Abstract
As a monophyletic group, the myxobacteria are known to produce a broad spectrum of secondary metabolites. However, the degree of metabolic diversity that can be found within a single species remains unexplored. The model species Myxococcus xanthus produces several metabolites also present in other myxobacterial species, but only one compound unique to M. xanthus has been found to date. Here, we compare the metabolite profiles of 98 M. xanthus strains that originate from 78 locations worldwide and include 20 centimeter-scale isolates from one location. This screen reveals a strikingly high level of intraspecific diversity in the M. xanthus secondary metabolome. The identification of 37 nonubiquitous candidate compounds greatly exceeds the small number of secondary metabolites previously known to derive from this species. These results suggest that M. xanthus may be a promising source of future natural products and that thorough intraspecific screens of other species could reveal many new compounds of interest.
Prokaryotes continue to provide important leads in the search for medically important bioactive natural products (1, 11). In particular, the myxobacteria have emerged as a promising source of natural products that exhibit highly diverse structures and biological activities (9). While most myxobacterial genera produce secondary metabolites, difficulties with cultivation and genetic manipulation hinder exploitation of the biosynthetic potential of many species. In contrast, the relatively fast growth and extensive genetic characterization of Myxococcus xanthus make the identification of natural products in this model species of particular interest.
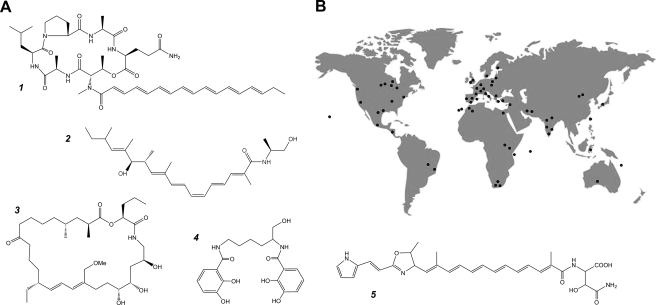
Myxococcus xanthus has been thoroughly investigated due to its amazing social behaviors, which include coordinated swarming over surfaces and a complex life cycle that culminates in the formation of multicellular fruiting bodies under starvation conditions (23). However, little attention has been paid to secondary metabolite production in this model species. M. xanthus strain DK1622 is the genotype most commonly studied with respect to social interactions and development. This strain has been found to produce several secondary metabolites, including the myxochromids (27), myxalamids (13), the antibiotic myxovirescin (8), and the siderophore myxochelin (15) (Fig. 1A). The molecular basis of synthesis for these compounds has been studied in some detail (25, 26, 30) and has been greatly facilitated by the availability of the DK1622 genome sequence (10). Very recently, a novel secondary metabolite class unique to M. xanthus, the DKxanthenes, was characterized and was shown to be essential for viable-spore formation (18). Additional reports of secondary metabolites from M. xanthus are limited to cittilin (20) and the antibiotics saframycin (12) and althiomycin (16).
FIG. 1.
(A) Structures of the secondary metabolites reported to derive from Myxococcus xanthus strain DK1622 to date. 1, myxochromid A2; 2, myxalamid A; 3, myxovirescin A; 4, myxochelin A; 5, DKxanthene-534. (B) Global distribution of sampling sites from which the 98 M. xanthus strains under investigation in this study originated. The predominance of subtropical and tropical zones with moderate-to-warm temperatures reflects the worldwide abundance pattern of many myxobacterial species (5). Multiple isolates from regional or local spatial ranges were summarized as single spots.
Most of these M. xanthus compounds are polyketides or nonribosomal peptides or hybrids thereof, and their biosynthesis is accomplished by large multienzyme complexes, the polyketide synthases (PKSs) or the nonribosomal peptide synthetases (NRPSs) (2), respectively. Genomic mining has revealed many putative natural product biosynthesis genes in the M. xanthus DK1622 genome, including at least 18 gene clusters encoding secondary metabolite assembly lines for PKSs, NRPSs, or hybrid compounds (10). This finding suggests that DK1622 should produce a substantial number of secondary metabolites, but only a few have actually been detected to date.
To search for additional secondary-metabolite diversity in M. xanthus, we screened 98 M. xanthus strains isolated from locations worldwide (Fig. 1B). We sought to determine whether the metabolic inventory of this species is largely present in DK1622 or whether there is significant potential for novel compound discoveries in other strains.
Multiple approaches have been used to screen microbes for novel natural products. While activity-guided methods have the potential to directly deliver candidate compounds for a specific biological activity, they are inherently biased due to the choice of targets in the activity assays. Chemical screening can rapidly reveal the presence of diverse compound classes in a set of samples but is nonetheless constrained by immense differences in the chromatographic and spectroscopic properties of natural products. For example, the still widely used method of UV/visible light detection by diode array detector-coupled liquid chromatography disproportionately reveals compounds with the most eye-catching absorption characteristics.
Liquid chromatography-coupled mass spectrometry (LC-MS) is now considered the most informative technique for analysis of bacterial secondary metabolite profiles and promises to detect the largest possible range of actual chemical diversity in a given sample. We therefore implemented a combination of fast chromatographic separation (UPLC), electrospray ionization (ESI), and high-resolution time of flight mass spectrometry (TOF-MS) as our primary screening platform (14). In addition to in-depth manual data examination, we use principal component analysis (PCA) in characterizing secondary metabolite profiles for the first time. Intriguingly, our analysis reveals that from a set of 98 M. xanthus isolates, candidate metabolites for 37 novel natural products were identified. Statistical analysis suggests that these 37 compounds are likely to represent ∼80% of the total natural secondary metabolome diversity in M. xanthus.
MATERIALS AND METHODS
Strains, fermentation conditions, and sample workup.
Strains were from the HZI collection (Helmholtz Centre for Infection Research, Germany; formerly GBF; 26 “Mxx” strains), from Dale Kaiser (5 “DK” strains), and from the collection of M.V. and G.J.V. (67 worldwide isolates) (29). The isolation of M. xanthus strains from a centimeter-scale population (including the 19 “A” strains and strain B5) in Tübingen, Germany, was described previously (28). All M. xanthus strains were fermented for 96 h in 50 ml liquid CTT medium (10 g of casitone, 10 ml of 0.8 M MgSO4, 10 ml of 1 M Tris-HCl, distilled H2O added to give 1 liter, and pH adjustment to 7.6 prior to autoclaving) on the same rotary shaker (Infors, Germany) at 200 rpm and 30°C in 250-ml Erlenmeyer flasks. The cultures were supplemented with 2% adsorber resin (Amberlite XAD-16; Sigma-Aldrich). After fermentation, cells and resin were harvested by centrifugation and extracted with methanol as described previously (26).
UPLC-coupled high-resolution ESI-TOF-MS.
LC-MS measurements were performed using a micrOTOF ESI-TOF mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) coupled to a UPLC system (Waters, Milford, MA) controlled by HyStar chromatography software (Bruker Daltonik GmbH). A mobile phase system consisting of water and acetonitrile (each containing 0.1% formic acid) was used, and separation was performed with an RP-18 column (50 by 2.1 mm, 1.7 μm particle size; Waters Acquity BEH). Data sets were acquired in positive electrospray (ESI) mode in a scan range from 100 to 1,200 m/z at a sampling rate of 2 Hz. Sodium formate solution was used for calibration and injected at the beginning of each chromatographic run. Quality control samples and blank runs were interspersed between the samples under investigation.
Data analysis and statistical interpretation.
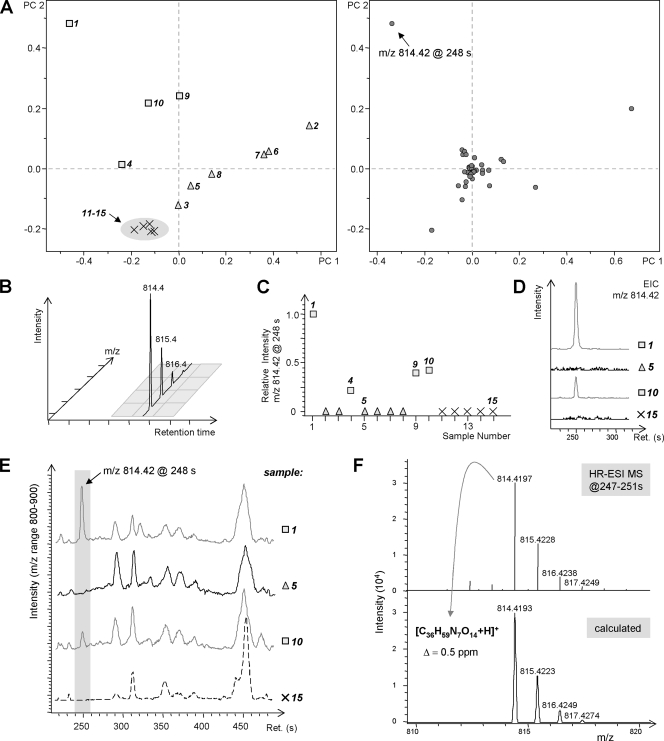
Data evaluation by PCA was performed with ProfileAnalysis, a program within the Metabolic Profiler software suite (Bruker Daltonik GmbH). The LC-MS data, commonly including retention time (RT), m/z, and intensity values, were prepared for PCA, using either one of two approaches: (i) classical bucketing, which involves the formation of rectangular “buckets,” comprising m/z and RT windows of adjustable size (these buckets summarize the intensities of all signals in the respective window [Fig. 2B]), or (ii) advanced bucketing, which identifies a chromatographic peak by detecting the signals at a certain m/z value in successive mass spectra as well as the corresponding isotope peaks and multiply charged ions from the same parent mass within a narrow tolerance interval. These signals are then defined as corresponding to one compound rather than being treated independently. This approach will be described in detail elsewhere. Further steps involved in PCA model generation are the transformation of RT-m/z pairs and intensity values into a covariance-based coordinate system and the calculation of so-called principal components (PCs), the number of which is chosen as sufficiently high to explain >95% of the variation in the data set (17). A pairwise plot of PCs yields a graphical representation, the score-and-loading plot, which consists of two diagrams: the first one reveals a grouping pattern of samples, and the second one serves to derive the observations (here, RT-m/z buckets) which are responsible for this pattern.
FIG. 2.
Identification of strain-specific compounds in M. xanthus extracts by UPLC-coupled ESI-TOF-MS and PCA. (A) Scores & Loadings plot for PCs 1 and 2 from an analysis comprising 10 randomly chosen samples (squares, triangles) and 5 biological replicates (crosses) of the DK1622 standard strain. The spatial arrangement in the left diagram indicates significant variation among these samples. Circles off the center in the right diagram represent signals (m/z-RT pairs termed “buckets”) which contribute to variation. Values along the axes represent the relative degrees and directions of correlation (dimensionless). (B) Illustration of the rectangular bucketing concept for preparation of LC-MS data prior to PCA. (C) Bucket statistics plot revealing that samples 1, 4, 9, and 10 contain the compound of 814.42 m/z with various abundances, while it is absent from the other samples. (D) EIC created for 814.42 m/z in four selected samples, demonstrating the qualitative and quantitative nature of the differences revealed by the PCA model (baselines in traces for samples 5 and 15 are upscaled by a factor of 10 for clarity). (E) BPCs in the m/z range of 800 to 900 for the same subset of samples. HR, high resolution. (F) Generation of a molecular formula proposal for a compound with an 814.4179 m/z by use of accurate m/z values from high-resolution mass spectra and evaluation by calculation of the Δm/z deviation and comparison to the simulated isotopic pattern.
The LC-MS data were generally integrated from 0.2 to 8.5 min and mass windows of 100 m/z, spanning the mass range from 300 to 1,200 m/z. The bucketing parameters were a Δm/z of 1 Da and a ΔRT of 20 s for conventional bucketing and a Δm/z of 20 mDa for advanced bucketing. Each data set was normalized to the total intensity in an analysis. Target screening was performed using QuantAnalysis software (Bruker Daltonik GmbH) for automatic peak integration on extracted ion chromatograms (EICs) created at a 10-mDa mass accuracy. Molecular formulae were calculated using the built-in “generate molecular formula” function in the DataAnalysis software suite, which implements the SigmaFit approach to rank proposals according to both mass deviation and isotopic pattern accuracy (19).
A diversity accumulation curve was calculated using the EstimateS software package (4). Compounds are regarded as chemical “species” in the context of this analysis, and a binary matrix listing their presence or absence in a sample (extract from one strain) was used as the input. The obtained curves represent the means of 100 accumulation randomizations. Estimates of the total number of compounds present were based on the abundance-based coverage estimator (3).
RESULTS
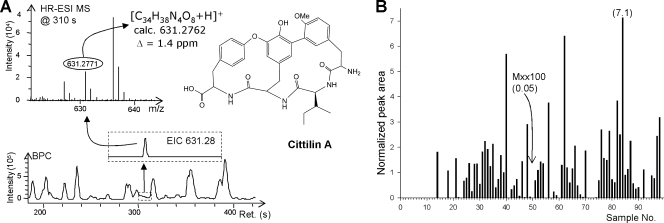
Extracts from 98 M. xanthus strains grown under standardized conditions were initially screened for the presence of all compounds known to date to derive from M. xanthus by using accurate m/z values for their pseudomolecular ions ([M+H]+) and RTs obtained from wild-type reference compounds. These analyses revealed that myxalamids, myxochromids, myxochelins, and DKxanthenes are produced by all strains under investigation, while myxovirescin and cittilin are found in large subsets of the extracts. Althiomycin and saframycin are extremely rare compounds, the former being produced by only two strains and the latter by none in this set (Table 1).
TABLE 1.
Secondary metabolites known to be produced by Myxococcus xanthus and properties used for their identification via UPLC-coupled high-resolution ESI-TOF-MS in this study
| Compound group and namea | Molecular formula |
m/z of [M+H]+
|
RT (min) | Example strain | No. of producers | |
|---|---|---|---|---|---|---|
| Calculated | Measured | |||||
| A | ||||||
| DKxanthene-534 | C29H34N4O6 | 535.2551 | 535.2553 | 7.52 | DK1622 | 98 |
| DKxanthene-518 | C29H34N4O5 | 519.2602 | 519.2605 | 7.65 | DK1622 | 98 |
| DKxanthene-560 | C31H36N4O6 | 561.2708 | 561.2704 | 7.63 | DK1622 | 98 |
| Myxalamid A | C26H41NO3 | 416.3159 | 416.3151 | 8.19 | DK1622 | 98 |
| Myxalamid B | C25H39NO3 | 402.3003 | 402.2998 | 8.15 | DK1622 | 98 |
| Myxalamid C | C24H37NO3 | 388.2846 | 388.2825 | 8.11 | DK1622 | 98 |
| Myxochelin A | C20H24N2O7 | 405.1656 | 405.1669 | 5.23 | DK1622 | 98 |
| Myxochelin B | C20H25N3O6 | 404.1816 | 404.1828 | 3.94 | DK1622 | 98 |
| Myxochromid A2 | C44H63N7O9 | 834.4760 | 834.4767 | 8.10 | DK1622 | 98 |
| Myxochromid A3 | C45H63N7O9 | 846.4760 | 846.4768 | 8.13 | DK1622 | 98 |
| Myxochromid A4 | C46H65N7O9 | 860.4916 | 860.4928 | 8.21 | DK1622 | 98 |
| B | ||||||
| Myxovirescin A | C35H61NO8 | 624.4470 | 624.4456 | 8.20 | DK1622 | 39 |
| Cittilin A | C34H38N4O8 | 631.2762 | 631.2771 | 5.17 | DK897 | 69 |
| Althiomycin | C16H17N5O6S2 | 440.0693 | 440.0706 | 4.51 | Mxx52 | 2 |
| Saframycin MX1 | C29H38N4O9 | 587.2712 | Mxx48 | 0 | ||
A, derivatives belonging to these four compound classes were detected in all extracts from 98 strains, including DK1622; B, known secondary metabolites with a nonubiquitous production pattern and number of strains which were found to produce these compounds.
Mining LC-MS data for novel M. xanthus metabolites.
The LC-MS chromatograms for random subsets comprising 10 samples each were compared manually by using base peak chromatograms (BPCs) in the mass range of 300 to 1,200 m/z at a window width of 100 m/z in order to detect differences between production profiles. Such subsets were also examined using PCA. PCA is a nonsupervised pattern recognition technique that aims to reveal groups of observations, trends, and outliers in a multivariate data set (17). It dramatically reduces the dimensionality of a data set while retaining relevant information in terms of variance. The output of a PCA model consists of a two-dimensional representation, the score-and-loading plot, which reveals a grouping pattern of samples and the observations (i.e., m/z signals at specific RTs) which determine this grouping pattern. In the following, the use of PCA to reveal significant differences between M. xanthus samples is illustrated (Fig. 2). Samples from replicate fermentations of M. xanthus DK1622 form a group in the Scores & Loadings plot (Fig. 2A, left, samples 11 to 15), while the other randomly chosen samples (no. 1 to 10) are scattered across the coordinate system, indicating that their compound profiles do not conform to that of the DK1622 reference group. The Scores & Loadings plot also gives information about the signals which are responsible for the grouping pattern of samples (Fig. 2A, right): as an example, the presence of a compound with an 814.42 m/z at an RT of 248 s (Fig. 2A, right, arrow) distinguishes the samples in the upper-left quadrant (no. 1, 4, 9, and 10) (Fig. 2A, left) from the other samples. This finding is confirmed by the bucket statistics plot (Fig. 2C) and is also reflected in EICs created for an 814.42 m/z (Fig. 2D): only samples with relative bucket intensities of >0 actually contain the compound with an 814.42 m/z. These observations can be independently verified using the raw LC-MS data by inspection of BPCs in the mass range of 800 to 900 m/z (Fig. 2E).
By use of the accurate m/z values from high-resolution measurements, molecular formulae were proposed for putative distinct compounds suggested by the above-described analyses (Fig. 2F). Naturally, this results in disproportionately high numbers of proposals as molecular weights increase. The proposals were ranked according to the mass positions and intensity ratios of isotope peaks in comparison to those in a calculated spectrum derived from a suggested molecular formula, implementing the SigmaFit approach (19). Elemental formulae were subsequently inspected manually, and obvious false positives (as determined by chemical considerations) were disregarded.
Compound proposals retained after both statistical analysis and manual examination were compiled into a list of 37 accurate m/z values, corresponding putative molecular formulae, and RTs (Table 2), and this list was used for a targeted analysis of all 98 datasets. This was achieved by automated generation of EICs, using the accurate m/z values of compounds, followed by peak integration and calculation of peak areas. Due to the high dynamic range and overall sensitivity of the instrumentation used, the presence or absence of a compound in a sample could be determined with high confidence and the relative production yields from all strains could be estimated (Fig. 3). The resulting binary matrix of 40 nonubiquitous compounds observed in all samples (Table 3) forms the basis of this study, enabling conclusions regarding the set of metabolites produced by one strain (hereafter referred to as a strain's “chemotype”) as well as the distribution of compounds across all samples.
TABLE 2.
Putative novel compounds not reported to derive from M. xanthus before which are revealed to be produced by subsets of the 98 strains under investigation in this study and the proposed molecular formulae according to high-resolution ESI-TOF-MS measurementsa
| Compound name | Molecular formula | m/z of [M+H] (charge) | RT (min) | Example strain | No. of producers |
|---|---|---|---|---|---|
| c382 | C18H27N3O6 | 382.1978 | 2.7 | DK897 | 49 |
| c448 | C45H69N9O10 | 448.7655 (2+) | 6.07 | Mxx149 | 76 |
| c455 | C20H30N4O8 | 455.2141 | 2.57 | Poland11 | 81 |
| c501 | C21H32N4O10 | 501.2192 | 2.88 | Poland11 | 48 |
| c505 | C47H72N14O11 | 505.2821 (2+) | 4.55 | Solanka10 | 1 |
| c506 | C45H74N10O16 | 506.271 (2+) | 3.47 | Sulawesi5 | 36 |
| c580 | C53H81N11O18 | 580.7964 (2+) | 4.26 | Massif1 | 51 |
| c587 | C54H83N11O18 | 587.8034 (2+) | 4.42 | Massif1 | 51 |
| c600 | C34H41N5O3S | 600.2999 | 1.6 | Mxx104 | 1 |
| c604 | C29H41N5O9 | 604.2974 | 3.34 | Mxx104 | 95 |
| c609 | C56H87N11O19 | 609.817 (2+) | 4.2 | B5 | 47 |
| c612 | C64H98N6O17 | 612.3562 (2+) | 7.08 | B5 | 95 |
| c615 | C26H42N6O9S | 615.2803 | 2.33 | Mxx104 | 11 |
| c621 | C26H48N6O11 | 621.3446 | 2.95 | NewJersey2 | 2 |
| c630 | C30H43N7O8 | 630.325 | 3.13 | A2 | 66 |
| c637 | C26H48N6O12 | 637.34 | 2.45 | NewJersey2 | 2 |
| c647 | C24H30N12O10 | 647.228 | 1.22 | Mxx73 | 2 |
| c651 | C27H50N6O12 | 651.3558 | 2.95 | Mxx121 | 39 |
| c663 | C67H87N7O21 | 663.8028 (2+) | 4.88 | GranSasso10 | 12 |
| c680 | C69H81N7O22 | 680.7786 (2+) | 4 | Iran9 | 8 |
| c683 | C31H50N6O11 | 683.3616 | 3.97 | Ser2 | 54 |
| c713 | C32H56N8O10 | 713.4184 | 3 | Mxx43 | 43 |
| c714 | C66H74N16O15S3 | 714.2429 (2+) | 4.08 | Solanka10 | 2 |
| c724 | C68H97N13O22 | 724.8503 (2+) | 3.3 | Mxx148 | 1 |
| c748 | C39H45N3O12 | 748.3077 | 4.33 | Mxx100 | 73 |
| c800 | C35H57N7O14 | 800.4034 | 3.5 | DK897 | 50 |
| c809 | C93H123N11O12S | 809.9635 (2+) | 7.5 | Sulawesi10 | 95 |
| c812 | C36H57N7O14 | 812.4029 | 3.68 | Serengeti2 | 48 |
| c813 | C36H61N8O13 | 813.4339 | 3.73 | Mxx73 | 2 |
| c814 | C36H59N7O14 | 814.4196 | 4.2 | DK897 | 47 |
| c832 | C84H125N15O20 | 832.9701 (2+) | 7.12 | DK836 | 85 |
| c857 | C42H64N8O11 | 857.4767 | 7.88 | BizenJP | 1 |
| c1124 | C51H81N17O12 | 1124.6301 | 6.47 | Serengeti2 | 92 |
| c1144 | C117H125N13O36 | 1144.9241 (2+) | 4.77 | Tor1 | 2 |
| c1160 | C54H81N9O19 | 1160.5746 | 4.06 | Niaux6 | 44 |
| c1162 | C53H83N11O18 | 1162.5989 | 3.72 | Mxx106 | 33 |
| c1174 | C55H83N9O19 | 1174.5909 | 4.42 | Niaux6 | 33 |
The m/z values of pseudomolecular ions are given for the 1+ charge state if not denoted as 2+. Elemental formulae are given for the neutral state.
FIG. 3.
Identification of cittilin A in extracts from M. xanthus Mxx100. (A) The sensitivity of the ESI-TOF screening platform enables reliable compound identification even in samples from very weak producers. HR, high resolution. (B) The obtained data are useful for a semiquantitative comparison of production yield in all 98 strains. Numbers in parentheses are yields relative to the average level from all 98 strains (which was set as 1).
TABLE 3.
Metabolite profiles of all 98 strains, listing the nonubiquitous compounds detected in their extracts as well as the number of producing strains for every compound and the number of compounds found in every sample
| Strain no. | Name | Presence of:
|
No. of compounds | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Myxovirescin A | Cittilin A | Althiomycin | c382 | c448 | c455 | c501 | c505 | c506 | c580 | c587 | c600 | c604 | c609 | c612 | c615 | c621 | c630 | c637 | c647 | c651 | c663 | c680 | c683 | c713 | c714 | c724 | c748 | c800 | c809 | c812 | c813 | c814 | c832 | c857 | c1124 | c1144 | c1160 | c1162 | c1174 | |||
| 1 | A0 | + | + | + | + | + | + | + | + | + | + | 10 | ||||||||||||||||||||||||||||||
| 2 | A12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 14 | ||||||||||||||||||||||||||
| 3 | A15 | + | + | + | + | + | + | + | + | 8 | ||||||||||||||||||||||||||||||||
| 4 | A17 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 5 | A2 | + | + | + | + | + | + | + | + | + | 9 | |||||||||||||||||||||||||||||||
| 6 | A23 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 14 | ||||||||||||||||||||||||||
| 7 | A3 | + | + | + | + | + | + | + | + | 8 | ||||||||||||||||||||||||||||||||
| 8 | A30 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 9 | A38 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 10 | A4 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 11 | A41 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 14 | ||||||||||||||||||||||||||
| 12 | A47 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 13 | A5 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 14 | A66 | + | + | + | + | + | + | + | + | + | + | 10 | ||||||||||||||||||||||||||||||
| 15 | A85 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 16 | A94 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 17 | A96 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 18 | A98 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 19 | |||||||||||||||||||||
| 19 | A9 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 20 | B5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 21 | BizenJP | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 22 | Bonn37 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 23 | Conero3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 24 | Conero8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 25 | CostaRica | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 26 | CSH6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 27 | DK1622 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 28 | DK801 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 20 | ||||||||||||||||||||
| 29 | DK816 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 19 | |||||||||||||||||||||
| 30 | DK836 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 24 | ||||||||||||||||
| 31 | DK897 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 20 | ||||||||||||||||||||
| 32 | Ferrara1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 33 | Ferrara10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 34 | Ferrara6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 18 | ||||||||||||||||||||||
| 35 | GranSasso10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 19 | |||||||||||||||||||||
| 36 | GranSasso5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 37 | Hlulume | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 38 | Houston | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 39 | Iran14 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 20 | ||||||||||||||||||||
| 40 | Iran9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 41 | Jura | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 42 | Kalalau | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 43 | Kohlhapur | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 44 | Malaga13 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 45 | Malaga15 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 46 | Massif1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 24 | ||||||||||||||||
| 47 | Massif6 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 48 | Michigan1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 49 | Mxx100 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 50 | Mxx104 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 51 | Mxx106 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 52 | Mxx122 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 53 | Mxx12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 54 | Mxx135 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 55 | Mxx143 | + | + | + | + | + | + | + | + | + | + | + | + | 13 | ||||||||||||||||||||||||||||
| 56 | Mxx144 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 19 | |||||||||||||||||||||
| 57 | Mxx147 | + | + | + | + | + | + | + | + | + | 9 | |||||||||||||||||||||||||||||||
| 58 | Mxx148 | + | + | + | + | + | + | + | + | + | + | 10 | ||||||||||||||||||||||||||||||
| 59 | Mxx149 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 60 | Mxx155 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 61 | Mxx15 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 62 | Mxx17 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 63 | Mxx23 | + | + | + | + | + | + | + | + | + | 9 | |||||||||||||||||||||||||||||||
| 64 | Mxx41 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 19 | |||||||||||||||||||||
| 65 | Mxx43 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 66 | Mxx46 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 67 | Mxx51 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 68 | Mxx52 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 17 | |||||||||||||||||||||||
| 69 | Mxx57 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 70 | Mxx62 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 71 | Mxx67 | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |||||||||||||||||||||||||||
| 72 | Mxx73 | + | + | + | + | + | + | + | 7 | |||||||||||||||||||||||||||||||||
| 73 | Mxx75 | + | + | + | + | + | + | + | 7 | |||||||||||||||||||||||||||||||||
| 74 | Mxx78 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 16 | ||||||||||||||||||||||||
| 75 | Mxx90 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 76 | Nei2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 77 | Nei6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22 | ||||||||||||||||||
| 78 | Niaux1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 79 | Niaux6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 80 | New Jersey1 | + | + | + | + | + | + | + | + | + | + | 10 | ||||||||||||||||||||||||||||||
| 81 | New Jersey2 | + | + | + | + | + | + | + | 7 | |||||||||||||||||||||||||||||||||
| 82 | Pali | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 14 | ||||||||||||||||||||||||||
| 83 | Poland11 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 84 | Poland3 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 85 | Poland7 | + | + | + | + | + | + | + | + | + | + | + | + | 12 | ||||||||||||||||||||||||||||
| 86 | Serengeti19 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 87 | Serengeti2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 16 | ||||||||||||||||||||||||
| 88 | Serengeti35 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| 89 | Serengeti8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 18 | ||||||||||||||||||||||
| 90 | Seychellen | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 16 | ||||||||||||||||||||||||
| 91 | Solanka10 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 92 | Solanka2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 24 | ||||||||||||||||
| 93 | Sulawesi10 | + | + | + | + | + | + | + | + | + | + | + | 11 | |||||||||||||||||||||||||||||
| 94 | Sulawesi5 | + | + | + | + | + | + | + | + | + | + | 10 | ||||||||||||||||||||||||||||||
| 95 | Tor1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 18 | ||||||||||||||||||||||
| 96 | TvarFI | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15 | |||||||||||||||||||||||||
| 97 | Wales | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 23 | |||||||||||||||||
| 98 | Warwick6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 21 | |||||||||||||||||||
| Total | 39 | 69 | 2 | 49 | 76 | 81 | 48 | 1 | 36 | 51 | 51 | 1 | 95 | 47 | 95 | 11 | 2 | 66 | 2 | 2 | 39 | 12 | 8 | 54 | 43 | 2 | 1 | 73 | 50 | 95 | 48 | 2 | 47 | 85 | 1 | 92 | 2 | 44 | 33 | 33 | ||
Diversity of M. xanthus metabolite profiles.
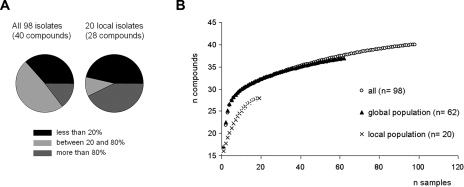
The number of nonubiquitous compounds produced by individual strains ranges from 6 to 24, with a mean of 16. One-third of the metabolites were classified as “rare” because they were found in less than 20% of the samples (Table 3 and Fig. 4A). The number of strains producing any given compound ranges between 1 and 95. Notably, 11 of the 40 observed compounds are found in only one or two extracts, and only 6 compounds are produced by more than 80% of the strains. Only nine chemotypes are exhibited by more than one strain, indicating that the metabolite profile of M. xanthus has greatly diversified across strains.
FIG. 4.
(A) Slices represent percentages of compounds which are produced by less than 20% (black), between 20 and 80% (light gray), or more than 80% (dark gray) of the strains in the respective sample sets. (B) Diversity accumulation curves, based on the distribution of 40 compounds in 98 samples as revealed in Table 3. Each data point represents the average number of discovered compounds that would result if n samples were taken (calculated as the mean of 100 randomizations).
Except for the 20 local isolates, there is no visually obvious relationship between a strain's origin and its metabolite pattern. To test this more rigorously, we compared the average degree of similarity within six meter-scale strain pairs (in which paired strains were isolated from the same meter-scale transect) with the average degree of similarity within six randomly chosen global strain pairs (in which paired strains were isolated from distant locations). Tellingly, the average degree of chemotype similarity among meter-scale isolate pairs was not significantly different from that among the global-scale pairs (two-tailed t test; P = 0.88). While larger samples of meter-scale isolates from more locations may reveal spatial structure in the distribution of chemotype diversity at this scale, the absence of a clear biogeographical signal in this analysis indicates that much mixing of global chemotypes occurs even at the meter scale.
Diversity accumulation curves were generated to estimate what proportion of global M. xanthus secondary metabolite diversity is likely to have been represented by our samples (Fig. 4B) and to compare global diversity with the diversity present at the centimeter scale. An accumulation curve is the product of both the diversity of a population and the sampling effort (4). If enough samples are taken, the same chemotypes will be sampled repeatedly and the accumulation curve will reach saturation, indicating that no more chemotypes are likely to be present. The shape of the accumulation curves (Fig. 4B) shows that most samples contain a large proportion of identified compounds. For example, on average, random sampling of just 10 strains would yield 30 of the 40 nonubiquitous compounds present in the total set. Nonetheless, the slopes of the global and total curves remain positive as sample size nears its maximum, suggesting that continued sampling effort should reveal more compounds.
Chemotype diversity on worldwide and local scales.
Our analysis reveals two major patterns. First, chemotype diversity in the local population is lower than that found globally (Fig. 4B), despite the facts that 18 distinct chemotypes are present in the centimeter-scale local set and that differences among the local chemotypes extend to known compounds (myxovirescin is not produced by all 20 local isolates, and cittilin was detected in only 3 samples). Two strains (A98 and B5) (Table 3, rows 18 and 20) account for much of the diversity in the local 20-strain subset, being the exclusive producers of 11 out of 28 observed compounds. Thus, although spatial structuring was not detected at the meter scale (see above), this larger data set shows reduced diversity at the centimeter scale.
Second, our data indicate that intensive sampling at individual sites can substantially increase the estimate of the total number of metabolites beyond that obtained from analysis of single samples from many sites. To examine this point, we made one estimate of total global richness in which we included only one sample per site in all cases where multiple samples had been taken from individual sites (e.g., Poland, Niaux, Ferrara, etc.) as well as all samples which were the sole isolates taken from their respective origin sites (n = 62). This set of strains yielded a global richness estimate of 41.7 metabolites (standard deviation, 0.73). However, analysis of all isolates, including those originating from the same site (Conero, Ferrara, Gran Sasso, Iran, Malago, Massif, Nei, Niaux, NJ, Poland, Serengeti, Solanka, Tübingen, and Sulawesi), yields a total global richness estimate of 45.1 metabolites (standard deviation, 1.37).
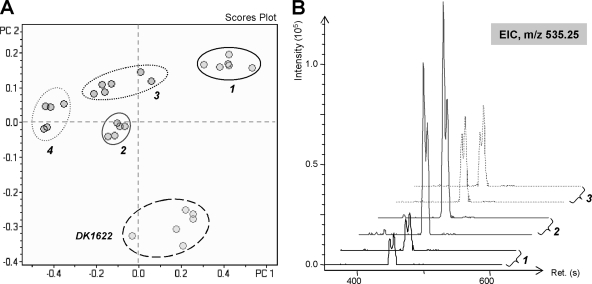
In order to both assess the effects of biological variation in the fermentation process and evaluate the use of a PCA model for in-depth examination of largely similar extracts, replicate fermentations of 4 “A” strains isolated in close proximity were performed and compared to replicate samples from the DK1622 model strain. The observed grouping pattern of samples in the Scores and Loadings plot (Fig. 5A) indicates that variation between replicate cultivations of the same strain is sufficiently small relative to systematic differences between the metabolite profiles of distinct isolates to allow clear detection of the latter. The compound most affecting variation among the strains was identified as DKxanthene-534, which was present in all isolates but which varied quantitatively in the amount present in the different strains. (Fig. 5B).
FIG. 5.
PCA for extracts from four M. xanthus strains isolated in close proximity and the DK1622 wild-type strain, using samples from replicate fermentations. (A) Grouping of samples reveals clear systematic differences between their metabolite profiles. The strains are as follows: 1, A23; 2, A47; 3, A96; and 4, A9. (B) Variation in the data set is largely due to quantitative differences in the production of DKxanthenes; representative EICs for an m/z of 535.25 (DKxanthene-534, C29H34N4O6) from high-resolution ESI LC-MS measurements are shown (two replicates for each strain). Ret., RT.
DISCUSSION
Myxobacteria are a rich source of natural products with fascinating structures and biological activities. However, the biosynthetic potentials of distinct myxobacterial genera are highly divergent, with half of all myxobacterial secondary metabolites isolated to date derived from the genus Sorangium (9). A myxobacterial metabolite profile has been described as a strain-specific rather than a species-specific property (20). While this is encouraging for the screening of less exploited myxobacterial species, like M. xanthus, it demands at the same time the implementation of a dereplication strategy for identifying natural products which are already known to derive from other myxobacteria. As an example, the siderophore myxochelin is produced by a wide variety of myxobacterial groups, including Myxococcus, Stigmatella and Sorangium species (15, 22, 24).
Using ESI-TOF-MS for the analysis of fermentation extracts of 98 strains from globally distributed locations, this study has revealed a high level of intraspecific diversity in M. xanthus metabolite profiles. All observations (high-resolution m/z signals and the derived molecular formula at a specific RT in an LC-MS data set) constituting a distinct difference between the metabolite profiles of two or more strains were regarded as indicating putative compounds of interest, regardless of potential biological activity or function. Molecular formulae derived from accurate mass measurements were used for the identification of known compounds from an in-house database and served as descriptors for putative novel compounds (Table 2). PCA was successfully used to detect significant differences between LC-MS datasets even in the presence of significant background noise. To our knowledge, this is the first use of PCA for the analysis of complex metabolite samples, such as myxobacterial extracts.
While previous screening projects that included multiple M. xanthus strains reported only a small number of compounds (7), we are confident that the sample size and screening methods used in this study have captured a substantial fraction of global M. xanthus metabolite diversity. Seven of the eight previously known metabolites were present among our samples (Table 1), with only saframycin (already known to be rare [12]) absent. An impressive 37 additional compounds that represent putative novel metabolites could be defined. Some of these compounds may represent novel derivatives of previously classified natural product families, as families such as the DKxanthenes and the myxalamids occur in at least 11 and 4 slightly differing chemical forms, respectively (13, 18).
Analysis of the M. xanthus DK1622 genome has revealed a high number of putative PKSs, NRPSs, or hybrid biosynthetic pathways, the products of which are not yet known. A recent study has provided evidence that many of the encoded enzymatic natural product assembly lines are actually active and expressed, since the corresponding peptides could be identified by mass spectrometric proteome analysis (21). We expect that comparative mining of the M. xanthus secondary metabolome as performed in this study can facilitate the definition of potential candidate compounds and help to disclose the products delivered by these biosynthetic enzymes in strains that possess the respective gene clusters. Studies to elucidate the function of each secondary metabolite pathway in strain DK1622 are currently in progress.
It is unlikely that all of the putative novel compounds listed in Table 2 can be structurally elucidated and directly correlated with a biosynthetic gene cluster. In principle, some of these compounds may represent artifacts resulting from metabolic differences between strains, although the mass range chosen for our analysis should mitigate this possibility. Alternatively, proposed molecular formulae might be incorrect because the detected compound is actually a breakdown product of labile compounds or a product of chemical reactions taking place during the workup procedure. However, the existence of distinct compounds with very similar occurrence patterns plus putative elemental compositions suggests that at least some are likely to be real secondary metabolites synthesized by PKS and/or NRPS biosynthetic routes. For example, compounds c5802+ and c5872+ as well as c800 and c814 (which display an m/z difference of 14 Da) are suggestive of biosynthetically meaningful structural differences and also have similar RTs (Tables 2 and 3). Furthermore, with respect to the above-mentioned genomic potential of M. xanthus DK1622, we find it intriguing that this strain produces 10 candidates from the list of putative novel compounds which we report here (Table 2). DK1622 is additionally shown for the first time to produce the known myxobacterial natural product cittilin (20).
The genetic population structure of M. xanthus has recently been studied at the centimeter scale, utilizing 78 local isolates from a 16- by 16-cm plot of soil, and it was concluded that M. xanthus cells are “surrounded in the soil by a wide range of genetically distinct conspecifics” (28). The picture that emerges from our analysis of metabolite profiles from 19 of these local isolates is fully consistent with this statement. Although the metabolite profiles of the local Tübingen isolates (Table 3, rows 1 to 19) are more similar to each other than they are to most of the global isolates, they nonetheless show that the sampled centimeter-scale population houses substantial metabolite diversity. Another similarity between the genetic analysis of Vos and Velicer (28) and our results here is the finding that the metabolite profile of strain A98 (Table 3, row 18), which had been identified as phylogenetic outgroup of most of the local isolates, differs remarkably from the profiles of those other isolates. Analysis of genetic diversity within versus between pairs of meter-scale isolates did not reveal spatial structure in the distribution of chemotype diversity at this scale.
Comparative metabolite profiling of replicate extracts from strains isolated in close proximity by use of a PCA model (Fig. 5) revealed quantitative variation in the levels of identical compounds as a major determinant of systematic differences between their overall production patterns. One compound revealed by this analysis was identified as a derivative within the DKxanthene family, representatives of which are essential for developmental sporulation (18). Other recent studies have shown that M. xanthus strains can exhibit intense intraspecific antagonism, with some strains being capable of preventing conspecific competitors from sporulating altogether (6). We hypothesize that asymmetries in secondary metabolite production may contribute to such social incompatibilities.
A more advanced understanding of M. xanthus metabolite profiles and their functional significance will likely require consideration of not only the absolute presence or absence of compounds but also their quantitative degree of production. In addition to the classically proposed role of hindering the growth of heterospecific competitors, such compounds might also serve intraspecific biological functions (e.g., for cell-cell communication) that are not revealed by standard activity-based screening efforts.
In conclusion, we have shown that the diversity of the M. xanthus secondary metabolome is significantly greater than suggested by previously identified compounds. The large number of putative novel compounds revealed by our screen correlates well with the high number of genes of unassigned specific function in the DK1622 genome that are predicted to encode proteins involved in secondary metabolite synthesis. This high level of diversity also suggests that a significant number of natural products in M. xanthus have yet to be discovered and characterized, some of which are likely to have structures originating from PKS- and NRPS-type biosynthetic machinery. The isolation, purification, and chemical characterization of target compounds defined in this study as well as the identification of their corresponding biosynthetic gene clusters are the subject of ongoing research.
Acknowledgments
We thank Rolf Jansen (HZI Braunschweig) for kindly providing reference substances.
Research in the laboratory of R.M. was funded by Deutsche Forschungsgemeinschaft (DFG) and Bundesministerium für Bildung und Forschung (BMB+F).
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Bode, H. B., and R. Müller. 2005. The impact of bacterial genomics on natural product research. Angew. Chem. Int. Ed. Engl. 44:6828-6846. [DOI] [PubMed] [Google Scholar]
- 2.Bode, H. B., and R. Müller. 2006. Analysis of myxobacterial secondary metabolism goes molecular. J. Ind. Microbiol. Biotechnol. 33:577-588. [DOI] [PubMed] [Google Scholar]
- 3.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 4.Colwell, R. K. 2006. EstimateS: statistical estimation of species richness and shared species from samples. Version 8 user's guide and application. http://purl.oclc.org/estimates.
- 5.Dawid, L. 2000. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24:403-427. [DOI] [PubMed] [Google Scholar]
- 6.Fiegna, F., and G. J. Velicer. 2005. Exploitative and hierarchical antagonism in a cooperative bacterium. PLoS Biol. 3:e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspari, F., Y. Paitan, M. Mainini, D. Losi, E. Z. Ron, and F. Marinelli. 2005. Myxobacteria isolated in Israel as potential source of new anti-infectives. J. Appl. Microbiol. 98:429-439. [DOI] [PubMed] [Google Scholar]
- 8.Gerth, K., H. Irschik, H. Reichenbach, and G. Höfle. 1982. The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J. Antibiot. 35:1454-1459. [DOI] [PubMed] [Google Scholar]
- 9.Gerth, K., S. Pradella, O. Perlova, S. Beyer, and R. Müller. 2003. Myxobacteria: proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J. Biotechnol. 106:233-253. [DOI] [PubMed] [Google Scholar]
- 10.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabley, S., and R. Thiericke. 1999. The impact of natural products on drug discovery, p. 3-37. In S. Grabley and R. Thiericke (ed.), Drug discovery from nature. Springer, Berlin, Germany.
- 12.Irschik, H., W. Trowitzsch-Kienast, K. Gerth, G. Höfle, and H. Reichenbach. 1988. Saframycin Mx1, a new natural saframycin isolated from a myxobacterium. J. Antibiot. 41:993-998. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, R., G. Reifenstahl, K. Gerth, H. Reichenbach, and G. Höfle. 1983. Antibiotika aus Gleitenden Bakterien, XV: Myxalamide A, B, C und D, eine Gruppe homologer Antibiotika aus Myxococcus xanthus Mx x12 (Myxobacterales). Liebigs Ann. Chem. 7:1081-1095. [Google Scholar]
- 14.Krug, D., G. Zurek, B. Schneider, C. Bässmann, and R. Müller. 2007. Analysis of secondary metabolites from myxobacteria using ESI-TOF-MS and PCA, p. 41-42. In A. Matheson (ed.), The applications book March 2007. LC-GC Europe, Chester, United Kingdom.
- 15.Kunze, B., N. Bedorf, W. Kohl, G. Höfle, and H. Reichenbach. 1989. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J. Antibiot. 42:14-17. [DOI] [PubMed] [Google Scholar]
- 16.Kunze, B., H. Reichenbach, H. Augustiniak, and G. Höfle. 1982. Isolation and identification of althiomycin from Cystobacter fuscus (Myxobacterales). J. Antibiot. 35:635-636. [DOI] [PubMed] [Google Scholar]
- 17.Lavine, B. K. 2000. Clustering and classification of analytical data, p. 1-20. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry: applications, theory, and instrumentation. Wiley & Sons, New York, NY.
- 18.Meiser, P., H. B. Bode, and R. Müller. 2006. DKxanthenes: novel secondary metabolites from the myxobacterium Myxococcus xanthus essential for sporulation. Proc. Natl. Acad. Sci. USA. 103:19128-19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojanperä, S., A. Pelander, M. Pelzing, I. Krebs, E. Vuori, and I. Ojanperä. 2006. Isotopic pattern and accurate mass determination in urine drug screening by liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 20:1161-1167. [DOI] [PubMed] [Google Scholar]
- 20.Reichenbach, H., and G. Höfle. 1999. Myxobacteria as producers of secondary metabolites, p. 149-179. In S. Grabley and R. Thiericke (ed.), Drug discovery from nature. Springer, Berlin, Germany.
- 21.Schley, C., M. O. Altmeyer, R. Swart, R. Müller, and C. G. Huber. 2006. Proteome analysis of Myxococcus xanthus by off-line two-dimensional chromatographic separation using monolithic poly-(styrene-divinylbenzene) columns combined with ion-trap tandem mass spectrometry. J. Proteome Res. 5:2760-2768. [DOI] [PubMed] [Google Scholar]
- 22.Schneiker, S., O. Perlova, O. Kaiser, K. Gerth, A. Alici, M. O. Altmeyer, D. Bartels, T. Bekel, S. Beyer, E. Bode, H. B. Bode, C. J. Bolten, J. V. Choudhuri, S. Doss, Y. A. Elnakady, B. Frank, L. Gaigalat, A. Goesmann, C. Groeger, F. Gross, L. Jelsbak, L. Jelsbak, J. Kalinowski, C. Kegler, T. Knauber, S. Konietzny, M. Kopp, L. Krause, D. Krug, B. Linke, T. Mahmud, R. Martinez-Arias, A. C. McHardy, M. Merai, F. Meyer, S. Mormann, J. Munoz-Dorado, J. Perez, S. Pradella, S. Rachid, G. Raddatz, F. Rosenau, C. Ruckert, F. Sasse, M. Scharfe, S. C. Schuster, G. Suen, A. Treuner-Lange, G. J. Velicer, F. J. Vorholter, K. J. Weissman, R. D. Welch, S. C. Wenzel, D. E. Whitworth, S. Wilhelm, C. Wittmann, H. Blöcker, A. Pühler, and R. Müller. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25:1281-1289. [DOI] [PubMed] [Google Scholar]
- 23.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silakowski, B., B. Kunze, G. Nordsiek, H. Blöcker, G. Höfle, and R. Müller. 2000. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur. J. Biochem. 267:6476-6485. [DOI] [PubMed] [Google Scholar]
- 25.Simunovic, V., and R. Müller. 2007. Mutational analysis of the myxovirescin biosynthetic gene cluster reveals novel insights into the functional elaboration of polyketide backbones. Chembiochem 8:1273-1280. [DOI] [PubMed] [Google Scholar]
- 26.Simunovic, V., J. Zapp, S. Rachid, D. Krug, P. Meiser, and R. Müller. 2006. Myxovirescin biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl CoA synthases and trans-acting acyltransferases. Chembiochem 7:1206-1220. [DOI] [PubMed] [Google Scholar]
- 27.Trowitzsch Kienast, W., K. Gerth, H. Reichenbach, and G. Höfle. 1993. Myxochromid A: Ein hochungesättigtes Lipopeptidlacton aus Myxococcus virescens. Liebigs Ann. Chem. 1993:1233-1237. [Google Scholar]
- 28.Vos, M., and G. J. Velicer. 2006. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl. Environ. Microbiol. 72:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos, M., and G. J. Velicer. 2008. Isolation by distance in the spore-forming soil bacterium Myxococcus xanthus. Curr. Biol. 18:386-391. [DOI] [PubMed] [Google Scholar]
- 30.Wenzel, S. C., P. Meiser, T. Binz, T. Mahmud, and R. Müller. 2006. Nonribosomal peptide biosynthesis: point mutations and module skipping lead to chemical diversity. Angew. Chem. Int. Ed. Engl. 45:2296-2301. [DOI] [PubMed] [Google Scholar]