from each other at the biome scale, with principal components 1 and 2 (PC1 and -2, respectively) accounting for 66% of the variance (Fig. 1). The samples from all three tropical evergreen streams were tightly clustered, both within and among streams. All three streams in the eastern deciduous biome displayed low within-stream variation but showed high among-stream variation, as West Creek samples clearly separated from samples from the other two streams. We observed tight clustering of all samples from McDonalds Branch in the southeastern coniferous biome, which also clustered with single samples from Shinns Branch and Mt. Misery. The latter two streams, however, displayed high within-stream variation.
FIG. 1.
PCA of variation in sedimentary microbial community composition in the nine streams by PLFA analysis. Each abbreviation represents an individual sample: E, eastern deciduous; T, tropical evergreen; S, southern coniferous; and 1, 2, and 3, individual streams within the biome. The percent variation explained by each axis is indicated in parentheses. Identified fatty acids had component loadings of >0.5 or <−0.5 and exerted strong influence on the pattern of variation among samples along the respective component axes.
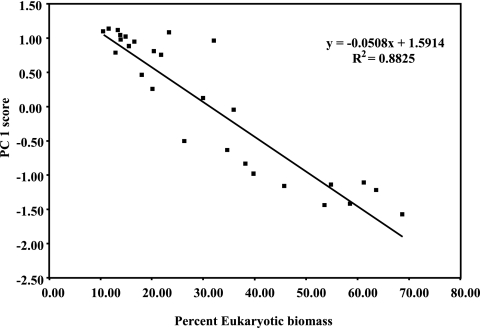
Factor loadings for individual fatty acids showed that levels of microeukaryotes in the samples drove much of the variation among the microbial communities. High abundances of fatty acids associated with algae were present in samples with negative PC1 factor scores, whereas high abundances of fatty acids associated with fungi were present in samples with negative PC2 factor scores. Samples with positive PC1 factor scores had high relative abundances of fatty acids associated with bacteria, and lipid markers for a mixed group of bacteria were present in samples with positive PC2 factor scores. The importance of the proportions of prokaryotes and eukaryotes within communities to the differences observed in microbial community structure was further indicated by the high correlation between PC1 factor scores and the percentage that microeukaryotes comprise of total microbial biomass (Fig. 2).
FIG. 2.
Relationship between PCA factor 1 score and the calculated percentage that microeukaryotes contribute to total microbial biomass for all stream samples.
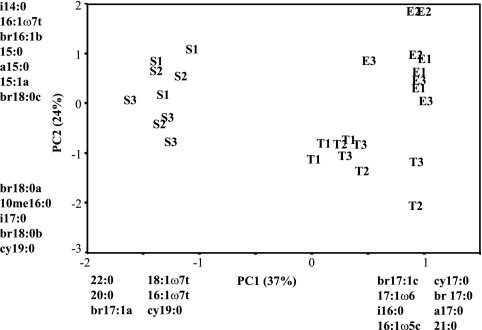
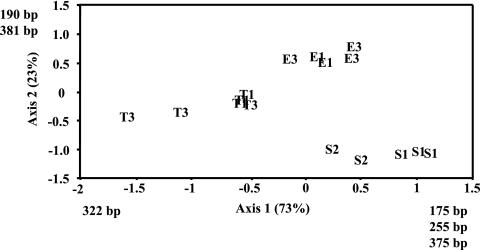
PCA of profiles using only PLFAs of bacterial origin further accentuated the separation of samples into three discrete groups. Each group contained all of the samples from one biome, and there was no discernible difference among samples from streams within a biome, indicating that each biome possessed a unique community of bacteria on streambed sediments (Fig. 3). An extensive literature search failed to produce the information necessary to allow assignment of signature fatty acid or functional group affinities to the fatty acids identified by PCA as critical to the observed components of variation. A strong within-biome control of streambed bacterial community structure was corroborated by the nondimensional scaling analysis of tRFLP profiles based on a data set of 16 restriction fragments (Fig. 4). Ordination axes 1 and 2 described 73% and 23% of the variation, respectively. After 50 iterations, the stress of the final solution was 7.5 and stable (Table 3). Axis 1 separated sediments from tropical evergreen streams from sediments from eastern deciduous and southeastern coniferous streams. Axis 2 further separated sediments from eastern deciduous and southeastern coniferous streams. Pearson's correlation coefficients of OTU with ordination axis indicated that 6 OTU contributed to defining the differences among biomes (Fig. 4). The communities from streams in the southeastern coniferous forest were positively correlated with OTU of 175, 255, and 375 bp on axis 1, and the tropical evergreen biome was correlated with a 322-bp OTU. OTU of 190 bp and 381 bp were correlated with communities from streams in the eastern deciduous biome. The OTU of 381 bp is most closely related to a plastid from a diatom (Haslea salstonica; AF514854; 98%), and the OTU of 190 bp is most closely related to an uncultured planctomycete (AJ290171; 93%) (31).
FIG. 3.
Patterns of variation in sedimentary bacterial community composition in the nine streams by PLFA analysis after removal of fatty acids assigned a priori to the functional group microeukaryotes (19) and those known to be common to both bacteria and microeukaryotes (58) from the PLFA profiles. Figure layout and abbreviations are as described in the legend to Fig. 1.
FIG. 4.
Patterns of variation in sedimentary microbial community composition in the nine streams by molecular analysis. Shown is a scatter plot of sample scores from nonmetric multidimensional scaling analysis, with each abbreviation representing an individual sample. The percent variation explained by each axis is represented in parentheses. Abbreviations are as described in the legend to Fig. 1. Identified fragment lengths had Pearson's correlation coefficients of >0.5 or <−0.5 and exerted strong influence on the pattern of variation among samples along the respective component axes. Samples not plotted were not sampled or had been sampled but lost in transport or processing.
TABLE 3.
Monte Carlo test of stress in relation to dimensionality
| Sediment axis | Stress result ina:
|
|||||
|---|---|---|---|---|---|---|
| Real data (40 runs)
|
Randomized data (50 runs)
|
|||||
| Minimum | Mean | Maximum | Minimum | Mean | Maximum | |
| 1 | 18.98 | 33.11 | 53.98 | 31.41 | 49.02 | 54.00 |
| 2 | 7.56 | 9.34 | 34.82 | 15.18 | 21.38 | 16.41 |
P = 0.02 by Monte Carlo test for comparison of 40 runs on the real data from the sedimentary communities with 50 runs on randomized data.
DISCUSSION
Bacterial community structure in the nine headwater streams displayed a distinct biome-level biogeography, even when streams within a biome displayed high within-stream (southeastern coniferous biome) or among-stream (eastern deciduous biome) variation in total microbial community structure. Much of that variation was related to the ratio of prokaryotic to eukaryotic biomass within individual samples. Field observations that springtime blooms of Tribonema were particularly abundant in McDonalds Branch (S1) and present in dense but patchy distributions within the other southeastern coniferous streams (S2 and S3) are consistent with the importance of chrysophyte algal biomarkers (18:4ω3, 20:5ω3) (11) in describing the variation in microbial community structure among samples from these streams (Fig. 1). Similarly, the separation of White Clay Creek (E1) and Birch Run (E2) from West Creek in the eastern deciduous biome is consistent with the twofold-greater eukaryote contribution to microbial biomass in E1 and E2 and the importance of a lipid marker for algal chlorophytes (18:3ω3) (11) in describing the variation in microbial community structure quantified by PC1 (Fig. 1). A high correlation of the ratio of prokaryotic to eukaryotic biomass to variation in microbial community structure quantified by PCA has been reported in microbial communities from shallow, subtidal marine sediments (22), stream sediments (36, 53), and freshwater reservoir sediments (51), but those studies were limited to comparisons within a single ecosystem where changes in community structures were attributed to seasonality. This is the first study, to our knowledge, in which the sampling regimen eliminated the seasonal component of variation and the prokaryotic/eukaryotic biomass ratio influenced microbial community structure at the biome level.
Despite the positive correlation between algal biomass and bacterial abundance reported for other stream communities (45) and observed in our study, it is clear that in our study streams algal biomass did not control bacterial community structure. In fact, when microeukaryotic lipids were removed from the analysis, the within- and among-stream variation for each biome was greatly reduced for those biomes with higher algal contributions (southern coniferous and eastern deciduous) and remained mostly unchanged for the highly shaded tropical evergreen biome streams with low levels of algal biomass. Clearly, the major variation in sedimentary bacterial community structure occurred at the biome level, and the variation among streams within a biome was comparable to the variation observed within individual streams. This pattern was evident in both the tRFLP and bacterial PLFA descriptions of community structure and supports the contention, based on a study of bacterial communities within coastal marine waters, that biogeographical patterns are generated by processes that are comparable across major global biomes (42).
Several studies have used PLFA profiles to investigate longitudinal variation within streams (53) or the impacts of anthropogenic stress on microbial community structure (e.g., see references 5 and 36), but we are unaware of any other studies comparing variation in sedimentary microbial and bacterial community structure across streams. Two studies have used the PLFA method to investigate spatial variation in community structure in marine sediments. Federle et al. (18) found significant spatial variation at the centimeter and 100-m levels within subtropical estuarine sediments. Baird et al. (2), using a replicated sampling scheme unprecedented for deep-sea research, found a similar pattern at a deep-sea site subjected to high-energy currents. Several studies have been conducted using molecular approaches to examine spatial variation in benthic bacterial community composition. Gao et al. (25) examined a single stream in each of nine states and reported high within-stream (upstream versus downstream) and among-stream variation. In some cases, variation could be correlated with differences in environmental conditions, but no overall geographic pattern was reported. Braker et al. (7) found distinct bacterial community structures, based upon unique distributions of 16S rRNA gene tRFLP, in marine sediments from Puget Sound and the Washington margin, although the study is based on four cores, limiting the information available to regional spatial scales. A study focused on denitrifying bacterial communities in marine sediments found the most variation in community composition at the kilometer scale (the largest examined), whereas relative abundance of similar OTU became important for differentiating communities at the centimeter scale (49). In our study, within-stream samples characterize variation at the meter scale and among-stream samples characterize variation at the kilometer scale. Clearly, the occurrence of the principal component of variation at the biome level indicates that the greatest variation in bacterial community structure in streams occurs at the largest spatial scales and suggests the development of biogeographic patterns within these communities.
The contrasting patterns of microbial and bacterial community structure suggest that microeukaryotes and bacteria are responding to different environmental determinants. The three geographic locations differ in important physical, chemical, and biological parameters, including terrestrial plant communities, which, in turn, lead to differences in the terrestrially derived, dissolved organic matter quantity and quality in stream water (24, 33). Which of these determinants are the proximate causes of the observed bacterial distribution is not known at this time.
The total microbial biomass for the eastern deciduous and tropical evergreen stream sediments was similar to total microbial biomass for several other temperate streams (36, 53) and within the range reported for freshwater sediments (12). Sedimentary biomass in White Clay Creek sediments was similar to that reported 15 years prior, while biomass in West Creek was elevated in the current study (6). In contrast, total microbial biomass for the southeastern coniferous stream sediments was higher than the range previously reported by Dobbs and Findlay (12) and comparable to that in sediments from a seasonally anaerobic, depositional zone within a freshwater reservoir (51). Bacterial abundances were also greatest in the highly organic sediments of the southeastern coniferous streams, and these values were similar to those observed for benthic organic matter in streams (17, 23). The discrepancy between direct microscopy and biochemical measurements of abundance was likely due to two issues: the necessity of converting biomass, measured as moles of phospholipid phosphate, to number of cells and the difficulty in obtaining a clean, debris-free preparation for microscopic counts. The conversion from biomass to cell number was based on cultured bacteria, particularly Escherichia coli (3). Assuming the average size of the cultured bacteria used in the study by Balkwill et al. (3) was a rod 1.5 μm in length and 1 μm in diameter, a 2.5 times reduction in size to 0.6 μm by 0.4 μm (length by diameter) would yield a biomass-to-cell number conversion factor that would our bring estimates of abundance for eastern deciduous and tropical evergreen stream sediments to unity. The lack of correlation between the two measures of abundance for southeastern coniferous stream sediments is likely due to the high organic content of the sediments causing difficulty in obtaining clean, debris-free preparations for microscopic counts.
Biogeography of aquatic bacteria is in its infancy, and a globally consistent pattern, if one exists, has yet to emerge. Different communities have been observed in a comparison of freshwater lakes and pelagic marine habitats (13), as well as for lakes and streams within an Arctic tundra catchment (9). Within coastal marine sediments, spatially isolated assemblages occur over scales of a few kilometers, with environmental conditions selecting for the individual assemblages (27). A review of studies from a wide range of terrestrial and aquatic environments concluded that free-living bacteria exhibit biogeographical patterns that are the result of both environmental selection and the legacies of historical events (37). It has been suggested that different groups of microbes may show very different biogeographies (14). In all of the recent investigations, streams have been largely ignored. The emergence of clear biome-level patterns in our study suggests that further investigations into the microbial communities present in low-order streams may provide clues to the physical, chemical, and biotic factors influencing the biogeography of bacteria.
Acknowledgments
Sherman Roberts, Michael Gentile, Raphael Morales, and Christian Collado assisted in sample collection and processing. Jen Mosher, Charles Dow, Heather Brooks, and Melanie Arnold assisted in data analysis.
This research was supported by NSF DEB 9904047 and DEB 0096276.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Aluwihare, L. I., and D. J. Repeta. 1999. A comparison of the chemical characteristics of oceanic DOM and extracellular DOM produced by marine algae. Mar. Ecol. Prog. Ser. 186:105-117. [Google Scholar]
- 2.Baird, B. H., D. E. Nivens, J. H. Parker, and D. C. White. 1985. The biomass, community structure, and spatial distribution of the sedimentary microbiota from a high-energy area of the deep sea. Deep-Sea Res. Part A 32:1089-1099. [Google Scholar]
- 3.Balkwill, D. L., F. R. Leach, J. T. Wilson, J. F. McNabb, and D. C. White. 1988. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, and direct counts in subsurface aquifer sediments. Microb. Ecol. 16:73-84. [DOI] [PubMed] [Google Scholar]
- 4.Battin, T. J., L. A. Kaplan, J. D. Newbold, X. Cheng, and C. Hansen. 2003. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 69:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-David, E. A., P. J. Holden, D. J. M. Stone, B. D. Harch, and L. J. Foster. 2004. The use of phospholipid fatty acid analysis to measure impact of acid rock drainage on microbial communities in sediments. Microb. Ecol. 48:300-315. [DOI] [PubMed] [Google Scholar]
- 6.Bott, T. L., and L. A. Kaplan. 1985. Bacterial biomass, metabolic state, and activity in stream sediments: relation to environmental variables and multiple assay comparisons. Appl. Environ. Microbiol. 50:508-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment polymorphism analysis of amplified nitrate reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, A. 2001. Ocean circulation, p. 208-209. In B. Heineman (ed.), Ocean circulation. Elsevier, Boston, MA.
- 9.Crump, B. C., H. E. Adams, J. E. Hobbie, and G. W. Kling. 2007. Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology 88:1365-1378. [DOI] [PubMed] [Google Scholar]
- 10.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkman, N. A., and J. C. Kromkamp. 2006. Phospholipid-derived fatty acids as chemotaxonomic markers for phytoplankton: application for inferring phytoplankton composition. Mar. Ecol. Prog. Ser. 324:113-125. [Google Scholar]
- 12.Dobbs, F. C., and R. H. Findlay. 1993. Detection of microbial-community response to disturbance, p. 347-358. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Current methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 13.Dolan, J. R. 2005. An introduction to the biogeography of aquatic microbes. Aquat. Microb. Ecol. 41:39-48. [Google Scholar]
- 14.Dolan, J. R. 2006. Microbial biogeography? J. Biogeogr. 33:199-200. [Google Scholar]
- 15.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fazi, S., S. Amalfitano, J. Pernthaler, and A. Puddu. 2005. Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ. Microbiol. 7:1633-1640. [DOI] [PubMed] [Google Scholar]
- 18.Federle, T. W., M. A. Hullar, R. J. Livingston, D. A. Meeter, and D. C. White. 1983. Spatial distribution of biochemical parameters indicating biomass and community composition of microbial assemblies in estuarine mud flat sediments. Appl. Environ. Microbiol. 45:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay, R. H. 2004. Determination of microbial community structure using phospholipid fatty acid profiles, p. 983-1004. In G. A. Kowalchuk, F. J. De Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. Van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 20.Findlay, R. H., and F. C. Dobbs. 1993. Quantitative description of microbial communities using lipid analysis, p. 271-284. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Current methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 21.Findlay, R. H., G. M. King, and L. Watling. 1989. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 55:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findlay, R. H., and L. Watling. 1998. Seasonal variation in the structure of a marine benthic microbial community. Microb. Ecol. 36:23-30. [DOI] [PubMed] [Google Scholar]
- 23.Findlay, S., J. Tank, S. Dye, H. M. Valett, P. J. Mulholland, W. H. McDowell, S. L. Johnson, S. K. Hamilton, J. Edmonds, W. K. Dodds, and W. B. Bowden. 2002. A cross-system comparison of bacterial and fungal biomass in detritus pools of headwater streams. Microb. Ecol. 43:55-66. [DOI] [PubMed] [Google Scholar]
- 24.Frazier, S. W., K. O. Nowack, K. M. Goins, F. S. Cannon, L. A. Kaplan, and P. G. Hatcher. 2003. Characterization of organic matter from natural waters using tetramethylamonium hydroxide thermochemolysis GC-MS. J. Anal. Appl. Pyrolysis 70:99-128. [Google Scholar]
- 25.Gao, X., O. A. Olapadel, and L. G. Leff. 2005. Comparison of benthic bacterial community composition in nine streams. Aquat. Microb. Ecol. 40:51-60. [Google Scholar]
- 26.Hagstrom, Å., T. Pommier, F. Rohwer, K. Simu, W. Stolte, D. Svensson, and U. L. Zweifel. 2002. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewson, I., M. E. Jacobson-Meyers, and J. A. Furhman. 2007. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, Southern California borderlands. Environ. Microbiol. 9:923-933. [DOI] [PubMed] [Google Scholar]
- 28.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horz, H.-P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hullar, M. A. J., L. A. Kaplan, and D. A. Stahl. 2006. Recurring seasonal dynamics of microbial communities in stream habitats. Appl. Environ. Microbiol. 72:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan, L. A. 1994. A field and laboratory procedure to collect, process, and preserve freshwater samples for dissolved organic carbon analysis. Limnol. Oceanog. 39:1470-1476. [Google Scholar]
- 33.Kim, S., L. A. Kaplan, R. Benner, and P. G. Hatcher. 2004. Hydrogen-deficient molecules in natural riverine water samples—evidence for the existence of black carbon in DOM. Mar. Chem. 92:225-234. [Google Scholar]
- 34.Kruskal, J. B. 1964. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psykometrika 29:115-129. [Google Scholar]
- 35.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 36.Langworthy, D. E., R. D. Stapleton, G. S. Sayler, and R. H. Findlay. 1998. Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl. Environ. Microbiol. 64:3422-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 38.McCune, B., and M. J. Mefford. 1999. PC-ORD. Multivariate Analysis of Ecological Data, version 4 ed. MjM Software Design, Gleneden Beach, OR.
- 39.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 40.Nold, S. C., and G. Zwart. 1998. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17-35. [Google Scholar]
- 41.Page, K. A., S. A. Connon, and S. J. Giovannoni. 2004. Representative freshwater bacterioplankton isolated from Crater Lake, Oregon. Appl. Environ. Microbiol. 70:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pommier, T., B. Canback, L. Riemann, K. H. Bostrom, K. Simu, P. Lundberg, A. Tunlid, and A. Hagstrom. 2007. Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16:867-880. [DOI] [PubMed] [Google Scholar]
- 43.Ramette, A., and J. M. Tiedje. 2006. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197-206. [DOI] [PubMed] [Google Scholar]
- 44.Ramette, A., and J. M. Tiedje. 2007. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. USA 104:2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rier, S. T., and R. J. Stevenson. 2001. Relation of environmental factors to density of epilithic lotic bacteria in 2 ecoregions. J. N. Am. Benthol. Soc. 20:520-532. [Google Scholar]
- 46.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y.-H. Rogers, L. I. Falcón, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Scala, D. J., and L. J. Kerkhoff. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauer, M., C. Kamenik, and M. W. Hahn. 2005. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71:5900-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smoot, J. C., and R. H. Findlay. 2001. Spatial and seasonal variation in a freshwater reservoir sedimentary microbial community as determined by phospholipid fatty acid analysis. Microb. Ecol. 42:350-358. [DOI] [PubMed] [Google Scholar]
- 52.Sokal, R. R., and F. J. Rolf. 1990. Biometry. W. H. Freeman and Co., San Francisco, CA.
- 53.Sutton, S. D., and R. H. Findlay. 2003. Sedimentary microbial community dynamics in a regulated stream: East Fork Little Miami River, Ohio. Environ. Microbiol. 5:256-266. [DOI] [PubMed] [Google Scholar]
- 54.Torsvik, V., K. Salte, R. Sørheim, and J. Goksøyr. 1990. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 56:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 56.Velji, M. I., and L. J. Albright. 1985. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can. J. Microbiol. 32:121-126. [Google Scholar]
- 57.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 58.White, D. C., H. C. Pinkart, and D. B. Ringelberg. 1997. Biomass measurements: biochemical approaches, p. 91-101. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, DC.
- 59.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]