Abstract
Species-specific identification of the major cooked and fresh poisonous mushrooms in Japan was performed using a real-time PCR system. Specific fluorescence signals were detected, and no nonspecific signals were detected. Therefore, we succeeded in developing a species-specific test for the identification of poisonous mushrooms within 1.5 h.
Mushrooms are identified by morphological characteristics; however, the morphological characteristics are inconsistent and unstable criteria because they are strongly influenced by the environmental conditions (2, 9). For this reason, poisonous wild mushrooms can be misidentified as edible mushrooms, and they can be eaten by mushroom hunters.
In general, rapid identification of poisonous mushroom species eaten by patients is required for proper medical treatment. However, cooked and eaten mushrooms do not retain their original shape. Therefore, techniques such as DNA-based identification, which do not depend on morphology, are required. Identification of wild and cultivated mushrooms and determination of genetic differences among basidiomycetes by using DNA techniques are usually performed by comparing the nucleotide sequences of amplified DNA fragments from nuclear and mitochondrial ribosomal DNAs (rDNAs) (1, 4, 6, 9, 10, 12, 13).
In this study, we tried to develop a rapid system of poisonous mushroom identification using a real-time PCR system. We herein propose a new strategy for mushroom identification in medical facilities.
Strains and culture conditions.
In this study, 4 poisonous mushroom species that are frequently eaten by mistake in Japan and 27 popular edible mushroom species (Tables 1 and 2) were used. The mycelia of most strains were grown on GP medium (7); the mycelia of Clitocybe and Lactarius strains were grown on modified Hamada's medium (14); and the mycelia of the Armillaria strains were grown on malt extract medium (5) for later DNA extraction.
TABLE 1.
Poisonous mushroom strains used in this study
| Species | Strain | Sourcea | Accession no. | Remarks |
|---|---|---|---|---|
| Omphalotus japonicus | NBRC 4931 | NBRC | AB301601 | Stock culture (mycelium) |
| NBRC 6992 | NBRC | Stock culture (mycelium) | ||
| NBRC 8341 | NBRC | Stock culture (mycelium) | ||
| NBRC 8342 | NBRC | Stock culture (mycelium) | ||
| NBRC 8616 | NBRC | Stock culture (mycelium) | ||
| NBRC 8765 | NBRC | Stock culture (mycelium); cultivated fruiting bodies were used for cooking and DNA extraction | ||
| NBRC 8917 | NBRC | Stock culture (mycelium) | ||
| NBRC 8918 | NBRC | Stock culture (mycelium) | ||
| NBRC 30243 | NBRC | Stock culture (mycelium) | ||
| Tottori Oj-1 | Tottori, Japan | Fruiting body isolated from the field | ||
| Tottori Oj-2 | Tottori, Japan | Fruiting body isolated from the field | ||
| Entoloma rhodopolius | Tottori Er-1 | Tottori, Japan | AB301602 | Fruiting body isolated from the field |
| Tottori Er-2 | Tottori, Japan | Fruiting body isolated from the field | ||
| Tottori Er-3 | Tottori, Japan | Fruiting body isolated from the field | ||
| Tottori Er-4 | Tottori, Japan | Fruiting body isolated from the field | ||
| Tottori Er-5 | Tottori, Japan | Fruiting body isolated from the field | ||
| Tricholoma ustale | NBRC32808 | NBRC | Stock culture (mycelium) | |
| NBRC32825 | NBRC | Stock culture (mycelium) | ||
| Tottori Tu-1 | Tottori Japan | Fruiting body isolated from the field | ||
| Tottori Tu-2 | Tottori Japan | Fruiting body isolated from the field | ||
| 611 | DDBJ | AB036894 | Only nucleotide sequence used | |
| Clitocybe acromelalga | NBRC30567 | NBRC | AB301606 | Stock culture (mycelium) |
NBRC, National Institute of Technology and Evaluation (NITE) Biological Resource Center, Japan; DDBJ, DNA Data Bank of Japan.
TABLE 2.
Edible mushroom strains used in this study
| Species | Strain | Sourcea | Accession no. | Remarks |
|---|---|---|---|---|
| Lentinula edodes | #5-8 | Tottori University | Stock culture (mycelium) | |
| 271 | DDBJ | DQ467886 | Only nucleotide sequence used | |
| Pleurotus ostreatus | Nichinou-Chusei | Tottori University | Stock culture (mycelium) | |
| S474 | DDBJ | AY540332 | Only nucleotide sequence used | |
| Panellus serotinus | NBRC 30264 | NBRC | Stock culture (mycelium) | |
| CBS 722.83 | DDBJ | AY265847 | Only nucleotide sequence used | |
| Entoloma sarcopus | Tottori Es-3 | Tottori, Japan | AB301603 | Fruiting body isolated from the field |
| Lyophyllum shimeji | OK1L | Kinki University | AB301604 | Stock culture (mycelium) |
| Lyophyllum decastes | NBRC33134 | NBRC | AB301605 | Stock culture (mycelium) |
| Armillaria mellea | NBRC31621 | NBRC | Stock culture (mycelium) | |
| M1 | DDBJ | AJ250051 | Only nucleotide sequence used | |
| Clitocybe clavipes | NBRC30524 | NBRC | AB301607 | Stock culture (mycelium) |
| Clitocybe gibba | NBRC100092 | NBRC | AB301608 | Stock culture (mycelium) |
| Lactarius akahatsu | NBRC33157 | NBRC | AB301609 | Stock culture (mycelium) |
| Lactarius chrysorrheus | NBRC32791 | NBRC | AB301610 | Stock culture (mycelium) |
| Lactarius hatsudake | NBRC32778 | NBRC | AB301611 | Stock culture (mycelium) |
| Tricholoma matsutake | JK | Kyoto, Japan | Fruiting body isolated from the field | |
| MC1 | DDBJ | AB036892 | Fruiting body isolated from the field | |
| Pholiota lubrica | NBRC32453 | NBRC | AB301612 | Stock culture (mycelium) |
| Cortinarius tenuipes | Nara Ct-1 | Nara, Japan | AB301613 | Only nucleotide sequence used |
| Auricularia auricula | TU-AA1 | Tottori University | Stock culture (mycelium) | |
| Agaricus bisporus | B100 | HFPRI | Stock culture (mycelium) | |
| Flammulina velutipes | HATUYUKI | Tottori university | Stock culture (mycelium) | |
| Grifola frondosa | MS1 | HFPRI | Stock culture (mycelium) | |
| Hypsizigus marmoreus | TU-HM1 | Tottori University | Stock culture (mycelium) | |
| Pholiota nameko | 3024 | Tottori University | Stock culture (mycelium) | |
| Pleurotus eryngii | MH006062 | Tottori University | Stock culture (mycelium) |
DDBJ, DNA Data Bank of Japan; NBRC, National Institute of Technology and Evaluation (NITE) Biological Resource Center, Japan; HFPRI, Hokkaido Forest Products Research Institute.
The fruiting body of Omphalotus japonicus was cultivated on sawdust medium (2.5 kg wet weight, 80% sawdust, 20% wheat bran, 65% final moisture content) in polypropylene bags. O. japonicus mycelia were inoculated to the medium and grown at room temperature for 2 months. At the end of this period, the bags were removed and the colonized substrates were irradiated with visible light (12 h dark and 12 h light) at room temperature for 2 months.
Cooking of fruiting bodies.
Fruiting bodies were cooked as follows. For baking, the pilei of fruiting bodies were cut into 3-cm square pieces and baked on both sides in an indoor electric grill (HPS-39G-T; Sanyo Electric Co., Osaka, Japan) without oil at 240°C for 2 min each. For stir-frying, the pilei of fruiting bodies were cut into 3-mm-thick slices and stir-fried with mixing on the oiled electric grill at 240°C for 2 min. For tempura-style cooking (deep-frying in tempura batter), the pilei of fruiting bodies were cut into 3-cm square pieces and battered. The battered mushroom pieces were deep-fried in oil at 180°C for 2 min. For boiling, the pilei of fruiting bodies were cut into 3-cm square pieces and boiled in water for 30, 60, 120, or 180 min.
DNA extraction from fruiting bodies.
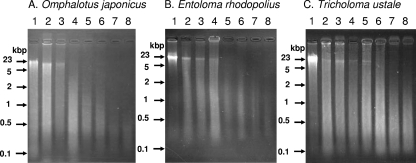
Fruiting body tissues and mycelia were frozen in liquid nitrogen and ground to a fine powder in a mortar and pestle. DNAs were extracted from 300 mg of frozen powder (3). Figure 1 compares the size distributions of DNA isolated from fresh and cooked mushrooms. A broad range of DNA sizes up to 23 kbp was consistently present in the fresh, baked, stir-fried, and tempura-style samples, and the sizes of DNAs extracted from boiled mushroom samples were smaller than those in the other samples. For DNA isolated from O. japonicus boiled for 120 and 180 min, only DNA of less than 500 bp in size was observed.
FIG. 1.
Electrophoretic analysis of extracted DNA. Samples were electrophoresed on 2.0% agarose gels and stained with ethidium bromide. Panels A, B, and C correspond to DNA purified from the fruiting bodies of O. japonicus, E. rhodopolius, and T. ustale, respectively. In each panel, lanes and samples are lane 1, fresh; lane 2, baked for 4 min; lane 3, stir-fried for 2 min; lane 4, tempura-style (deep-fried in tempura batter for 2 min); lanes 5 to 8, boiled for 30, 60, 120, or 180 min, respectively. The size markers are EcoRI/HindIII double digests of lambda DNA and 100-bp ladders.
Primer design for species-specific primers.
DNA sequencing of the rDNA ITS region (12) was carried out as previously described (8). The nucleotide sequences of the rDNA ITS region of four poisonous mushrooms and of edible mushrooms that are morphologically similar were aligned, and four oligonucleotide primer pairs were designed for the specific amplification of O. japonicus, Entoloma rhodopolius, Clitocybe acromelalga, and Tricholoma ustale rDNA (Table 3), because these poisonous mushrooms have been responsible for approximately 85% of the mushroom poisoning events in Japan over the last five years.
TABLE 3.
Species-specific primers for each poisonous mushroom species
| Targeted species | Primer
|
Product length (bp) | ||||
|---|---|---|---|---|---|---|
| Name | Length (bp) | Sequence (5′-3′) | Positionb | Tm (°C)c | ||
| O. japonicus | OJSP-F | 19 | GTGCACGTTTCCTTTCAAT | 59-77 | 60.12 | 107 |
| OJSP-R | 20 | AGAATCATCAACAGAGCTGC | 165-146 | 59.55 | ||
| E. rhodopolius | ERSP-F | 22 | TTTGAGAACTGCTGTGAAAATC | 126-148 | 60.81 | 110 |
| ERSP-R | 23 | GGCACAAAGTCCCTATATGTTTA | 235-213 | 60.61 | ||
| C. acromelalga | CASP-F | 19 | GGTGCACACCTGATAACCA | 53-71 | 62.09 | 108 |
| CASP-R | 20 | AGCTTAAGCTTTCGCACCAG | 160-141 | 63.39 | ||
| T. ustale | TUSP-F | 22 | TAGTAGGGACCTCTGTTGCCTT | 501-522 | 62.77 | 80 |
| TUSP-R | 25 | AACCTCCAATTTAAAGCTGCTTCAC | 580-556 | 66.06 | ||
| Universala | ITS1 | 19 | TCCGTAGGTGAACCTGCGG | Variable | 68.4 | Variable |
| ITS2 | 20 | GCTGCGTTCTTCATCGATGC | 68.13 | |||
| ITS3 | 20 | GCATCGATGAAGAACGCAGC | Variable | 68.13 | Variable | |
| ITS4 | 20 | TCCTCCGCTTATTGATATGC | 61.49 | |||
White et al. (12).
Positions of nucleotides correspond with sequence of ITS region in each species. The nucleotide sequences were published in the DDBJ nucleotide sequence database under accession numbers AB301601, AB301602, AB301606, and AB036894 respectively.
Tm, melting temperature. Values were calculated with oligonucleotide calculator software (Sigma-Aldrich, Tokyo, Japan) on the web site (http://www.genosys.jp/adt/SGJ_NN_3rd.html).
Species-specific detection.
PCR amplifications were performed using four species-specific primer pairs, using genomic DNAs from twenty-seven mushroom species as templates. DNA was also extracted from 300 mg of frozen powder using a GeneAll plant SV mini kit (GeneAll Biotechnology, Seoul, Korea) in accordance with the manufacturer's recommendations. Real-time PCR was carried out using Sybr green real-time PCR master mix (Toyobo Co., Osaka, Japan) with 2 μM forward and reverse primers for detection of O. japonicus and T. ustale and 0.4 μM forward and reverse primers for detection of E. rhodopolius and C. acromelalga. Real-time PCR was carried out in a 10-μl reaction mixture volume containing 5 μl of the master mix, about 2.5 ng of extracted genomic DNA, and appropriate primers. Thermocycling was performed using a LineGene real-time thermal cycler (BioFlux, Tokyo, Japan). The reaction was performed for 20 cycles, and the following cycling profile was used. The first denaturing step was at 95°C for 1 min, and then the PCR cycles were 5 s of denaturation at 95°C, 5 s of annealing at 65°C, and then 15 s of extension at 72°C. Fluorescence data were acquired during the elongation step in every cycle.
When the specific primer pairs were used, increases in fluorescence intensity were detected only for the targeted species (data not shown). Thus, we concluded that the designated primers were species specific for each poisonous mushroom. In order to confirm the universality of the four species-specific primer pairs for each species, PCR amplification was performed using genomic DNAs from 11 strains of O. japonicus, 5 strains of E. rhodopolius, and 4 strains of T. ustale (data not shown).
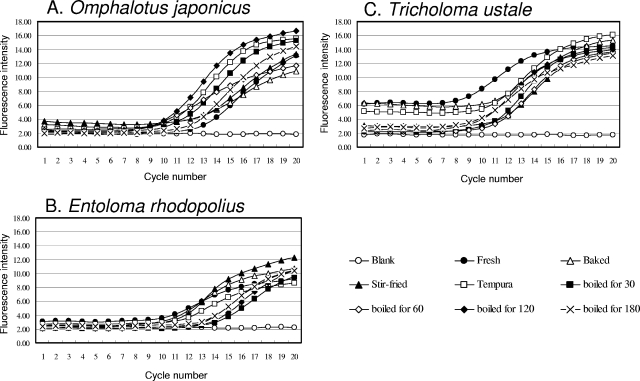
To confirm the amplification of specific DNA fragments of cooked mushrooms, real-time PCR was carried out. The results are shown in Fig. 2. For each primer set used for amplifying DNA from the poisonous mushroom species, species-specific detection was successful. In our proposed system, less than 1 h is required for detection via real-time PCR. However, in order to perfect the system we developed in this study, more specific primers should be designed in further studies.
FIG. 2.
Fluorescence intensity profile during real-time PCR with species-specific primer pairs using genomic DNAs from cooked and uncooked mushrooms as templates. (A) Amplified with OJSP-F/OJSP-R using genomic DNA of O. japonicus. (B) Amplified with ERSP-F/ERSP-R using genomic DNA of E. rhodopolius. (C) Amplified with CASP-F/CASP-R using genomic DNA of T. ustale. Blank, no genomic DNA; Fresh, genomic DNA from fresh fruiting body; Baked, baked for 4 min; Stir-fried, stir-fried for 2 min; Tempura, deep-fried in tempura batter for 2 min.
Nucleotide sequence accession numbers.
The nucleotide sequence data obtained in this study have been deposited in the DDBJ nucleotide sequence database under accession numbers AB301601 to AB301613.
Acknowledgments
This research was partially supported by the Japanese Ministry of Education, Science, Sports, and Culture via a grant-in-aid for scientific research (C), 18580164, 2006-2007.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Bao, D. P., T. Aimi, and Y. Kitamoto. 2005. Cladistic relationships among the Pleurotus ostreatus complex, the Pleurotus pulmonarius complex, and Pleurotus eryngii based on the mitochondrial small subunit ribosomal DNA sequence analysis. J. Wood Sci. 51:77-82. [Google Scholar]
- 2.Bresinsky, A., O. Hilber, and H. P. Molitoris. 1977. The genus Pleurotus as an aid for understanding the concept of species in Basidiomycetes. Bibl. Mycol. 61:229-258. [Google Scholar]
- 3.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Reporter 1:19-21. [Google Scholar]
- 4.Gonzalez, P., and J. Labarere. 1998. Sequence and secondary structure of the mitochondrial small-subunit rRNA V4, V6, and V9 domains reveal highly species-specific variations within the genus Agrocybe. Appl. Environ. Microbiol. 64:4149-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwa′sna, H. 2001. Fungi in the rhizosphere of common oak and its stumps and their possible effect on infection by Armillaria. Appl. Soil Ecol. 17:215-227. [Google Scholar]
- 6.Lee, J. S., M. O. Lim, K. Y. Cho, J. H. Cho, S. Y. Chang, and D. H. Nam. 2006. Identification of medicinal mushroom species based on nuclear large subunit rDNA sequences. J. Microbiol. 44:29-34. [PubMed] [Google Scholar]
- 7.Ohnishi, Y., M. Nagase, T. Ichiyanagi, Y. Kitamoto, and T. Aimi. 2007. Transcriptional regulation of two cellobiohydrolase encoding genes (cel1 and cel2) from the wood-degrading basidiomycete Polyporus arcularius. Appl. Microbiol. Biotechnol. 76:1069-1078. [DOI] [PubMed] [Google Scholar]
- 8.Palapala, V. A., T. Aimi, S. Inatomi, and T. Morinaga. 2002. ITS-PCR-RFLP method for the distinguishing commercial cultivars of edible mushroom, Flammulina velutipes. J. Food Sci. 67:2486-2490. [Google Scholar]
- 9.Petersen, R. H., and K. W. Hughes. 1999. Species and speciation in mushrooms? Development of a species concept poses difficulties. Bio. Sci. 49:440-452. [Google Scholar]
- 10.Schmidt, O., and U. Moreth. 2000. Species-specific PCR primers in the rDNA-ITS region as a diagnostic tool for Serpula lacrymans. Mycol. Res. 104:69-72. [Google Scholar]
- 11.Schnabel, G., J. S. Ash, and P. K. Bryson. 2005. Identification and characterization of Armillaria tabescens from the southeastern United States. Mycol. Res. 109:1208-1222. [DOI] [PubMed] [Google Scholar]
- 12.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, NY.
- 13.Wu, Q. X., G. M. Mueller, F. M. Lutzoni, Y. Q. Huang, and S. Y. Guo. 2000. Phylogenetic and biogeographic relationships of eastern Asian and eastern North American disjunct Suillus species (fungi) as inferred from nuclear ribosomal RNA ITS sequences. Mol. Phylogenet. Evol. 17:37-47. [DOI] [PubMed] [Google Scholar]
- 14.Yamada, A., K. Maeda, H. Kobayashi, and H. Murata. 2006. Ectomycorrhizal symbiosis in vitro between Tricholoma matsutake and Pinus densiflora seedlings that resembles naturally occurring ‘shiro.’ Mycorrhiza 16:111-116. [DOI] [PubMed] [Google Scholar]