Abstract
Burkholderia cepacia complex strains are genetically related but phenotypically diverse organisms that are important opportunistic pathogens in patients with cystic fibrosis (CF,) as well as pathogens of onion and banana, colonizers of the rhizospheres of many plant species, and common inhabitants of bulk soil. Genotypic identification and pathogenicity characterization were performed on B. cepacia complex isolates from the rhizosphere of onion and organic soils in Michigan. A total of 3,798 putative B. cepacia complex isolates were recovered on Pseudomonas cepacia azelaic acid tryptamine and trypan blue tetracycline semiselective media during the 2004 growing season from six commercial onion fields located in two counties in Michigan. Putative B. cepacia complex isolates were identified by hybridization to a 16S rRNA gene probe, followed by duplex PCR using primers targeted to the 16S rRNA gene and recA sequences and restriction fragment length polymorphism analysis of the recA sequence. A total of 1,290 isolates, 980 rhizosphere and 310 soil isolates, were assigned to the species B. cepacia (160), B. cenocepacia (480), B. ambifaria (623), and B. pyrrocinia (27). The majority of isolates identified as B. cepacia (85%), B. cenocepacia (90%), and B. ambifaria (76%) were pathogenic in a detached onion bulb scale assay and caused symptoms of water soaking, maceration, and/or necrosis. A phylogenetic analysis of recA sequences from representative B. cepacia complex type and panel strains, along with isolates collected in this study, revealed that the B. cenocepacia isolates associated with onion grouped within the III-B lineage and that some strains were closely related to strain AU1054, which was isolated from a CF patient. This study revealed that multiple B. cepacia complex species colonize the onion rhizosphere and have the potential to cause sour skin rot disease of onion. In addition, the onion rhizosphere is a natural habitat and a potential environmental source of B. cenocepacia.
Burkholderia (formerly Pseudomonas) cepacia was first described by W. H. Burkholder in 1950 as the phytopathogen responsible for causing sour skin, a bacterial soft-rotting disease of onion in New York (8). B. cepacia and two isolates of the related species B. cenocepacia have since been isolated from diseased onion bulbs in Japan (46), and B. cenocepacia has been isolated as the causal agent of fingertip rot disease of banana (26). Similarly, B. cepacia emerged as an important opportunistic human pathogen in the 1970s and 1980s following isolation from the respiratory tracts of patients with cystic fibrosis (CF) or chronic granulomatous disease (23, 32, 44). In addition, a variety of Burkholderia spp. have been characterized as important environmental strains, with phenotypes that include biological control of plant root-infecting fungi, plant growth promotion, nitrogen fixation, and biodegradation of recalcitrant compounds in soil (36).
During the last decade, advances in genetic techniques have facilitated a reassessment of the B. cepacia species framed against the background that organisms known as B. cepacia included plant pathogens, human pathogens, biological-control strains, and strains active in biodegradation. The results from these genetic analyses revealed that bacteria formerly identified as B. cepacia represent a complex of genetically related yet phenotypically diverse strains collectively referred to as the B. cepacia complex (3). The B. cepacia complex currently encompasses at least nine distinct species, B. cepacia, B. multivorans, B. cenocepacia, B. stabilis, B. vietnamiensis, B. dolosa, B. ambifaria, B. anthina, and B. pyrrocinia (11, 12, 31, 37, 50). Strains from each of these species have been isolated from human CF patients, and all except B. dolosa have been isolated from the plant rhizosphere or soil (10). Currently, the species most commonly associated with clinical infection are B. cenocepacia and B. multivorans (10, 32). The species associated with natural occurrences of plant disease are B. cepacia and B. cenocepacia on onion and B. cenocepacia on banana (20, 24).
Since members of most species of the B. cepacia complex occur naturally in the environment and are opportunistic human pathogens, the source of strains infecting humans has been a subject of intense interest (2, 5, 22). The environmental distribution of B. cepacia complex strains includes the rhizospheres of several agricultural-crop plants, soil in woodland and urban habitats, and river water (1, 13, 15, 16, 34, 39, 41, 55). These results may implicate environmental sources as a natural niche for potentially human-pathogenic strains. This hypothesis was reinforced by Lipuma et al. (27) with genetic data linking the identities of CF patient isolates to isolates recovered from the environment. Concomitant with these results are epidemiological studies indicating that patient-to-patient acquisition of B. cepacia complex strains decreased dramatically after the introduction of infection control measures (32, 51). Thus, new acquisitions could be due to strains originating from the environment.
The risk posed to humans by naturally occurring strains of the B. cepacia complex requires further study. For example, the prevalence of each of the nine species in natural environmental settings remains to be determined by systematic study. Rhizosphere distribution studies have been conducted on maize and rice (15, 38, 55), both plant species that are not subject to infection by B. cepacia complex strains. In contrast, onion is a host of B. cepacia; however, only one study has examined the occurrence of B. cepacia populations associated with decayed onions and onion field soil (54), and this work was done prior to the current changes in the taxonomy of the species. Thus, the occurrence and potential pathogenicity of other species of the B. cepacia complex in the onion rhizosphere have not been fully evaluated. We hypothesized that because onion is a host for plant-pathogenic strains of B. cepacia, the onion rhizosphere would harbor populations of B. cepacia and possibly other B. cepacia complex species that were distinct from those previously found in the rhizospheres of maize and rice. In addition, we wondered if isolates of any other B. cepacia complex species that colonized the onion rhizosphere would also be pathogenic to onion. The aim of this work was to isolate, identify, and determine the pathogenicity of B. cepacia complex strains from onion field soil and the rhizosphere of onion and to genetically compare these strains with reference strains from humans and the rhizospheres of other plants.
MATERIALS AND METHODS
Sample collection, bacterial isolation, and culture conditions.
Bulk soil and rhizosphere (soil loosely associated with onion roots) samples were collected on four and three sampling dates, respectively, from each of six commercial onion fields in Michigan during the 2004 onion-growing season. According to the U.S. Department of Agriculture Natural Resources Conservation Service Websoil Survey (http://websoilsurvey.nrcs.usda.gov), the soil type from each field sampled was a combination of Houghton, Adrian, and/or Palmes muck high-organic-matter soil. Bulk soil samples were taken on 6 June (populations 1 to 3, located in Eaton county) and 14 June (populations 4 to 6, located in Allegan county). In subsequent samplings, both bulk soil and rhizosphere samples were taken. Populations 1 to 3 were sampled on 21 June (onion phenology, two-leaf seedling stage), 14 July (immature-bulb stage), and 31 August (mature-bulb stage). Populations 4 to 6 were sampled on 13 July (immature-bulb stage), 2 August (immature-bulb stage), and 26 August (mature-bulb stage). Global positioning satellite (GPSMAP 76S; Garmin International, Olathe, KS) coordinates were utilized so that subsamples (five per field) could be taken from the same locations at each of the sampling dates. Soil was collected using a standard soil probe that was surface sterilized between samplings. The onion roots were collected by gently removing whole onion plants from the soil. Each sample was placed into a sterile plastic bag and transported to the laboratory on ice for processing.
Bacterial isolation and selection of putative B. cepacia complex isolates.
Soil and root samples were weighed (2-g fresh weight) and separately placed in 20 ml prechilled buffer (0.1 M potassium phosphate [pH 7.0], 0.1% peptone), and bacterial cells were dislodged by sonication for 7 min in an ultrasonic bath (model 250T; VWR Scientific, Houston, TX). Samples (0.1 ml) from appropriate serial dilutions of the sonicate were plated on trypan blue tetracycline (TB-T) (21) and P. cepacia azelaic acid tryptamine (PCAT) (7) semiselective media. The plates were incubated at 29°C for 96 h, following which 50 putative B. cepacia complex colonies were chosen (if present) for further analyses from each of the sampling substrates (soil and rhizosphere) and medium type (TB-T and PCAT) isolation combinations from the four sampling dates. The method of selecting bacterial isolates involved placing the isolation plates on a 50-square numbered grid; a single colony from each of the 50 grids (if present) was chosen randomly for further testing. The total number of subpopulation samples was 84 (six populations times seven sampling substrates times two media). Thus, a potential maximum of 4,200 (84 × 50) putative B. cepacia complex colonies could have been isolated if at least 1 colony was present in each grid on each sampling plate. A final collection of 3,794 isolates was maintained for characterization. The bacterial isolates were maintained as stock cultures in 20% glycerol at −80°C.
Identification of isolates.
A multistep hybridization and PCR method was used for identification of isolates to the individual B. cepacia complex species level. The isolates were grown on King's B (KB) agar medium (25) for 48 h at 29°C. Cells from a single colony were removed from the plates using a sterile toothpick and placed into 100 μl of lysis buffer (0.05 M KCl, 0.01 M Tris-HCl, 1% Tween 20, pH 8.3) in a sterile microcentrifuge tube. Genomic DNA for PCRs was prepared by lysing the cells at 100°C for 15 min. B. cepacia complex isolates were identified using hybridization with a 16S probe followed by a recA and 16S rRNA gene-targeted multiplex PCR assay as previously described (41, 42). Final identification to the species level was done using a restriction fragment length polymorphism analysis of PCR-amplified recA as described previously (31, 37). The sequences of the recA primers (primers BCR1 and BCR2) and 16S rRNA gene primers (primers CeMV and Burk3) were as described previously (41). For DNA-DNA dot blot hybridization, 2 μl of cell lysate was spotted onto Hybond-N+ membranes (Amersham, Piscataway, NJ), and the DNA was cross-linked to the membranes using a Stratalinker (Stratagene, La Jolla, CA). Probe labeling, hybridizations, posthybridization washes, and detection of hybridizing sequences were conducted as described previously (41). The positive control strains used in hybridizations were gifts from C. F. Gonzalez and included B. cepacia ATCC 25416, B. cenocepacia K56-2, B. multivorans ATCC 17616, and B. stabilis DB01. The negative control strains used in hybridizations were Escherichia coli DH10B, Pseudomonas aeruginosa PAO1, and Pseudomonas syringae 7B12 and DC3000.
Onion tissue maceration model.
Yellow globe onions (Allium cepa) grown in Michigan were used throughout this study in pathogenicity assays. The thin, papery covering, along with the outer bulb scale, of each onion was removed prior to quartering the bulbs with a sterile knife. The quartered individual onion scales were placed in a sterile aluminum pan (22.9 cm by 33.0 cm) containing two sheets of sterile Whatman no. 1 filter paper premoistened with 90 ml of sterile distilled water. The bacterial isolates used in the assays were grown overnight in KB broth. Individual onion scales were wounded on the inner surface with a sterile pipette tip (1- to 200-μl volume), and 5 μl of bacterial culture (107 CFU/ml) was inoculated into the wound. The onion scales were incubated at 30°C for 48 h. The degree of maceration was estimated by probing with a toothpick. A rating scale of 0 to 3 was used to indicate the degree of tissue maceration. A rating of 0 indicated no maceration, 1 indicated 1 to 33% macerated tissue area, 2 indicated 34% to 66% macerated tissue area, and 3 indicated 67% to 100% macerated tissue area. Inoculations with each isolate were replicated three times, and an average rating was tabulated. Negative controls consisted of no wounding and inoculation with B. cepacia strain ATCC 25416, and wounding and inoculation with KB broth. The positive control consisted of wounding and inoculation with B. cepacia strain ATCC 25416.
Phylogenetic analysis.
The recA gene was chosen for phylogenetic analyses of B. cepacia complex species, as that sequence had been used previously for the same purpose by other research groups (37, 41). A representative sample of 26 B. cepacia complex isolates from onion was chosen for analysis. We initially focused our isolate characterization on population 6 because that population contained the largest number of B. cepacia complex isolates and isolates from all four species. We later included randomly selected isolates representative of each population and sampling date so that each population and sampling date was represented in the final collection. In addition, recA gene sequences of 17 reference B. cepacia complex strains were obtained from GenBank. Descriptions of all of these strains are given in Table 1. The oligonucleotide primers BCR1 and BCR2 (39) were used to amplify the 1,041-bp recA coding sequence. The PCR methodology and DNA fragment purification were as described previously (29); all DNA sequencing was done at the Research Technology Support Facility at Michigan State University.
TABLE 1.
Onion rhizosphere populations of B. cepacia complex species recovered on PCAT and TB-T media on three sampling dates in 2004
| Speciesa | No. recovered on sampling dateb
|
|||||
|---|---|---|---|---|---|---|
| Day 172 | Day 194 | Day 195 | Day 214 | Day 243 | Day 238 | |
| Population 1 | ||||||
| B. cepacia | 0 | 0 | 16 | |||
| B. cenocepacia IIIB | 0 | 57 | 38 | |||
| B. ambifaria | 6 | 21 | 12 | |||
| B. pyrrocinia | 0 | 0 | 0 | |||
| Total B. cepacia complex | 6 | 78 | 66 | |||
| Population 2 | ||||||
| B. cepacia | 0 | 0 | 0 | |||
| B. cenocepacia IIIB | 0 | 44 | 33 | |||
| B. ambifaria | 82 | 35 | 26 | |||
| B. pyrrocinia | 0 | 0 | 0 | |||
| Total B. cepacia complex | 82 | 79 | 59 | |||
| Population 3 | ||||||
| B. cepacia | 0 | 0 | 0 | |||
| B. cenocepacia IIIB | 13 | 90 | 30 | |||
| B. ambifaria | 28 | 4 | 35 | |||
| B. pyrrocinia | 0 | 0 | 0 | |||
| Total B. cepacia complex | 41 | 94 | 65 | |||
| Population 4 | ||||||
| B. cepacia | 0 | 0 | 13 | |||
| B. cenocepacia IIIB | 0 | 0 | 0 | |||
| B. ambifaria | 0 | 0 | 1 | |||
| B. pyrrocinia | 0 | 0 | 0 | |||
| Total B. cepacia complex | 0 | 0 | 14 | |||
| Population 5 | ||||||
| B. cepacia | 1 | 18 | 17 | |||
| B. cenocepacia IIIB | 0 | 14 | 7 | |||
| B. ambifaria | 67 | 25 | 16 | |||
| B. pyrrocinia | 0 | 0 | 0 | |||
| Total B. cepacia complex | 68 | 57 | 40 | |||
| Population 6 | ||||||
| B. cepacia | 18 | 30 | 16 | |||
| B. cenocepacia IIIB | 0 | 53 | 60 | |||
| B. ambifaria | 36 | 10 | 27 | |||
| B. pyrrocinia | 16 | 0 | 0 | |||
| Total B. cepacia complex | 70 | 93 | 109 | |||
Nucleotide sequences were aligned by eye using MacClade (30). Phylogenetic analyses were performed using PAUP*4.0b10 (47) via neighbor joining (NJ) and maximum parsimony (MP). NJ trees were generated using the algorithm of Saitou and Nei (45) with Kimura two-parameter distances. MP analysis employed heuristic searches with 10 sequences of random taxon addition and tree bisection and reconnection branch swapping. Support for groups obtained in phylogenetic analyses was assessed by bootstrap analysis (1,000 pseudoreplicates) using PAUP*4.0. Sequences from 17 reference strains obtained from GenBank were also utilized in the analysis, and the recA sequence from B. pseudomallei strain K96243T was used as the outgroup.
Statistical analyses.
Fisher's exact test for count data, analyses of variance (ANOVA), Tukey's honestly significant difference test, and multivariate linear regressions were performed with the statistical package R (39a; http://www.R-project.org./). In multiple regressions, isolate count data (Tables 1 and 2) were found to deviate from normal distributions and were then square-root transformed prior to performing statistical calculations (40).
TABLE 2.
Number of isolates, identification by species, and recovery on each of two selective media of B. cepacia complex isolates from soil and the rhizosphere of onion from six populations in Michigan
| Species | Population | No. of isolatesa from:
|
No. of isolates on:
|
||
|---|---|---|---|---|---|
| Soil | Rhizosphere | PCAT | TB-T | ||
| B. cepacia | |||||
| 1 | 1 | 16 | 2 | 15 | |
| 2 | 4 | 0 | 1 | 3 | |
| 3 | 2 | 0 | 0 | 2 | |
| 4 | 26 | 13 | 4 | 35 | |
| 5 | 13 | 36 | 15 | 34 | |
| 6 | 6 | 43 | 11 | 38 | |
| Total | 52 | 108 | 33 | 127 | |
| B. cenocepacia | |||||
| 1 | 24 | 95 | 70 | 49 | |
| 2 | 0 | 77 | 52 | 25 | |
| 3 | 25 | 128 | 74 | 79 | |
| 4 | 0 | 0 | 0 | 0 | |
| 5 | 8 | 8 | 10 | 6 | |
| 6 | 2 | 113 | 90 | 25 | |
| Total | 59 | 421 | 296 | 184 | |
| B. ambifaria | |||||
| 1 | 4 | 39 | 7 | 36 | |
| 2 | 28 | 143 | 44 | 127 | |
| 3 | 35 | 73 | 18 | 90 | |
| 4 | 1 | 1 | 0 | 2 | |
| 5 | 72 | 106 | 32 | 146 | |
| 6 | 48 | 73 | 24 | 97 | |
| Total | 188 | 435 | 125 | 498 | |
| B. pyrrocinia | |||||
| 1 | 0 | 0 | 0 | 0 | |
| 2 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 0 | |
| 6 | 11 | 16 | 9 | 18 | |
| Total | 11 | 16 | 9 | 18 | |
RESULTS
Bacterial isolation and isolate identification.
Totals of 310 and 980 presumptive B. cepacia complex isolates were recovered from onion soil and the onion rhizosphere, respectively. The ratio of B. cepacia complex isolates identified to total bacterial isolates selected from the PCAT and TB-T plates was 1,290/3,798 (34%). Isolate identification to the species level was done by comparing restriction fragment patterns of the recA gene fragment (data not shown), and the Burkholderia species identified in this study were B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia. Specific recA restriction patterns were also utilized to determine that all of the B. cenocepacia isolates recovered belonged to the previously described III-B lineage (31).
Recovery of B. cepacia complex isolates from bulk soil and the onion rhizosphere.
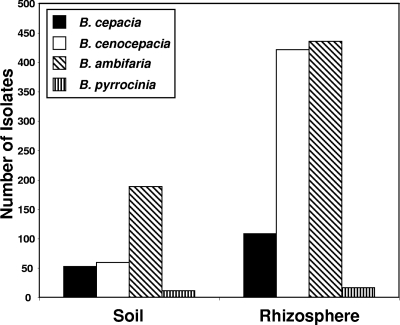
B. cepacia complex isolates were less prevalent in bulk soil samples than in the rhizosphere, as the soil isolates comprised 24% of the 1,290 total B. cepacia complex isolates. The predominant species present in bulk soil was B. ambifaria, representing 61% of the total soil isolates, with B. cepacia, B. cenocepacia, and B. pyrrocinia representing 17%, 19%, and 3%, respectively (Fig. 1). B. ambifaria was also the most common organism recovered from the rhizosphere but was recovered in proportions similar to those of B. cenocepacia (44% and 43%, respectively, of the total) (Fig. 1), which differed from the proportions of these organisms in bulk soil. Isolates of the onion pathogen B. cepacia were also recovered from the onion rhizosphere (11% of the total), and B. pyrrocinia was recovered from only one site (soil and rhizosphere) and represented 2% of the total isolates (Fig. 1).
FIG. 1.
Total numbers of B. cepacia complex isolates (B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia) recovered from field soil and the rhizosphere of onion from six populations in Michigan over three sampling dates in 2004.
A numerical analysis of rhizosphere isolates recovered from each field population indicated differences in the total numbers and frequencies of species per population, as well as differences according to the sampling date. For example, at least 150 B. cepacia complex isolates were recovered from five of the six populations, but only 14 isolates were recovered from population 4 (Table 1). B. ambifaria was commonly observed at all three sampling times, but only 13 isolates of B. cenocepacia (all from population 3) were recovered at the first sampling, when the onion root mass was very small, as the roots were in initial stages of development (Table 1). At the second sampling, when the onion roots were extensively developed, there was a spike in recovery of B. cenocepacia in all populations except population 4, while the numbers of B. ambifaria isolates decreased in most cases (Table 1).
A summary analysis of the frequency of isolation of each species from bulk soil and the onion rhizosphere from each field population is shown in Table 2. Populations were used as a factor, with six modalities. The effects of populations on the number of isolates recovered from soil or rhizosphere was determined by multiple linear regression analyses, and the statistical significance was determined by using 1,000 permutations of the data. The population origin had no significant effect on the number of isolates recovered from soil (F = 0.141; P = 0.758) nor on that recovered from the rhizosphere (F = 0.230; P = 0.542). Interestingly, species had a significant effect on the number of isolates recovered from both soil (F = 0.661; P = 0.015) and rhizosphere (F = 0.861; P = 0.003). For each species, a significant difference was found between the number of isolates retrieved from soil and rhizosphere: B. cepacia (Fisher's exact test), P = 7.611−10; B. cenocepacia, P = 9.114−12; and B. ambifaria, P = 2.885−8. In each case, there were significantly more isolates recovered from the rhizosphere than from soil (Table 2).
Effects of location and sampling time on the composition of B. cepacia complex populations.
Statistical analyses were performed on populations grouped by field location, with group 1 comprising populations 1 to 3 and group 2 comprising populations 4 to 6. There was a significant association of population with sampling date in the number of recovered B. cepacia complex isolates in group 1 (Fisher's exact test, P = 1.926−13). The detailed analysis revealed that only isolate counts at day 195 and day 243 were not significantly different from each other across the three populations (P = 0.693), whereas all other pairwise comparisons were highly significant (P ≪ 0.05). A significant association of population with sampling date in the number of recovered B. cepacia complex isolates was also observed in group 2 (Fisher's exact test, P = 2.349−8). All pairwise comparisons among sampling dates produced significant differences among populations (P ≪ 0.05), and all pairwise comparisons of populations were significant, except between populations 4 and 5 (P = 0.058).
To test the effects of the sampling time on B. cepacia complex species recovery, each population was treated as a replicate for statistical analyses. B. pyrrocinia was removed from the subsequent analyses because there were data only on day 194 in population 6. ANOVA were used to determine if the factor time had an influence on the isolate recovery rate for each separate species. The sampling date was significant only to explain B. cenocepacia isolate counts (ANOVA, 5 df; F = 4.272; P = 0.0183), marginally significant for B. cepacia isolate counts (F = 2.552; P = 0.085), and not significant to explain B. ambifaria isolate counts (F = 0.4309; P = 0.819). Tukey's honestly significant tests were further applied to the B. cenocpacia data to determine which dates provided significant differences in the mean counts. All pairwise comparisons of mean differences were not significantly different from each other, except between dates 172 and 214 (mean difference = 63.7; P = 0.022) and between dates 194 and 214 (mean difference = 59.3; P = 0.034).
Frequencies of recovery of B. cepacia complex isolates on PCAT and TB-T selective media.
A numerical analysis of the isolation frequencies for the four B. cepacia complex species indicated a differential recovery of each species on PCAT and TB-T selective media (Table 2). Both B. cepacia and B. ambifaria were preferentially recovered on TB-T medium, with 79% and 80% of the total isolates, respectively, of the two species obtained from TB-T (Table 2). Similar results were observed for B. pyrrocinia, although only 27 total isolates were obtained in this study (Table 2). In contrast, B. cenocepacia was recovered more efficiently on PCAT medium, with 62% of the total isolates obtained on this medium (Table 2).
Onion pathogenesis.
Each of the 1,290 B. cepacia complex isolates identified in this study was assayed for pathogenicity on detached onion bulb scales (Fig. 2). After inoculation and incubation at 30°C for 48 h, the percent area of macerated tissue was estimated, and a rating of 0 to 3 was given. Isolates of each of the species examined (B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia) were determined to be pathogenic on onion bulb scales, and only 14 of 1,290 total isolates were nonpathogenic (Table 3). The large majority of isolates tested were highly virulent, with pathogenicity ratings of 2 or 3 indicating maceration of 34 to 100% of the onion bulb tissue (Table 3). In addition to water-soaking and maceration symptoms, necrosis was observed with a small percentage of the B. cepacia isolates. A total of 86% of isolates of B. cepacia (137/160) were highly virulent (rating, 2 to 3) (Fig. 2) in this model; this frequency was actually surpassed by B. cenocepacia, where 90% (434/480) of the isolates were highly virulent (Table 3). A slightly reduced percentage of B. ambifaria isolates (76%; 476/623) and only 15% (4/27) of B. pyrrocinia isolates were highly virulent (Table 3).
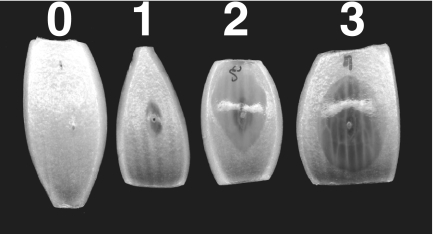
FIG. 2.
Rating system for the onion pathogenesis assay used to type B. cepacia complex isolates. Onion slices were wounded, inoculated with 5 μl of bacterial culture (107 CFU/ml), and incubated at 30°C for 48 h. A rating scale of 0 to 3 was used to indicate the degree of tissue maceration. A rating of 0 indicated no maceration, 1 indicated 1 to 33% macerated tissue area, 2 indicated 34% to 66% macerated tissue area, and 3 indicated 67% to 100% macerated tissue area.
TABLE 3.
Pathogenicities of B. cepacia complex isolates from soil and the rhizosphere of onion in Michigan
| Species | No. of isolates | No. of isolates with an onion pathogenicity rating ofa:
|
|||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| B. cepacia | 160 | 0 | 23 | 52 | 85 |
| B. cenocepacia | 480 | 0 | 46 | 174 | 260 |
| B. ambifaria | 623 | 4 | 143 | 223 | 253 |
| B. pyrrocinia | 27 | 10 | 13 | 3 | 1 |
The positive control strain B. cepacia ATCC 25416 was included in each pathogenicity assay and consistently produced a pathogenicity rating of 3.
Phylogenetic analysis.
The recA genes from 26 B. cepacia complex isolates collected in this study were amplified by PCR and sequenced. Phylogenetic analysis of 1,041 aligned nucleotide positions of the recA gene was carried out using NJ and MP (Table 4). The two techniques yielded essentially identical results, and the NJ tree is shown in Fig. 3. The NJ and MP trees were characterized by the presence of five well-supported major lineages, corresponding to the four recognized species (B. cenocepacia III-B, B. cepacia, B. pyrrocinia, and B. ambifaria) plus a fifth lineage comprised of three additional B. cenocepacia (III-A) strains. The relationships of these five major groups to each other in our analyses are the same as that observed by Payne et al. (37), with the exception that in our analysis, the clade containing B. cenocepacia J2315 was basal to the B. cepacia, the B. pyrrocinia, and the other B. cenocepacia clades (instead of being a sister to B. ambifaria).
TABLE 4.
Strains of B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia utilized for a phylogenetic analysis of recA sequences
| Species and straina | Source or relevant information | GenBank accession no. of recA sequence | Source or reference | |
|---|---|---|---|---|
| B. cepacia | ||||
| 3ST4 | Onion field soil | EU079008 | This study | |
| 4ST9 | Onion field soil | EU079004 | This study | |
| 5RT136 | Onion rhizosphere | EU079007 | This study | |
| 5RT161 | Onion rhizosphere | EU079011 | This study | |
| 6RP83 | Onion rhizosphere | EU079006 | This study | |
| 6RT68 | Onion rhizosphere | EU079005 | This study | |
| 6RT141 | Onion rhizosphere | EU079009 | This study | |
| 6SP128 | Onion field soil | EU079010 | This study | |
| KBC-3 | Maize rhizosphere | AY769897 | 40 | |
| ATCC 25416 | Onion | AF143786 | 31 | |
| B. cenocepacia | ||||
| 1RP102 | Onion rhizosphere | EU079000 | This study | |
| 3RP82 | Onion rhizosphere | EU078998 | This study | |
| 6RP111 | Onion rhizosphere | EU079003 | This study | |
| 6RT111 | Onion rhizosphere | EU078999 | This study | |
| 6RT130 | Onion rhizosphere | EU079001 | This study | |
| 6RT131 | Onion rhizosphere | EU079002 | This study | |
| AU1054 | Human pathogen | NC_008060.1 | 27 | |
| B9 | Banana, Taiwan | AY598028 | 26 | |
| CA2 | Banana, Taiwan | DQ664181 | 26 | |
| GQ52 | Banana, Taiwan | DQ664182 | 26 | |
| HI2424 | Soil | NC_008542 | 27 | |
| J2315 | Human pathogen, United Kingdom | −b | 31 | |
| KBC-4 | Maize rhizosphere | AY769898 | 40 | |
| K56-2 | Human pathogen, Canada | AF143779 | 31 | |
| MC0-3 | Maize rhizosphere | NZ_AAVA01000001 in silico
|
||
| MQ41 | Banana, Taiwan | DQ664183 | 26 | |
| PC184 | Human pathogen, United States | AF143784 | 31 | |
| B. ambifaria | ||||
| 2SP165 | Onion field soil | EU079012 | This study | |
| 5RT130 | Onion rhizosphere | EU079013 | This study | |
| 5SP7 | Onion field soil | EU079017 | This study | |
| 6RT55 | Onion rhizosphere | EU079016 | This study | |
| 6SP146 | Onion field soil | EU079018 | This study | |
| 6ST151 | Onion field soil | EU079014 | This study | |
| 6ST196 | Onion field soil | EU079015 | This study | |
| AMMD | Pea rhizosphere | NC_008390 | 36 | |
| AU0212 | Human isolate | AF456005 | 37 | |
| KBC-7 | Maize rhizosphere | AY769901 | 40 | |
| B. pyrrocinia | ||||
| 6RP78 | Onion rhizosphere | EU079020 | This study | |
| 6RT51 | Onion rhizosphere | EU079019 | This study | |
| 6RT94 | Onion rhizosphere | EU079021 | This study | |
| 6ST17 | Onion field soil | EU079023 | This study | |
| 6ST128 | Onion field soil | EU079022 | This study | |
| ATCC 39277 | Soil, United States | AF143801 | 14 | |
| BC011 | Water, United States | AF456045 | 14 | |
Codes for identification of B. cepacia complex strains from this study are as follows. The first number represents the populations, R or S refers to recovery from rhizosphere or soil, P or T refers to recovery on PCAT or TB-T medium, and the last number refers to the sampling date (1 to 50, first sampling date; 51 to 100, second sampling date; 101 to 150, third sampling date; 151 to 200, fourth sampling date).
Draft sequence information for B. cenocepacia J2315 recA is available from http://www.sanger.ac.uk/Projects/B_cenocepacia/.
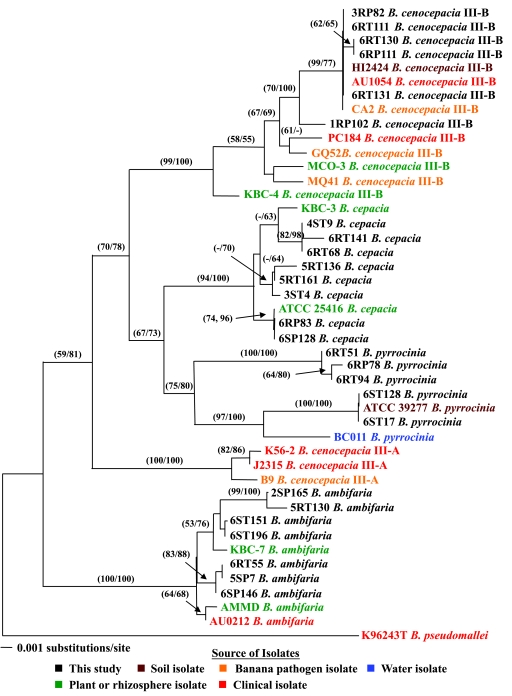
FIG. 3.
Phylogenetic analysis of the recA data set from 45 B. cepacia complex strains with B. pseudomallei K96243T as the outgroup (descriptions of isolates and reference strains are in Table 4). A color key is used to designate the source of isolates used in this analysis. The naming convention for the isolates from this study (e.g., 3RP82) is defined in a footnote to Table 4. The tree shown was obtained by NJ using Kimura two-parameter distances. The numbers in parentheses (MP/NJ) on the branches show the MP and NJ bootstrap values (>50).
In total, four to six polymorphic recA alleles were identified from the onion isolates in this study (Fig. 3). The onion isolates recovered in the study each fell within one of the major clades. For example, 1RP102, 6RT131, 6RP111, 6RT130, 6RT111, and 3RP82 all fell within the well-supported B. cenocepacia (III-B) clade, which includes the human pathogens B. cenocepacia AU1054 and PC184 (Fig. 3). The onion isolates of B. cepacia, B. pyrrocinia, and B. ambifaria were all grouped similarly with the reference strains and, in some cases, shared identical DNA sequences (Fig. 3).
DISCUSSION
Three B. cepacia complex species, B. cepacia, B. cenocepacia, and B. ambifaria, were regularly isolated and a fourth, B. pyrrocinia, was sporadically isolated from the onion rhizospheres and onion field soils from several locations in Michigan. B. cenocepacia and B. ambifaria were the most prevalent species recovered in this study, representing 85.5% of the total (Table 1). This result is somewhat surprising considering that B. cepacia has historically been the species most often associated with onion rot diseases (53) and the association of B. cenocepacia with the onion rhizosphere had not been previously documented. In this study, only 11.2% of the onion rhizosphere isolates were B. cepacia, which is close to the 20.4% recovery of B. cepacia reported from the maize rhizosphere (41). Infection of onion occurs as bacteria invade wounds in young leaves and move downward through leaves into the corresponding scales in the bulb (24). Splashing water from rain or overhead irrigation is thought to disseminate bacterial inoculum from the soil to the necks of onion plants and plays a key role in the level of disease incidence (49). The linkage between rhizosphere populations and onion disease has not been fully elucidated, but presumably the increased populations of pathogenic B. cepacia complex bacteria around developing onion leaves and bulbs provide bacteria that can be disseminated to wounds by splashing water.
The plant rhizosphere appears to represent an important ecological niche for B. cepacia complex bacteria, as these organisms have been isolated from or shown to colonize the rhizospheres of diverse hosts, including perennial ryegrass, pea, cotton, and coffee (48). The high frequency of B. ambifaria in onion field soil and in the onion rhizosphere adds to a body of knowledge that this species comprises strains that are adapted to the soil environment and are effective colonizers of the rhizospheres of many plant species (16, 42). Likewise, B. cenocepacia strains have also been isolated from soil and the rhizospheres of plants, including maize, wheat, and lupine (1). The environmental prevalence of B. cenocepacia is potentially clinically significant in that an epidemic CF strain has also been isolated from onion field soil from New York (27).
In this study, significantly more B. cenocepacia bacteria were recovered from rhizosphere than from soil samples, and analyses of the temporal distribution also indicated a significant association between the sampling time and recovery of B. cenocepacia. This species was most commonly isolated after the development of the onion rhizosphere and was extremely rare at the first sampling time, when onion roots were in an initial stage of growth (Table 1). The temporal results suggest that B. cenocepacia is efficiently selected from soil by the onion rhizosphere and that the organism can then readily colonize this habitat. A comparison of distribution analyses from other studies indicated that B. cenocepacia was widely prevalent in the rhizospheres of wheat and lupine (1) but represented only a small fraction of the recovered B. cepacia complex strains from the maize rhizosphere (3.2% of the total) (41) and was not recovered from the rice rhizosphere (55). However, in another study, B. cenocepacia represented 70.5% of the isolates from the maize rhizosphere from field sites in Italy (17). It should be noted that a follow-up analysis of some of these isolates using multilocus sequence typing indicated that only 7% were B. cenocepacia, with another 39% belonging to potentially new B. cepacia complex groups that were closely related to but distinct from B. cenocepacia (15). It is unknown how plant species and environmental factors influence the rhizosphere colonization of B. cenocepacia. The prevalence of the species in the onion rhizosphere, coupled with the knowledge that isolates of B. cenocepacia from diseased onions were shown to be pathogenic on onion in a prior study (46), implies that host factors unique to the onion plant may play an important role in the colonization of onion by B. cenocepacia.
The numbers of B. cepacia complex isolates from five of six onion field sampling locations in this study were relatively similar. However, sampling location 4 yielded only 14 total B. cepacia complex isolates, all obtained at the third sampling time (Table 1). The factors resulting in this much reduced occurrence are currently unknown. In Michigan, onion crops are sometimes rotated in the following season with crops such as maize, radish, or soybean. If B. cepacia complex bacteria do not colonize the rhizospheres of these hosts as readily as that of onion, the overall soil populations could decrease over time. The factors resulting in the reduced B. cepacia complex populations at sampling location 4 could reveal important insights into the ecology of these organisms in managed agricultural habitats.
In this study, we found that various B. cepacia complex species were differentially recovered on the PCAT and TB-T selective media. Our results are similar to those of Dalmastri et al. (17) in this regard, with approximately four times more B. cepacia and B. ambifaria bacteria recovered on TB-T than on PCAT medium and an increased number of B. cenocepacia bacteria recovered on PCAT medium. Although neither of these media is sufficiently selective to eliminate other organisms, as shown in this study and in previous studies (21, 34), we recommend that both PCAT and TB-T media be used in combination in population analyses of B. cepacia complex from environmental sources.
From a total of 1,290 B. cepacia complex isolates examined in an onion pathogenicity assay, only 14 were incapable of causing water-soaking and maceration symptoms, while 1,051 (81.5%) caused significant symptoms with a disease rating of 2 or 3 (Table 3). To the best of our knowledge, this is the first report of B. ambifaria and B. pyrrocinia causing water-soaking and maceration symptoms in onion bulb scale tissue. The results of many studies have shown that strains of B. cepacia and B. cenocepacia are capable of causing water soaking and onion tissue maceration in laboratory inoculations (9, 18, 52, 53). Many clinical B. cenocepacia strains isolated from CF patients are nonpathogenic on onion (6, 19, 53), although some CF strains, including known epidemic strains, have been shown to cause onion maceration (9). In addition, one strain of B. vietnamiensis was reported to produce the plant tissue water-soaking phenotype on onion (18).
Phylogenetic analysis, based on the recA sequence, was used to identify four well-supported major groups, which corresponded to recognized species as reported previously (37). Onion isolates of B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia clustered with reference strains in each of the species. A recent phylogenetic analysis using multilocus sequence typing included five sublineages of B. cenocepacia, with the human pathogen strains AU1054 and PC184 placed in lineage III-B and J2315 and K56-2 placed in lineage III-A (3). The recA sequence from AU1054 was identical or closely similar to those of several of the onion isolates from this study. The determination of whether onion isolates such as 6RT131 are potential human pathogens is a current focus in our laboratory.
The recA sequences from onion isolates of B. ambifaria analyzed in this study were similar to those of the reference strains KBC-7, isolated from the maize rhizosphere; AMMD, isolated from the pea rhizosphere; and AU0212, a human isolate from a CF patient (12). However, of the B. ambifaria isolates examined, only the isolates from onion macerated onion tissue in a pathogenicity assay, while the reference strains AMMD and KBC-7 did not (data not shown). A major virulence factor in onion pathogenicity is the presence of a polygalacturonase enzyme involved in tissue maceration that is encoded by the plasmid-borne pehA gene (20). It is possible that virulence determinants are disseminated among B. cepacia complex bacteria via horizontal transfer in the onion rhizosphere.
The results of many gene distribution studies have shown that species such as B. cenocepacia are comprised of subpopulations that differ in the carriage of specific virulence genes associated with human or animal pathogenesis (reviewed in reference 32). In some cases, rhizosphere or plant pathogen isolates of B. cenocepacia have also been examined for the distribution of markers, such as the B. cepacia epidemic strain marker and cable pilin gene (4, 26). The genetic distinction between plant pathogen and human pathogen appears to be blurred in species such as B. cenocepacia, and the relatively large genome sizes of these organisms seem to enable the carriage of a diverse gene set conferring fitness in a wide range of ecological niches. Studies of the genetic requirement for plant rhizosphere colonization in B. cepacia complex species have been initiated (35), and this topic has been examined in related organisms, such as P. aeruginosa and Pseudomonas putida (28, 33, 43). The role of the plant host in the selection of rhizosphere-colonizing B. cepacia complex strains and the maintenance of specific virulence genes also remain to be elucidated.
In summary, we determined that B. cepacia, B. cenocepacia, and B. ambifaria were the dominant B. cepacia complex species present in the onion rhizosphere and onion field soil in Michigan. The vast majority of isolates from each of these species and a few isolates of B. pyrrocinia were pathogenic in an onion disease assay, causing water soaking and tissue maceration. While B. cenocepacia collectively includes organisms that are opportunistic human pathogens and pathogens of plants, including onion and banana, B. ambifaria and B. pyrrocinia had not been previously reported as potential plant pathogens. The genetic content, distribution, and mobility of gene sequences involved in virulence are variable within the B. cepacia complex. Our current investigations focus on the identification of gene sets important for rhizosphere colonization and plant pathogenesis and the ecological factors that influence the maintenance of these determinants in environmental B. cepacia complex populations.
Acknowledgments
This work was supported by grants from the Michigan State University Project GREEEN, the Michigan State University Center for Microbial Pathogenesis, and the Michigan Agricultural Experiment Station.
We thank Jim Tiedje for useful discussions.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Balandreau, J., V. Viallard, B. Cournoyer, T. Coeyne, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, A., E. Mahenthiralingam, P. Drevinek, P. Vandamme, J. R. Govan, D. J. Waine, J. J. LiPuma, L. Chiarini, C. Dalmastri, D. A. Henry, D. P. Speert, D. Honeybourne, M. C. J. Maiden, and C. G. Dowson. 2007. Environmental Burkholderia cepacia complex isolates in human infections. Emerg. Infect. Dis. 13:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. J. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevivino, A., S. Tabacchoni, L. Chiarini, M. V. Carusi, M. Del Gallo, and P. Visca. 1994. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology 140:1069-1076. [DOI] [PubMed] [Google Scholar]
- 7.Burbage, D. A., and M. Sasser. 1982. A medium selective for Pseudomonas cepacia. Phytopathology 72:706. [Google Scholar]
- 8.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-118. [Google Scholar]
- 9.Butler, S. L., C. J. Doherty, J. E. Hughes, J. W. Nelson, and J. R. W. Govan. 1995. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J. Clin. Microbiol. 33:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiarini, L., A. Bevivino, C. Dalmastri, S. Tabacchioni, and P. Visca. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277-286. [DOI] [PubMed] [Google Scholar]
- 11.Coeyne, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroucke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI: a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 12.Coeyne, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov.: a novel member of the Burkholderia cepacia complex comprising biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 13.Coeyne, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 14.Coeyne, T., P. Vandamme, J. J. LiPuma, J. R. W. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalmastri, C., A. Baldwin, S. Tabacchioni, A. Bevivino, E. Mahenthiralingam, L. Chiarini, and C. Dowson. 2007. Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ. Microbiol. 9:1632-1639. [DOI] [PubMed] [Google Scholar]
- 16.Dalmastri, C., A. Fiore, C. Alisi, A. Bevivino, S. Tabacchioni, G. Giuliano, A. R. Sprocati, L. Segre, E. Mahenthiralingam, L. Chiarini, and P. Vandamme. 2003. A rhizospheric Burkholderia cepacia complex population: genotypic and phenotypic diversity of Burkholderia cenocepacia and Burkholderia ambifaria. FEMS Microbiol. Ecol. 46:179-187. [DOI] [PubMed] [Google Scholar]
- 17.Dalmastri, C., L. Pirone, S. Tabacchioni, A. Bevivino, and L. Chiarini. 2005. Efficacy of species-specific recA PCR tests in the identification of Burkholderia cepacia complex environmental isolates. FEMS Microbiol. Lett. 246:39-45. [DOI] [PubMed] [Google Scholar]
- 18.Engledow, A. S., E. G. Medrano, E. Mahenthiralingam, J. J. LiPuma, and C. F. Gonzalez. 2004. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype in Burkholderia cenocepacia. J. Bacteriol. 186:6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, C. F., and A. K. Vidaver. 1979. Bacteriocin, plasmid and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. J. Gen. Microbiol. 110:161-170. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez, C. F., E. A. Pettit, V. A. Valadez, and E. Provin. 1997. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol. Plant-Microbe Interact. 10:840-851. [DOI] [PubMed] [Google Scholar]
- 21.Hagedorn, C., W. D. Gould, T. R. Bardinelli, and D. R. Gustavson. 1987. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl. Environ. Microbiol. 53:2265-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, A., J. Govan, and R. Goldstein. 1998. Agricultural use of Burkholderia (Pseudomonas) cepacia: a threat to human health? Emerg. Infect. Dis. 4:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isles, A. L., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 24.Kawamoto, S. O., and J. W. Lorbeer. 1974. Infection of onion leaves by Pseudomonas cepacia. Phytopathology 64:1440-1445. [Google Scholar]
- 25.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the detection of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 26.Lee, Y.-A., and C.-W. Chan. 2007. Molecular typing and presence of genetic markers among strains of banana finger-tip rot pathogen, Burkholderia cenocepacia, in Taiwan. Phytopathology 97:195-201. [DOI] [PubMed] [Google Scholar]
- 27.Lipuma, J. J., T. Spilker, T. Coeyne, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 28.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 29.Ma, Z., T. J. Proffer, J. L. Jacobs, and G. W. Sundin. 2006. Overexpression of the 14α-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 72:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddison, D. R., and W. P. Maddison. 2003. MacClade: analysis of phylogeny and character analysis, version 4.06. Sinauer, Sunderland, MA.
- 31.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 33.Mark, G. L., J. M. Dow, P. D. Kiely, H. Higgins, J. Haynes, C. Baysse, A. Abbas, T. Foley, A. Franks, J. Morrissey, and F. O'Gara. 2005. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA 48:17454-17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, S. C. M., J. J. LiPuma, and J. L. Parke. 2002. Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Sullivan, L. A., A. J. Weightman, T. H. Jones, A. M. Marchbank, J. M. Tiedje, and E. Mahenthiralingam. 2007. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ. Microbiol. 9:1017-1034. [DOI] [PubMed] [Google Scholar]
- 36.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 37.Payne, G. W., P. Vandamme, S. H. Morgan, J. J. LiPuma, T. Coeyne, A. J. Weightman, T. H. Jones, and E. Mahenthiralingam. 2005. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71:3917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne, G. W., A. Ramette, H. L. Rose, A. J. Weightman, T. H. Jones, J. M. Tiedje, and E. Mahenthiralingam. 2006. Application of a recA gene-based identification approach to the maize rhizosphere reveals novel diversity in Burkholderia species. FEMS Microbiol. Lett. 259:126-132. [DOI] [PubMed] [Google Scholar]
- 39.Pirone, L., L. Chiarini, C. Dalmastri, A. Bevivino, and S. Tabacchioni. 2005. Detection of cultured and uncultured Burkholderia cepacia complex bacteria naturally occurring in the maize rhizosphere. Environ. Microbiol. 7:1734-1742. [DOI] [PubMed] [Google Scholar]
- 39a.R Development Core Team. 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 40.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramette, A., and J. M. Tiedje. 2007. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. USA 104:2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos-Gonzalez, M. I., M. J. Campos, and J. L. Ramos. 2005. Analysis of Pseudomonas putida KT2440 gene expression in the maize rhizosphere: in vitro expression technology capture and identification of root-activated promoters. J. Bacteriol. 187:4033-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenstein, B. J., and D. E. Hall. 1980. Pneumonia and septicemia due to Pseudomonas cepacia in a patient with cystic fibrosis. Johns Hopkins Med. J. 147:188-189. [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Sotokawa, N., and Y. Takikawa. 2004. Occurrence of bacterial rot of onion bulbs caused by Burkholderia cepacia in Japan. J. Gen. Plant Pathol. 70:348-352. [Google Scholar]
- 47.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 48.Tabacchioni, S., A. Bevivino, C. Dalmastri, and L. Chiarini. 2002. Burkholderia cepacia complex in the rhizosphere: a minireview. Ann. Microbiol. 52:103-117. [Google Scholar]
- 49.Teviotdale, B. L., R. M. Davis, J. P. Guerard, and D. H. Harper. 1989. Effect of irrigation management on sour skin of onion. Plant Dis. 73:819-822. [Google Scholar]
- 50.Vandamme, P., D. Henry, T. Coeyne, S. Nuzla, M. Vancanneyt, J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia: two additional Burkholderia cepacia complex bacteria may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, N. M., A. A. Casano, L. P. Manangan, R. L. Sinkowitz-Cochran, and W. R. Jarvis. 2002. Risk factors for Burkholderia cepacia colonization and infection among cystic fibrosis patients. J. Pediatr. 141:512-517. [DOI] [PubMed] [Google Scholar]
- 52.Wigley, P., and N. F. Burton. 1999. Genotypic and phenotypic relationships in Burkholderia cepacia isolated from cystic fibrosis patients and the environment. J. Appl. Microbiol. 86:460-468. [DOI] [PubMed] [Google Scholar]
- 53.Yohalem, D. S., and J. W. Lorbeer. 1994. Intraspecific metabolic diversity among strains of Burkholderia cepacia isolated from decayed onions, soils, and the clinical environment. Antonie van Leeuwenhoek 65:111-131. [DOI] [PubMed] [Google Scholar]
- 54.Yohalem, D. S., and J. W. Lorbeer. 1997. Distribution of Burkholderia cepacia phenotypes by niche, method of isolation and pathogenicity to onion. Ann. Appl. Biol. 130:467-479. [Google Scholar]
- 55.Zhang, L., and G. Xie. 2007. Diversity and distribution of Burkholderia cepacia complex in the rhizosphere of rice and maize. FEMS Microbiol. Lett. 266:231-235. [DOI] [PubMed] [Google Scholar]