Abstract
In order to elucidate the potential mechanisms of U(VI) reduction for the optimization of bioremediation strategies, the structure-function relationships of microbial communities were investigated in microcosms of subsurface materials cocontaminated with radionuclides and nitrate. A polyphasic approach was used to assess the functional diversity of microbial populations likely to catalyze electron flow under conditions proposed for in situ uranium bioremediation. The addition of ethanol and glucose as supplemental electron donors stimulated microbial nitrate and Fe(III) reduction as the predominant terminal electron-accepting processes (TEAPs). U(VI), Fe(III), and sulfate reduction overlapped in the glucose treatment, whereas U(VI) reduction was concurrent with sulfate reduction but preceded Fe(III) reduction in the ethanol treatments. Phyllosilicate clays were shown to be the major source of Fe(III) for microbial respiration by using variable-temperature Mössbauer spectroscopy. Nitrate- and Fe(III)-reducing bacteria (FeRB) were abundant throughout the shifts in TEAPs observed in biostimulated microcosms and were affiliated with the genera Geobacter, Tolumonas, Clostridium, Arthrobacter, Dechloromonas, and Pseudomonas. Up to two orders of magnitude higher counts of FeRB and enhanced U(VI) removal were observed in ethanol-amended treatments compared to the results in glucose-amended treatments. Quantification of citrate synthase (gltA) levels demonstrated a stimulation of Geobacteraceae activity during metal reduction in carbon-amended microcosms, with the highest expression observed in the glucose treatment. Phylogenetic analysis indicated that the active FeRB share high sequence identity with Geobacteraceae members cultivated from contaminated subsurface environments. Our results show that the functional diversity of populations capable of U(VI) reduction is dependent upon the choice of electron donor.
Uranium contamination in subsurface environments is a widespread problem at mining and milling sites across North America, South America, and Eastern Europe (1). Uranium in the oxidized state, U(VI), is highly soluble and toxic and thus is a potential contaminant to local drinking-water supplies (46). Nitrate is often a cocontaminant with U(VI) as a result of the use of nitric acid in the processing of uranium and uranium-bearing waste (6, 45). Oxidized uranium can be immobilized in contaminated groundwater through the reduction of U(VI) to insoluble U(IV) by indirect (abiotic) and direct (enzymatic) processes catalyzed by microorganisms. Current remediation practices favor the stimulation of reductive uranium immobilization catalyzed by indigenous microbial communities along with natural attenuation and monitoring (5, 24, 40, 44, 65, 68, 69). Microbial uranium reduction activity in contaminated subsurface environments is often limited by carbon or electron donor availability (13, 24, 44, 69). Previous studies have indicated that U(VI) reduction does not proceed until nitrate is depleted (13, 16, 24, 44, 68, 69), as high nitrate concentrations inhibit the reduction of U(VI) by serving as a competing and more energetically favorable terminal electron acceptor for microorganisms (11, 16). The fate and transport of uranium in groundwater are also strongly linked through sorption and precipitation processes to the bioreduction of Fe minerals, including oxides, layer-silicate clay minerals, and sulfides (7, 23, 53).
In order to appropriately design U(VI) bioremediation strategies, the potential function and phylogenetic structure of indigenous subsurface microbial communities must be further understood (24, 34, 46). Conflicting evidence has been presented on which microbial groups, Fe(III)- or sulfate-reducing bacteria (FeRB or SRB), effectively catalyze the reductive immobilization of U(VI) in the presence of amended electron donors (5, 44, 69). The addition of acetate to the subsurface at a uranium-contaminated site in Rifle, Colorado, initially stimulated FeRB within the family Geobacteraceae to reduce U(VI) (5, 65). However, with long-term acetate addition, SRB within the family Desulfobacteraceae, which are not capable of U(VI) reduction, increased in abundance and a concomitant reoxidation of U(IV) was observed (5, 65). At a uranium-contaminated site in Oak Ridge, Tennessee, in situ and laboratory-based experiments successfully employed ethanol amendments to stimulate denitrification followed by the reduction of U(VI) by indigenous microbial communities (13, 24, 44, 48, 50, 57, 68). In these studies, ethanol amendments stimulated both SRB and FeRB, with SRB likely catalyzing the reduction of U(VI). This suggests that the potential for bioremediation will be affected by the choice of electron donor amendment through effects on the functional diversity of U(VI)-reducing microbial populations. As uranium reduction is dependent on the depletion of nitrate, the microbial populations mediating nitrate reduction are also critical to the design of bioremediation strategies. Although nitrate-reducing bacteria (NRB) have been studied extensively in subsurface environments (2, 15, 19, 24, 56, 58, 70), the mechanisms controlling the in situ metabolism of NRB remain poorly understood.
The dynamics of microbial populations capable of U(VI) reduction in subsurface sediments are poorly understood, and the differences in the microbial community dynamics during bioremediation have not been explored. Based on the results of previous studies (13, 44, 49, 57, 68, 69), we hypothesized that the activity of nitrate- and Fe(III)-reducing microbial populations, catalyzing the reductive immobilization of U(VI) in subsurface radionuclide-contaminated sediments, would be dependent on the choice of electron donor. The objectives of the present study were (i) to characterize structure-function relationships for microbial groups likely to catalyze or limit U(VI) reduction in radionuclide-contaminated sediments and (ii) to further develop a proxy for the metabolic activity of FeRB. Microbial activity was assessed by monitoring terminal electron-accepting processes (TEAPs), electron donor utilization, and Fe(III) mineral transformations in microcosms conducted with subsurface materials cocontaminated with high levels of U(VI) and nitrate. In parallel, microbial functional groups (i.e., NRB and FeRB) were enumerated and characterized using a combination of cultivation-dependent and -independent methods.
MATERIALS AND METHODS
Site and sample description.
The study was conducted at the Oak Ridge Field Research Center (ORFRC) of the U.S. Department of Energy's Environmental Remediation Sciences Program (ERSP), which is located adjacent to the Y-12 industrial complex within the Oak Ridge National Laboratory (ORNL) reservation in Oak Ridge, Tennessee. Waste products from uranium enrichment processes at the Y-12 complex were collected and stored in four unlined ponds, the S-3 ponds, until 1988 when they were pumped and capped by a parking lot (6). Subsurface groundwater flow created a contaminant plume originating from the S-3 pond site that currently extends approximately 7 km along a geological strike east and west of the ponds to a depth of >150 m (6). The Area 2 experimental plot within the ORFRC is located west, down gradient, of the S-3 pond site and is characterized by a pH of 6.5, up to 8 mM sulfate, 1 to 26 mM nitrate, and 0.5 to 4.5 μM uranium. For a detailed site description, refer to the ORFRC Web page (http://www.esd.ornl.gov/orifrc/).
Sediments were sampled from borehole FB094 within ORFRC Area 2, 6.7 to 7.2 m below the surface, on 19 September 2005 using a Geoprobe equipped with polyurethane sleeves lining the corer. Cores (5.6 cm in diameter and 91 cm in length) were aseptically sectioned under strictly anoxic conditions in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, Michigan) and stored anaerobically in gas-tight containers at 4°C prior to overnight shipment to Florida State University. Groundwater samples were withdrawn from Area 2 well FW209 and collected in airtight bottles for shipment.
Microcosm design and sampling.
Sediment from each core section was pooled and homogenized under aseptic conditions in an N2-filled glove bag. Microcosms were constructed by loading 60 g of homogenized sediment into 250-ml sterile Pyrex bottles, diluting the sediment 1:5 (wt/vol) with 240 ml of site groundwater, and capping with sterile rubber stoppers and plastic screw caps with apertures. The microcosms were neutralized with sterile, anoxic 1 M NaHCO3 to a final pH of 6.93. Three replicate microcosms were established for each of the following treatments: amendment with ethanol, amendment with glucose, and unamended for use as a control. Ethanol and glucose were added from sterile, anoxic stocks to final concentrations of 20 and 10 mM, respectively. All microcosms were shaken at 150 rpm for 1 h, flushed with sterile N2 for 20 min, and incubated statically in the dark at 30°C.
Each of the nine microcosms was sampled by removing ∼5 ml every other day using a sterile syringe equipped with an 18-gauge needle under a H2/N2 atmosphere in a Coy anaerobic chamber. The Fe(II) content in the water and soil suspension of each microcosm was determined immediately by adding 0.2 ml of the microcosm sample to 0.5 M HCl. Pore waters were extracted from the microcosm samples by centrifugation in the Coy chamber at 5,000 × g for 5 min, followed by filtration through a 0.22-μm nylon syringe filter. On days 0, 4, and 21, samples from each replicate microcosm were collected and pooled for cultivation-independent community characterization. The samples were centrifuged at 7,000 × g for 7 min, the supernatant discarded, and the solid phase frozen at −80°C until nucleic acid extraction.
Pore-water and solid-phase geochemistry.
The pore-water nitrate concentrations were determined colorimetrically as previously described (8), and the total sulfate concentrations were determined using ion chromatography. Pore-water samples for uranium analysis were acidified with nitric acid in the Coy chamber and frozen at −20°C until kinetic phosphorescence analysis using a KPA-11 analyzer (Chem-Chek Instruments, Richland, Washington). Pore water for the determination of carbon substrate utilization was filtered into 1.8-ml glass vials, capped with a Teflon-coated butyl-rubber septum, and stored at −20°C. Aliphatic acids, alcohols, and sugars were determined with a Hewlett Packard 1090 series II high-performance liquid chromatograph (32). Wet chemical extractions were used to determine the poorly crystalline Fe oxide minerals by extracting triplicate microcosm samples in 0.5 M HCl for 1 h under anoxic conditions in the Coy chamber (31). The HCl-extractable Fe(II) of filtered extracts was quantified colorimetrically by analysis in ferrozine buffer (50 mM HEPES, 0.1% ferrozine, pH 7) (31).
Mössbauer spectroscopy.
Microcosm samples for Mössbauer spectroscopy were collected under an H2/N2 atmosphere in the Coy chamber into glass serum bottles and then tightly capped with butyl-rubber stoppers and aluminum crimp seals. Mössbauer spectra were obtained by using a Web Research, Inc., spectrometer equipped with a Janis Model SHI-850-5 closed-cycle cryostat, operating at a liquid He (nominally 4.2 to 6 K), liquid N2 (nominally 77 K), or room (nominally 298 K) temperature (63). The gamma-ray source was 57Co with anominal 50-mCi activity dispersed at 10% in an Rh-foil matrix. Mössbauer hyperfine parameters (δ, isomer shift; Δ, quadrupole splitting; and Bhf, magnetic hyperfine field) were calculated using a least-squares fitting program assuming Lorentzian line shapes. The isomer shift, velocity scale, and magnetic hyperfine fields were calibrated relative to a 0.7-μm-thick Fe(0) foil measured at room temperature. Texture effects were assumed to be absent.
Bacterial enrichment and enumeration.
FeRB and NRB were enumerated by the three-tube most-probable number (MPN) technique using 10-fold serial dilutions of sediment or microcosm material in growth medium. The medium for FeRB was prepared according to methods described previously (52) for neutrophilic organisms (pH 7). For NRB, the medium was prepared as above with sodium nitrate added to a 25 mM final concentration. Carbon substrates (glucose and ethanol) were added from anoxic, sterile stock solutions to a 20 mM final concentration, and tubes were purged with 9:1 N2-CO2 prior to sealing with a butyl-rubber stopper. Pooled microcosm samples collected on days 7 and 21 of the incubation were used to inoculate MPN dilution series with iron or nitrate as the electron acceptor and the same carbon source as the source microcosm. Microcosm samples from the unamended treatment, as well as homogenized FB094 sediment, were also used to inoculate MPN dilution series for both carbon substrates with each electron acceptor. The tubes were incubated at 30°C in the dark for 3 months. Positive growth was scored by visual screening of turbidity, and electron acceptor utilization was verified in the highest positive dilutions by colorimetric quantification as described above. MPN values and 95% confidence intervals were calculated from standard MPN tables (3). Samples from the highest positive dilutions were collected and frozen at −80°C for nucleic acid-based community characterization.
DNA extraction and small-subunit (SSU) rRNA gene clone library construction.
Microbial community DNA was extracted from microcosm samples using a phenol-chloroform procedure adapted from Scala and Kerkhof (55a). The main deviations from the original method included the use of liquid nitrogen and a 55°C water bath for the freeze/thaw steps. DNA extracts were purified by using a single high-salt-ethidium bromide-phenol extraction step, as described by Lovell and Piceno (33). DNA was extracted from MPN cultures by using an Ultra Clean soil DNA kit according to the manufacturer's instructions (Mo Bio Laboratories, Solana Beach, California). Samples (∼1 ml) from the three highest positive dilutions of each MPN dilution series were centrifuged at 7,000 × g for 7 min, and the cell pellets were combined for use in the extraction.
DNA was PCR amplified using the Bacteria domain-specific SSU rRNA gene primers 27F (5′-AGR GTT TGA TCM TGG CTC AG-3′) (26) and 1392R (5′-ACG GGC GGT GTG TAC-3′) (67). PCR products were cloned, screened using restriction fragment length polymorphism (RFLP) analysis, sequenced, and analyzed as previously described (2). Phylogenetic analysis of clone sequences and reference sequences from the GenBank database available from the National Center for Biotechnology Information (4) was performed with the ARB software package (39).
Terminal-RFLP (TRFLP) analysis.
SSU rRNA genes were amplified from DNA extracts as described above except that the 27F primer was labeled at the 5′ end with 6-carboxyfluorescein (Applied Biosystems, Foster City, California). Fluorescently labeled PCR products were precipitated with 0.5 volume 7.5 M ammonium acetate and 3 volumes of 100% ethanol, followed by a 10-min incubation at room temperature and centrifugation at 20,000 × g at 4°C for 15 min. The resuspended PCR product was quantified by image analysis of an electrophoresis gel as described previously (55a). PCR product in the amount of 15 ng was digested with the restriction enzyme MnlI (New England Biolabs, Beverly, Massachusetts) for 6 h at 37°C, purified, and run on an ABI 310 genetic analyzer (Applied Biosystems, Foster City, California) using Genescan software as described previously (43). For the comparative analysis, all fingerprints were normalized to the same total peak area and the small peaks representing <1% of the total area were excluded from further analysis.
Real-time PCR quantification of mRNA transcripts.
Total RNA and mRNA were extracted from a total of 3.9 ml of microcosm sample according to previously described methods (10). The gltA-specific primers used for mRNA real-time PCR quantification were CS375nF (5′-AAC AAG ATG RCM GCC TGG G-3′) and CS598nR (5′-TCR TGG TCG GAR TGG AGA AT-3′) (this study; modified from reference 21). cDNA synthesis was performed using the method described by Chin et al. (9, 10) with 0.5 μg template mRNA and CS598nR incubated at 52°C for 60 min, followed by enzyme inactivation at 70°C for 15 min. cDNA was purified, quantified and prepared for real-time PCR quantification using Sybr green and primers CS375nF and CS598nR (9, 10). Quantitative analysis of the cDNA was carried out with an Applied Biosystems 7500 real-time PCR system using 7500 real-time PCR system Sequence Detection Software (version 1.3.1) (9, 10). Thermocycling was performed with an initial activation step at 50°C for 2 min and denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 1 min, and elongation at 65°C for 30 s, with a final extension step at 65°C for 6 min. The detection limits of the PCR assays were determined (9), and they were found to have a minimum sensitivity of 100 to 101 target molecules per reaction. The precision and reproducibility of the quantification were carefully optimized, and correct lengths of PCR products were verified (9).
Phylogenetic analysis of partial gltA cDNA clone sequences.
A clone library was generated with reverse transcription-PCR amplicons (224-bp length amplified with primers CS375nF and CS5985nR) obtained from mRNA isolated from microcosm sediment samples according the methods described above. The resulting nucleotide gltA sequences were compared to the GenBank protein databases according to the BLASTx algorithm (4), and the amino acid sequences were aligned with reference sequences using Clustal W (64). The resulting 75-amino-acid alignment was used to infer phylogenetic trees using neighbor-joining methods.
Nucleotide sequence accession numbers.
Sequences generated in this study were deposited in the GenBank database under the accession numbers EU236219 to EU236255 (SSU rRNA gene clones) and EU352200 to EU352205 (gltA cDNA clones).
RESULTS
Microbial activity in sediment microcosms.
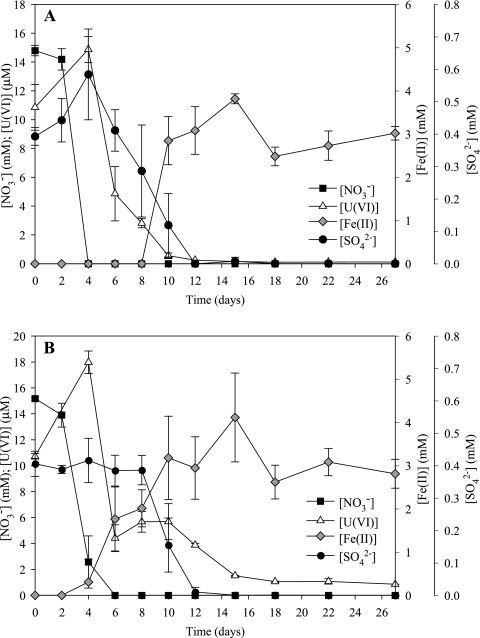
Microcosms were constructed with ORFRC Area 2 subsurface materials cocontaminated with uranium (10 μM initial concentration) and nitrate (15 mM initial concentration). Microbial activity was stimulated in microcosms amended with either ethanol or glucose. Little to no activity was observed in the unamended treatment (data not shown). Nitrate was rapidly reduced in the carbon-amended treatments beginning on day 2 and was completely consumed by day 4 and day 6 in the ethanol and glucose treatments, respectively (Fig. 1). In all treatments, the initial U(VI) concentration was approximately 10 μM and increased from day 4 to 15 to 18 μM (Fig. 1). In the ethanol treatment, uranium and sulfate reduction began on day 4 and both electron acceptors were completely reduced by day 12 (Fig. 1A). Fe(III) reduction [accumulation of Fe(II)] was first measured in the ethanol treatment on day 10, overlapping with the end of sulfate and uranium reduction (Fig. 1A). In the glucose treatment, the onset of uranium and iron reduction appeared to overlap with the depletion of nitrate (Fig. 1B). Fe(II) accumulation was first measured in the glucose treatment on day 6 of the incubation and in the ethanol treatment ceased by day 18. Uranium reduction began on day 4, and U(VI) was incompletely reduced to a final concentration of 1 μM by day 15 with no further U(VI) reduction detected (Fig. 1B). Sulfate was depleted in the microcosm between days 8 and 12, concurrent with uranium and iron reduction (Fig. 1B).
FIG. 1.
Electron acceptor usage in ethanol-amended (A) and glucose-amended (B) microcosms. Values are averages ± standard deviations of the results for triplicate microcosms.
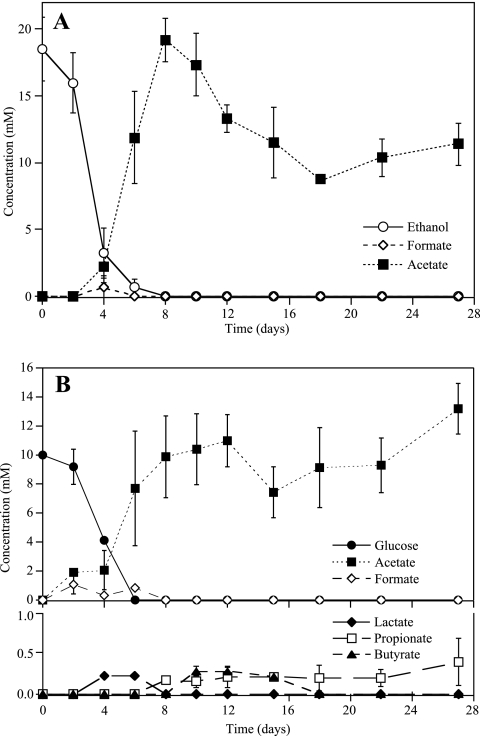
Measurable consumption of supplemental ethanol and glucose in microcosms occurred in conjunction with the onset of nitrate reduction. Supplemental ethanol was completely consumed by day 8, in conjunction with uranium and sulfate reduction, with transient formation of formate observed and acetate accumulating as end product (Fig. 2A). Acetate accumulated to a concentration of 20 mM on day 8 and slowly decreased during the remainder of the incubation. The consumption of acetate may have been linked to the reduction of iron in the ethanol treatment. In the glucose treatment, glucose was fully consumed by day 6 and acetate was the primary fermentation product accumulating, whereas lactate, butyrate, and propionate appeared transiently (Fig. 2B).
FIG. 2.
Electron donor utilization in ethanol-amended (A) and glucose-amended (B) microcosms as determined by using high performance liquid chromatography. Values are averages ± standard deviations of the results for triplicate microcosms.
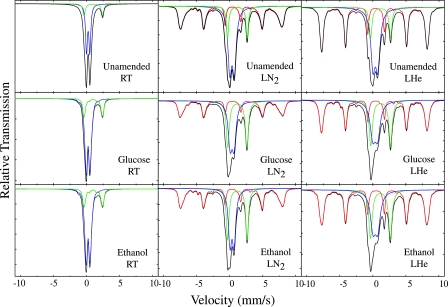
The form and speciation of iron-bearing minerals were determined for microcosm samples from the final time point of the experiment (day 27) for each of the treatments (ethanol, glucose, and unamended) using Mössbauer spectroscopy (Fig. 3 and Table 1). The results of spectra at room temperature (298 K) exhibited no sextet pattern, thus excluding the presence of nonsuperparamagnetic hematite, magnetite, and maghemite in ORFRC Area 2 sediments both before and after biostimulation (Fig. 3). All microcosm samples contained peaks for Fe(III) and Fe(II) in a silicate phase, and the peak areas for Fe(II) were greater in carbon-amended microcosms than in the unamended treatment. Spectra at 77 K revealed that the amount of Fe(II) in the unamended treatment was 14.6% of the total iron, which increased to 27.9 and 29.0% of total Fe in the ethanol and glucose treatments, respectively (Table 1). A poorly ordered or Al-substituted goethite phase was also present in all samples, and it was partially dissolved by microbial activity, as indicated by the decrease in sextet area between the unamended (49% of total Fe) and carbon-amended (40% of total Fe) samples. The increase in the percentage of Fe(II) was at the expense of Fe(III) in silicate, which decreased from 42% (at 77 K) in the unamended treatment to 32% (at 77 K) in the carbon-amended treatments (Table 1).
FIG. 3.
Mössbauer spectra of ORFRC ethanol- or glucose-amended or unamended microcosm samples measured at room temperature (RT; nominally 298 K), liquid nitrogen temperature (LN2; nominally 77 K), and liquid helium temperature (LHe; nominally 4.2 to 6 K). Curves in red are Fe(III) (oxyhydr)oxides, in green are Fe(II) in phyllosilicates, in blue are Fe(III) in phyllosilicates, and in black are the sum of all components (fitted experimental spectrum).
TABLE 1.
Mössbauer parameters of microcosm samplesa
| Sample | Temp (K) | Fe stateb or phase | δ (mm/s) | Δ (mm/s) | Bhf (T) | Γ (mm/s) | RA (%) |
|---|---|---|---|---|---|---|---|
| Unamended | RT | Fe(III) | 0.35 | 0.54 | 83 | ||
| Fe(II) | 1.13 | 2.84 | 17 | ||||
| LN2 | Fe(III) | 0.48 | 0.68 | 0.68 | 44 | ||
| Fe(II) | 1.27 | 2.97 | 0.42 | 15 | |||
| Goethite | 0.45 | −0.27 | 47 | 41 | |||
| LHe | Fe(III) | 0.49 | 0.73 | 0.68 | 39 | ||
| Fe(II) | 1.27 | 3.02 | 0.50 | 13 | |||
| Goethite | 0.47 | −0.25 | 49.5 | 48 | |||
| Glucose | RT | Fe(III) | 0.34 | 0.52 | 77 | ||
| Fe(II) | 1.09 | 1.97 | 23 | ||||
| LN2 | Fe(III) | 0.48 | 0.59 | 0.71 | 34 | ||
| Fe(II) | 1.27 | 2.90 | 0.55 | 31 | |||
| Goethite | 0.50 | −0.26 | 48.4 | 35 | |||
| LHe | Fe(III) | 0.47 | 0.73 | 0.96 | 31 | ||
| Fe(II) | 1.30 | 2.99 | 0.60 | 26 | |||
| Goethite | 0.47 | −0.28 | 49.5 | 43 | |||
| Ethanol | RT | Fe(III) | 0.35 | 0.53 | 77 | ||
| Fe(II) | 1.09 | 1.97 | 23 | ||||
| LN2 | Fe(III) | 0.47 | 0.60 | 0.60 | 34 | ||
| Fe(II) | 1.28 | 2.94 | 0.49 | 28 | |||
| Goethite | 0.49 | −0.19 | 47.7 | 38 | |||
| LHe | Fe(III) | 0.50 | 0.71 | 0.94 | 29 | ||
| Fe(II) | 1.26 | 3.00 | 0.62 | 26 | |||
| Goethite | 0.47 | −0.26 | 49.5 | 45 |
δ, isomer shift; Δ, quadrupole splitting; Γ, line full width at half maximum; Bhf, magnetic hyperfine field; RA, relative peak area. Errors in the isomer shift and quadrupole splitting are ±0.01 mm/s, in the hyperfine field ±0.5 T, and in the relative area ±1%.
Fe(III) and Fe(II) are ferric and ferrous iron, respectively, in the phyllosilicate phase.
Enumeration of microbial groups of dominant TEAPs.
NRB and FeRB microbial functional groups were enumerated using an MPN approach during the nitrate- (day 7) and iron-reduction (day 21) phases of the carbon-amended microcosms. NRB and FeRB were enumerated in the unamended control microcosms and the homogenized FB094 sediment samples with either ethanol or glucose added as the electron donor. The abundance of NRB in the microcosms on days 7 and 21 of the incubation was 109 cells ml−1 in both the ethanol and glucose treatments. The abundance of nitrate reducers in the glucose treatment at day 21 could not be determined, because it was not possible to distinguish whether positive growth was a result of nitrate respiration or glucose fermentation. The enumeration of NRB in the unamended microcosm and homogenized FB094 sediment revealed a greater abundance in cultures with glucose as the carbon substrate than in those amended with ethanol (Table 2).
TABLE 2.
Enumeration of iron- and nitrate-reducing bacteria in carbon-amended and -unamended microcosms and homogenized FB094 core sediment
| Inoculum, day | MPN carbon substrate | MPN (cells g−1) ofa:
|
|
|---|---|---|---|
| FeRBb | NRBc | ||
| Glucose microcosms, day 7 | Glucose | 3.2 × 106 (7.0 × 105-1.5 × 107) | 4.5 × 109 (9.5 × 108-2.1 × 1010) |
| Ethanol microcosms, day 7 | Ethanol | 1.0 × 109 (2.2 × 108-4.7 × 109) | 2.5 × 109 (5.5 × 108-1.2 × 1010) |
| Glucose microcosms, day 21 | Glucose | 2.0 × 107 (4.3 × 106-9.5 × 107) | ND |
| Ethanol microcosms, day 21 | Ethanol | 2.0 × 108 (4.3 × 107-9.50 × 108) | 2.0 × 109 (4.3 × 108-9.5 × 109) |
| Unamended microcosms, day 21 | Glucose | 1.2 × 102 (25-5.5 × 102) | 1.0 × 108 (2.2 × 107-4.7 × 108) |
| Ethanol | 1.0 × 104 (2.2 × 103-4.7 × 104) | 4.5 × 107 (9.5 × 106-2.1 × 108) | |
| Homogenized FB094 sediment | Glucose | 2.1 × 103 (4.5 × 102-9.8 × 103) | 4.0 × 106 (8.6 × 105-1.9 × 107) |
| Ethanol | 1.5 × 104 (3.2 × 103-7.0 × 104) | 4.0 × 103 (8.6 × 102-1.9 × 104) | |
Values represent abundance in cells g−1 sediment wet weight, determined in three replicate MPN serial dilutions after 3 months of incubation at 30°C. Values in parentheses represent the range of the MPN values within 95% certainty. ND, not determined.
MPN estimates for FeRB grown with FeOOH as an electron acceptor.
MPN estimates for NRB grown with nitrate as the electron acceptor.
In all MPN series, FeRB were more abundant in the ethanol treatment than in the glucose treatment (Table 2). Fe(III)-reducing cultures grown on ethanol produced a black, magnetic precipitate, whereas those grown on glucose did not. During the nitrate (day 7) and iron reduction (day 21) phases of the incubation, the abundance of FeRB in the glucose and ethanol treatments was between 106 and 109 cells g−1 sediment (wet weight) (Table 2). The enumeration of FeRB in the unamended microcosm and homogenized FB094 sediment showed counts of less than 104 cells ml−1; however, the abundance was an order of magnitude lower in the unamended microcosm than in the core material (Table 2).
Phylogenetic analysis of clone libraries.
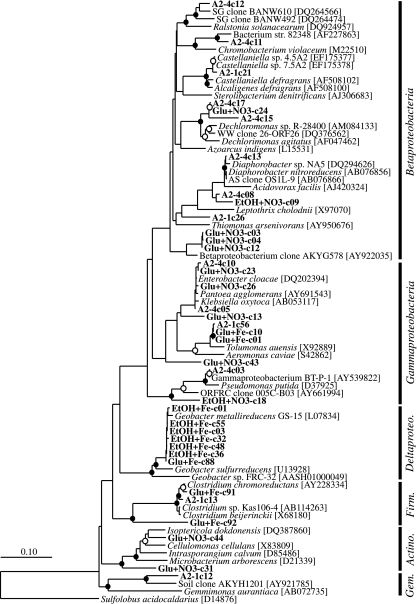
DNA extracts from microcosm samples were PCR amplified using Bacteria domain-specific primers and then pooled to construct a single clone library representing the community throughout the incubation. The highest positive dilutions from each MPN series were pooled for DNA extraction, PCR amplification, and clone library construction, resulting in four libraries for each set of cultivation conditions (ethanol and nitrate [EtOH+NO3−], glucose and nitrate [Glu+NO3−], ethanol and iron [EtOH+Fe], and glucose and iron [Glu+Fe]). A total of 74 microcosm- and 227 MPN-derived clones were screened using RFLP analysis, and a representative clone was sequenced for each phylotype. The 37 phylotypes sequenced from the microcosm- and MPN-derived clone libraries were most closely related to members of the Proteobacteria (classes Beta-, Delta- and Gammaproteobacteria), Gemmatimonadetes, Firmicutes, and Actinobacteria lineages.
Microcosm-derived SSU rRNA gene clones grouped into 14 different phylotypes, and the most frequently detected lineages were the Beta- (50% of total clones) and Gammaproteobacteria (44.6% of total clones), with the Gemmatimonadetes and Firmicutes phyla each comprising 2.7% of total clones (Table 3). Microcosm-derived clone A2-1c21 was most closely related (98% sequence similarity) to the Betaproteobacteria nitrate-reducing isolate Castellaniella sp. 4.5A2 (61) obtained from the ERSP-ORFRC site. The six phylotypes detected in the EtOH+Fe MPN-derived clone library were most closely related (99% identical) to Geobacter metallireducens (35) within the Deltaproteobacteria (Table 3 and Fig. 4). Four of the phylotypes detected in the Glu+Fe MPN-derived clone library grouped within the Firmicutes and Gammaproteobacteria, representing 2.9 and 95.7% of total clones, respectively. The remaining two phylotypes, Glu+Fe-c01 and Glu+Fe-c10, were related to the fermentative bacterium Tolumonas auensis (17), within the Aeromonadaceae family of the Gammaproteobacteria (Table 3 and Fig. 4).
TABLE 3.
Summary of phylogenetic affiliation and distribution of SSU rRNA gene clones from clone libraries of microcosm samples and MPN dilution series for FeRB and NRB
| Phylogenetic group | Clone designation | Nearest relative (GenBank accession no.) | % Similarity | Total clones | No. of related clones in sample from culture condition(s) of:
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Microcosm | EtOH+Fe | Glu+Fe | Glu+NO3 | EtOH+NO3 | |||||
| Betaproteobacteria | A2-4c13 | Diaphorobacter sp. strain NA5 (DQ294626) | 99 | 21 | 10 | 0 | 0 | 1 | 10 |
| A2-4c08 | AS clone OS1L-9 (AB076866) | 98 | 28 | 12 | 0 | 0 | 1 | 15 | |
| A2-4c12 | SG clone BANW610 (DQ264566) | 96 | 5 | 4 | 0 | 0 | 1 | 0 | |
| A2-4c11 | Bacterium strain 82348 (AF227863) | 96 | 3 | 3 | 0 | 0 | 0 | 0 | |
| A2-1c21 | Castellaniella sp. strain 4.5A2 (EF175377) | 98 | 2 | 2 | 0 | 0 | 0 | 0 | |
| A2-4c15 | Dechloromonas sp. strain R-28400 (AM084133) | 96 | 2 | 2 | 0 | 0 | 0 | 0 | |
| A2-4c17 | WW clone 26-ORF26 (DQ376562) | 93 | 2 | 2 | 0 | 0 | 0 | 0 | |
| A2-1c26 | SG clone BANW492 (DQ264474) | 89 | 2 | 2 | 0 | 0 | 0 | 0 | |
| Glu+NO3-c03 | Betaproteobacterium clone AKYG578 (AY922035) | 96 | 12 | 0 | 0 | 0 | 12 | 0 | |
| Glu+NO3-c24 | Dechloromonas sp. strain R-28400 | 96 | 2 | 0 | 0 | 0 | 2 | 0 | |
| Glu+NO3-c12 | Betaproteobacterium clone AKYG578 (AY922035) | 96 | 4 | 0 | 0 | 0 | 4 | 0 | |
| Glu+NO3-c04 | Betaproteobacterium clone AKYG578 (AY922035) | 96 | 2 | 0 | 0 | 0 | 2 | 0 | |
| EtOH+NO3-c09 | Diaphorobacter nitroreducens (AB076856) | 99 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Deltaproteobacteria | EtOH+Fe-c01 | Geobacter metallireducens GS-15 (CP000148) | 99 | 31 | 0 | 31 | 0 | 0 | 0 |
| EtOH+Fe-c03 | Geobacter metallireducens GS-15 (CP000148) | 99 | 33 | 0 | 33 | 0 | 0 | 0 | |
| EtOH+Fe-c32 | Geobacter metallireducens GS-15 (CP000148) | 99 | 1 | 0 | 1 | 0 | 0 | 0 | |
| EtOH+Fe-c36 | Geobacter metallireducens GS-15 (CP000148) | 99 | 1 | 0 | 1 | 0 | 0 | 0 | |
| EtOH+Fe-c48 | Geobacter metallireducens GS-15 (CP000148) | 99 | 1 | 0 | 1 | 0 | 0 | 0 | |
| EtOH+Fe-c55 | Geobacter metallireducens GS-15 (CP000148) | 99 | 1 | 0 | 1 | 0 | 0 | 0 | |
| Glu+Fe-c88 | Geobacter metallireducens GS-15 (CP000148) | 99 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Gammaproteobacteria | A2-4c05 | Enterobacter cloacae strain B5 (DQ202394) | 97 | 16 | 16 | 0 | 0 | 0 | 0 |
| Glu+Fe-c10 | Tolumonas auensis (X92889) | 96 | 1 | 0 | 0 | 1 | 0 | 0 | |
| A2-4c10 | Enterobacter sp. strain MACL08B (EF198245) | 99 | 11 | 8 | 0 | 0 | 3 | 0 | |
| A2-1c56 | Tolumonas auensis (X92889) | 96 | 1 | 1 | 0 | 0 | 0 | 0 | |
| A2-4c03 | Gammaproteobacterium BT-P-1 (AY539822) | 98 | 12 | 8 | 0 | 0 | 0 | 4 | |
| Glu+NO3-c43 | Klebsiella oxytoca (AB053117) | 99 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Glu+NO3-c23 | Enterobacter cloacae strain B5 (DQ202394) | 99 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Glu+NO3-c26 | Enterobacter cloacae strain B5 (DQ202394) | 99 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Glu+NO3-c13 | Pantoea agglomerans (AY691543) | 99 | 1 | 0 | 0 | 0 | 1 | 0 | |
| EtOH+NO3-c18 | NABIR-FRC clone 005C-B03 (AY661994) | 96 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Glu+Fe-c01 | Tolumonas auensis (X92889) | 96 | 83 | 0 | 0 | 66 | 17 | 0 | |
| Firmicutes | Glu+Fe-c92 | Clostridium sp. strain Kas106-4 (AB114263) | 97 | 1 | 0 | 0 | 1 | 0 | 0 |
| Glu+Fe-c91 | Clostridium chromoreductans (AY228334) | 97 | 1 | 0 | 0 | 1 | 0 | 0 | |
| A2-1c13 | Clostridium beijerinckii (X68180) | 98 | 2 | 2 | 0 | 0 | 0 | 0 | |
| Gemmatimonadetes | A2-1c12 | Soil clone AKYH1201 (AY921785) | 93 | 11 | 2 | 0 | 0 | 0 | 9 |
| Actinobacteria | Glu+NO3-c44 | Isoptericola dokdonensis strain DS-3 (DQ387860) | 95 | 1 | 0 | 0 | 0 | 1 | 0 |
| Glu+NO3-c31 | Isoptericola dokdonensis strain DS-3 (DQ387860) | 95 | 1 | 0 | 0 | 0 | 1 | 0 | |
FIG. 4.
Phylogenetic tree of SSU rRNA gene clone sequences (indicated by boldface type) and environmental clone reference sequences as determined by neighbor-joining methods incorporating Jukes-Cantor distance correction. Sulfolobus acidocaldarius was used as the outgroup. One thousand bootstrap analyses were conducted, and branch points supported by bootstrap resampling are indicated by filled circles (bootstrap values of >90%) and open circles (bootstrap values of >75%). The scale bar indicates 0.1 change per nucleotide position. Gem., Gemmatimonadetes; Actino., Actinobacteria; Firm., Firmicutes.
In nitrate-reducing MPN-derived clone libraries, the most frequently detected lineages were the Beta (47.9 and 65.0% of clones cultivated with Glu+NO3− and EtOH+NO3−, respectively)- and Gammaproteobacteria (50.0 and 12.5% of clones cultivated with Glu+NO3−and EtOH+NO3−, respectively) (Table 3 and Fig. 4). The seven Betaproteobacteria-related phylotypes grouped within the Alcaligenaceae, Neisseriaceae, Rhodocyclaceae, Comamonadaceae, and Oxalobacteraceae families. Phylotype EtOH+NO3−-c09 had 99% sequence similarity to the Comamonadaceae nitrate-reducing isolate Diaphorobacter nitroreducens (28), whereas phylotype Glu+NO3−-c24 had 96% sequence similarity to the Rhodocyclaceae isolate Dechloromonas sp. strain R-28400 (20) (Table 3 and Fig. 4). The eight Gammaproteobacteria-related phylotypes grouped in the Enterobacteriaceae, Pseudomonadaceae, and Aeromonadaceae families. Enterobacteriaceae-related phylotypes included Glu+NO3−-c13, which was 99% similar to the metal- and nitrate-reducing isolate Pantoea agglomerans (18); phylotypes Glu+NO3−-c23 and Glu+NO3−-c26, with 99% sequence similarity to Enterobacter cloacae (GenBank accession number DQ202394); and Glu+NO3−-c43, which was related to Klebsiella oxytoca (94% sequence similarity) (GenBank accession number AB053117) (Fig. 4). Phylotypes related to the Pseudomonadaceae family included EtOH-NO3-c18, which was most closely related to the ERSP-ORFRC clone 005C-B03 (15) and phylotype A2-4c03, related to gammaproteobacterium BT-P-1 (GenBank accession number AY539822). The two phylotypes related to the Actinobacteria phylum represented 2.1% of total clones in the Glu+NO3− library, whereas the remaining 22.5% of the EtOH+NO3− phylotypes were related to the Gemmatimonadetes phylum (Table 3).
TRFLP analysis of cultivated microorganisms.
SSU rRNA gene sequences derived from MPN and microcosm clone libraries constructed in this study were added to a database of all SSU rRNA genes generated from previous work at the ORFRC site. All sequences containing the 27F forward primer were digested in silico using the MnlI recognition site to determine the size of the 5′-terminal fragment (TRF). Currently, the ORFRC sequence database consists of >2,000 SSU rRNA gene sequences and >850 of the sequences are of the appropriate size to generate TRF sizes using in silico digestion. The TRF length was then matched to the peak sizes from TRFLP profiles to identify the phylotypes present in each MPN sample.
The TRFLP peaks detected in nitrate-reducing enrichment cultures were consistent with the TRF sizes expected for microorganisms belonging to the Proteobacteria (classes Beta-, Delta-, and Gammaproteobacteria), Chloroflexi, Bacteroidetes, Gemmatimonadetes, Firmicutes, and Actinobacteria phyla (Table 4). It is interesting to note that TRF of sequences from the Gemmatimonadetes phylum were only detected in the NRB cultures with glucose supplemented as an electron donor. TRFLP profiles from iron-reducing enrichments revealed that the community was comprised of members of the Proteobacteria (classes Alpha-, Beta-, Delta-, and Gammaproteobacteria), Chloroflexi, Bacteroidetes, Firmicutes, and Actinobacteria phyla (Table 4). Members of the Alphaproteobacteria were only detected in FeRB cultures supplemented with either glucose or ethanol as the electron donor. TRFLP peaks identified to the Betaproteobacteria (families Oxalobacteraceae, Comamonadaceae, and Rhodocyclaceae) were observed in ethanol- and glucose-amended NRB and FeRB MPN cultures (Table 4). Peaks observed in both NRB and FeRB cultures matched to the Oxalobacteraceae-related microcosm-derived clone A2-4c12 and the Dechloromonas-related phylotypes A2-4c15 and Glu+NO3−-c24 (Table 3). The Comamonadaceae family was unique to FeRB cultures, and peaks matched the TRF size of the Diaphorobacter-related microcosm-derived clone A2-1c26 (Table 3). Peaks identified to the Gammaproteobacteria in the NRB and FeRB MPN cultures (Table 4) matched in silico digests of microcosm- and MPN-derived clones related to members of the Enterobacteriaceae, Pseudomonadaceae, and Xanthomonadaceae families (Fig. 4). These peaks represented a large percentage of the peak area (∼15 to 34%) in glucose cultures inoculated from the unamended microcosms and homogenized sediment samples. Deltaproteobacteria-related TRFLP peaks were predominantly detected in FeRB cultures, with the highest abundance in ethanol-amended cultures, and peaks were identified as Anaeromyxobacter and Geobacter genera (Table 4). In NRB and FeRB MPN series, peaks were related to the Actinobacteria, with the most frequently detected peak related to the genus Arthrobacter in the Micrococcaceae family (Table 4). Firmicutes-identified peaks, representing the Clostridiaceae and Staphylococcaceae families, were found in all culture conditions, and the abundance of this group varied throughout the incubation depending on the electron donor.
TABLE 4.
Phylogenetic affiliation of identified TRFLP peak area in the highest positive dilutions for MPN dilution series for FeRB and NRB
| Phylogenetic group | % Total peak area for MPN dilution series with culture conditions ofa:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EtOH+NO3
|
Glu+NO3
|
EtOH+Fe
|
Glu+Fe
|
|||||||||||||
| Day 7 | Day 21 | Unam | Sed | Day 7 | Day 21 | Unam | Sed | Day 7 | Day 21 | Unam | Sed | Day 7 | Day 21 | Unam | Sed | |
| Alphaproteobacteria | - | - | - | - | - | - | - | - | 6.9 | 1.6 | - | - | - | 2.2 | - | 1.9 |
| Betaproteobacteria | 2.3 | - | - | 25.9 | 13 | 1.2 | - | 17.8 | 1.6 | 7.6 | 7.6 | 50.4 | 3.5 | 17.8 | 2.7 | 5.8 |
| Deltaproteobacteria | 1.8 | - | - | - | - | - | - | - | 1.1 | 44.4 | 4.9 | 21.8 | - | 16.6 | 1.5 | 2.6 |
| Gammaproteobacteria | - | 19.2 | 24.1 | - | 16.2 | 2.1 | 32.6 | 19.3 | 1.6 | - | 1.8 | 1.1 | 1.4 | - | 15.3 | 33.5 |
| Firmicutes | 4.8 | - | - | - | 5.1 | 10.5 | 1.8 | 19.3 | 43.4 | - | - | 11.2 | 21.6 | 2.2 | 5.8 | 8.3 |
| Actinobacteria | 25.5 | 7.4 | - | - | 1.6 | - | 21.9 | - | - | - | 9.4 | - | 8.8 | 3.5 | 5.9 | 13.7 |
| Chloroflexi | 4.1 | 5.8 | 7.8 | - | 2.1 | - | 5.7 | - | - | - | 1.2 | - | - | - | - | - |
| Gemmatimonadetes | - | - | - | - | 23.5 | - | 24.2 | - | - | - | - | - | - | - | - | - |
| Bacteroidetes | 2.9 | 4.3 | 5.7 | 26.5 | 2.1 | - | 4.2 | - | - | - | - | - | 4.6 | 3.3 | - | - |
TRFLP peaks were identified by in silico digests with MnlI of ORFRC clone sequences. Unam, unamended control microcosm; Sed, homogenized FB094 sediment; -, none detected.
Quantification of functional gene expression.
The metabolic activity of FeRB was related to the levels of mRNA transcripts for the Geobacteraceae-specific citrate synthase (gltA) gene (21), a key gene involved in the incorporation of acetate into the tricarboxylic acid cycle. The levels of gltA transcription correlated with the depletion of supplemental electron donors and the accumulation of acetate in sediment microcosms after 27 days of incubation. The number of gltA transcripts was higher in the glucose-amended microcosms (7.1 × 106 ± [mean ± standard deviation] 1.5 × 106 gltA copies μg−1 mRNA) than in the ethanol-amended microcosms (4.8 × 106 ± 1.5 × 106 gltA copies μg−1 mRNA). The levels of gltA transcripts were 5.3 × 104 ± 5.1 × 103 gltA copies μg−1 mRNA in the unamended microcosms. Phylogenetic analysis of partial gltA cDNA clone sequences indicated that the sequences were most closely related to the members of the Geobacteraceae family Geobacter bemidjiensis, isolated from petroleum-contaminated aquifer sediment (47); Geobacter metallireducens, isolated from freshwater sediments (36); Geobacter uraniireducens, isolated from uranium-contaminated sediments in Rifle, CO (5); and Geobacter strain FRC-32, isolated from the ORFRC site (data not shown).
DISCUSSION
In situ uranium bioremediation is focused on biostimulating indigenous microorganisms through a combination of pH neutralization and the addition of large amounts of electron donor. Glucose and ethanol have been previously demonstrated to stimulate microbial U(VI) reduction in the field (24, 44, 69) and are the favored electron donors for bioremediation strategies. However, few data on the dynamics of microbial populations capable of U(VI) reduction in subsurface sediments are available, and the differences in the microbial community dynamics for proposed electron donors have not been explored. Microbial community dynamics were investigated in microcosms constructed with uranium-contaminated sediment and groundwater from the ORFRC, unamended and amended separately with glucose or ethanol. We observed distinct differences in the structure and function of microbial communities capable of U(VI) reduction that were stimulated with glucose or ethanol. Ethanol promoted more-complete reduction of U(VI) than the glucose treatment, and members of the Proteobacteria, Firmicutes, and Actinobacteria played a significant role in mediating processes important to the immobilization of U(VI) in contaminated sediments.
The activity of microbial communities in ORFRC subsurface sediments is limited by carbon availability, and metal reduction is inhibited by the high concentrations of nitrate cocontaminants (6, 13, 25, 44-46, 49). Consistent with the results of prior studies, the addition of supplemental electron donors glucose and ethanol to microcosms successfully stimulated microbial activity. Metal reduction was not detectable until nitrate was largely removed, and U(VI) concentrations increased during nitrate reduction, likely due to abiotic oxidation of U(IV) by nitrate reduction intermediates (16, 57-59). FeRB were also affected by the high concentrations of nitrate (15 mM) present in ORFRC groundwater, as seen in the lower abundance of this group in nonbiostimulated treatments where nitrate had not been depleted.
In this study, we provide evidence that the choice of supplemental electron donors for biostimulation can have a major impact on the activity and functional diversity of subsurface microbial communities capable of U(VI) reduction. The succession of TEAPs, important for uranium reduction, differed significantly between glucose- and ethanol-amended microcosms. Regardless of the treatment, the majority of the electron donor was incompletely oxidized in the microcosms, with acetate accumulating as the primary end product. In contrast to the ethanol microcosms, we detected transient products of carbon oxidation in the glucose treatment, indicating that in addition to respiratory pathways, a portion of the supplemental glucose was oxidized through fermentation. Numerous pathways of glucose fermentation exist that can generate acetate as an end product and can support a more-diverse microbial community. Indeed, we observed that the abundance and diversity of microorganisms were higher in glucose-amended microcosms and that U(VI) and Fe(III) were reduced concurrently before the complete removal of nitrate. We hypothesize that the addition of glucose stimulated a mixed population of NRB and fermentative microorganisms that shunt electrons to metals. NRB and FeRB cultivated from microcosms amended with glucose were predominantly related to the Proteobacteria, Actinobacteria, and Firmicutes phyla. Taxa within these groups, such as Tolumonas spp. and Clostridium spp., have been shown to ferment glucose to acetate, and Clostridium spp. have been shown to shunt electrons to Fe(III). Evidence has also been presented for the functional diversity of Arthrobacter spp. (Actinobacteria) and Pantoea agglomerans (Gammaproteobacteria). Arthrobacter spp. utilize glucose, can reduce nitrate (14), and bioprecipitate U(VI) (42), whereas P. agglomerans can couple growth to the reduction of nitrate and metals (18). The overlap in potential function and detection of these microbial groups could account for the observed overlap in TEAPs and supports our inference that U(VI) and Fe(III) reduction was due to the activity of both dissimilatory metal-reducing and fermentative bacteria. In addition, members of the Gemmatimonadetes phylum comprised a large portion of NRB in the glucose treatment and, currently, little is known about the functional role of this group. This phylum is represented by a single isolated strain and uncultured representatives from multiple terrestrial and aquatic habitats (71). Therefore, future work should explore the role of this group in the bioremediation of nitrate- and uranium-contaminated subsurface environments.
The addition of ethanol as a supplemental electron donor promoted rapid and effective U(VI) reduction and selected for a population of anaerobic respiratory microorganisms within the Proteobacteria, Firmicutes, and Actinobacteria phyla. In the ethanol treatment, nitrate was fully consumed 2 days earlier than in the glucose treatment, in conjunction with incomplete oxidation of ethanol. The removal of nitrate allowed for a shift to U(VI) reduction that overlapped with sulfate reduction. Concurrent reduction of sulfate and U(VI) prior to the onset of Fe(III) reduction has been observed in biostimulation studies at the ORFRC (44, 49, 69), and laboratory studies of SRB have shown that the rate of uranium reduction is enhanced in the presence of sulfate (55, 62). Therefore, we propose that the overlap of U(VI) and sulfate reduction is due to the activity of SRB that couple the oxidation of ethanol to U(VI) reduction. This is based on the assumption that all U(VI) reduction observed was due to biotic processes; however, we cannot rule out the possibility that abiotic reduction was occurring due to the accumulation of sulfides or Fe(II) from TEAPs. The results of previous studies have shown that abiotic reduction of U(VI) is much slower than the enzymatic reaction (1, 37, 38), and therefore, it is unlikely that the U(VI) reduction observed in the microcosms is solely due to abiotic reactions. Although we did not enumerate SRB, it is reasonable to assume based on the initial nitrate concentration (15 mM) that SRB were in low abundance and out-competed by NRB. The concurrence of sulfate and uranium reduction coupled to ethanol oxidation to acetate emphasizes the potential importance of SRB as a key functional group in ethanol-biostimulated sediments.
The taxa detected in the ethanol-amended treatment were related to known dissimilatory NRB and FeRB in the Proteobacteria, Actinobacteria, and Firmicutes phyla. A higher abundance of Actinobacteria and Firmicutes phyla was observed during the early part of the ethanol microcosm incubation, which suggests that these groups are actively involved in nitrate reduction and subsequent uranium reduction. In ethanol-amended microcosms, the NRB detected were primarily related to Gamma- and Betaproteobacteria taxa that are known to reduce nitrate and metals. The Gammaproteobacteria genus Pseudomonas was detected throughout the time points of our study, and Pseudomonas spp. grow with nitrate as an electron acceptor and enzymatically catalyze the reduction of U(VI) to U(IV) (18, 66). The Betaproteobacteria genera Dechloromonas, Diaphorobacter, Acidovorax, and Castellaniella were abundant during the time points of shifting TEAPs in our microcosms, and these groups were detected previously in ORFRC subsurface sediments (2, 15, 48, 61). Taxa within the Dechloromonas and Diaphorobacter genera reduce nitrate (22, 28, 29) and are likely responsible for mediating nitrate reduction in the carbon-amended microcosms. Dechloromonas spp. have been shown to reduce sulfate (22), and we suggest that this group is responsible for the overlap in sulfate and U(VI) reduction observed in the ethanol-amended microcosms. The denitrifying bacteria of the genera Acidovorax and Castellaniella have been isolated from the ORFRC site (49, 61), and the Acidovorax isolate can reduce U(VI) (49). U(VI) and nitrate reduction can also be performed by the dissimilatory FeRB groups Anaeromyxobacter and Geobacter (Deltaproteobacteria). The highest abundance of FeRB was observed in the ethanol-amended treatment, and the microbial community was dominated by Deltaproteobacteria taxa. Environmental clone sequences closely related to members of the Deltaproteobacteria were shown to increase after biostimulation of the ORFRC subsurface with glucose or ethanol (24, 48, 51). The detection of Proteobacteria, Actinobacteria, and Firmicutes taxa in cultivation-independent studies in groundwater and sediments at the ORFRC (2, 24, 44, 48, 49, 51, 68, 69) led previous groups to infer their physiological role in bioremediation. By coupling geochemical analysis with community characterization, we provide further evidence that these taxa are active during bioremediation and are likely involved in nitrate, U(VI), and Fe(III) reduction.
Iron(III)-containing minerals are considered important electron acceptors for microbial respiration and can affect the fate and transport of uranium contaminants by acting as sorbents of U(VI) (7, 23, 53). Analysis of iron mineralogy indicated that phyllosilicates and goethite were reduced only in the carbon-amended microcosms, and phyllosilicate clay minerals were the predominant form of Fe(III)-containing minerals metabolized. This is consistent with the observations that pure cultures of FeRB readily respire model Fe(III)-rich phyllosilicate clays (30) and that the structural Fe(III) in phyllosilicates is likely to be reduced prior to Fe (oxyhydr)oxides. Clay minerals are highly reactive soil components and account for a large fraction of the Fe-containing minerals available for microbial respiration in subsurface sediments. Here we provide evidence that FeRB from uranium-contaminated subsurface sediments are capable of coupling the reduction of Fe(III)-rich clays to the oxidation of acetate which has accumulated as an end product of incomplete glucose and ethanol oxidation. Acetate supports the growth of Geobacteraceae species in enrichment cultures supplemented with Fe(III)-rich clay minerals (30), and the oxidation of acetate in this group proceeds through a complete citric acid cycle which is controlled by the citrate synthase protein (21). By quantifying the levels of citrate synthase (gltA) transcripts, we demonstrated a stimulation of Geobacteraceae activity during metal reduction in carbon-amended microcosms, with the highest expression observed in the glucose treatment. The active FeRB were most closely related to Geobacteraceae members cultivated from contaminated subsurface environments (5, 36, 47, 65) and shown to be important members of FeRB consortia in sediments (12, 36, 54, 60). By targeting a metabolic gene unique to the Geobacteraceae family, we were able to directly link the transformation of specific Fe(III) minerals with the oxidation of acetate by FeRB. We reiterate that Fe(III)-rich clay minerals comprise an important electron acceptor available for dissimilatory metal-reducing metabolism in terrestrial subsurface sediments. The presence and activity of FeRB are important to the long-term stability of U(IV) in the subsurface due to the potential for U(IV) oxidation by Fe(III) minerals (55, 58, 59).
Ethanol and glucose have been considered the primary electron donors to stimulate the reductive immobilization of U(VI) in bioremediation practices at the ORFRC. The results of this study revealed that the addition of ethanol stimulated rapid nitrate reduction followed by the complete reduction of U(VI) concurrent with sulfate reduction. We therefore propose that ethanol is a more-appropriate electron donor than glucose for future in situ bioremediation practices and that SRB are most likely a critical microbial functional group involved in the reductive immobilization of U(VI). The accumulation of acetate, followed by slow oxidation, may serve as a long-term electron donor to support the maintenance of a reduced subsurface environment, thereby promoting the insolubility of U(IV) species. The microbial community within the ethanol treatment was composed of members of the Proteobacteria, Actinobacteria, and Firmicutes, including phylogenetic groups not previously associated with uranium bioremediation, e.g., Arthrobacter and Dechloromonas. Biostimulation promoted the reduction of Fe(III)-rich clay materials and the transcription of citrate synthase genes unique to FeRB within the family Geobacteraceae. Members of the Geobacteraceae family are substantial contributors to metal reduction in uranium-contaminated subsurface sediments and are important for the long-term stability of reduced uranium. We speculate that these groups are well adapted for the extremes of contaminated subsurface sediments and are good targets for effective in situ uranium bioremediation.
Acknowledgments
This research was supported by the Office of Science (BER), U.S. Department of Energy grant no. DE-FG02-07ER64373, and by the Integrated Field-Scale Subsurface Research Challenge at Oak Ridge, operated by the Environmental Sciences Division, Oak Ridge National Laboratory (ORNL), under U.S. Department of Energy contract no. DE-AC05-00OR22725. J.E.K. was partially supported by a fellowship from the Hanse Wissenschaftskolleg and the Max Planck Society during the preparation of the manuscript.
We thank Lisa Fagan for sediment sampling and collaboration, Andrew Madden for providing the U(VI) analyses, and Lora McGuinness, Jonathon Delgardio, and Nicole Evans for their assistance with microcosm analysis.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Abdelouas, A., W. Lutze, and H. E. Nuttall. 1999. Uranium contamination in the subsurface; characterization and remediation. Rev. Mineral. Geochem. 38:433-473. [Google Scholar]
- 2.Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. [DOI] [PubMed] [Google Scholar]
- 3.Alef, K. 1991. Methodenhandbuch Bodenmikrobiologie: Aktivitaten, Biomasse, Differenzierung, p. 44-49. Ecomed, Landsberg/Lech, Germany.
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, S. C. 2001. Waste characteristics of the former S-3 ponds and outline of uranium chemistry relevant to NABIR Field Research Center studies, report no. ORNL/TM-2001/27. NABIR Field Research Center, Oak Ridge, TN.
- 7.Catalano, J. G., and J. G. E. Brown. 2005. Uranyl adsorption onto montmorillonite: evaluation of binding sites and carbonate complexation. Geochim. Cosmochim. Acta 69:2995. [Google Scholar]
- 8.Cataldo, D. A., L. E. Schrader, and V. L. Youngs. 1974. Analysis by digestion and colorimetric assay of total nitrogen in plant tissues high in nitrate. Crop Sci. 14:854-856. [Google Scholar]
- 9.Chin, K.-J., A. Esteve-Nunez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, K.-J., M. Sharma, L. Russell, K. O'Neill, and D. Lovley. 2007. Quantifying expression of a dissimilatory (bi)sulfite reductase gene in petroleum-contaminated marine harbor sediments. Microb. Ecol. 55:489-499. [DOI] [PubMed] [Google Scholar]
- 11.DiChristina, T. J. 1992. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J. Bacteriol. 174:1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duff, M. C., J. U. Coughlin, and D. B. Hunter. 2002. Uranium co-precipitation with iron oxide minerals. Geochim. Cosmochim. Acta 66:3533-3547. [Google Scholar]
- 13.Edwards, L., K. Kusel, H. Drake, and J. E. Kostka. 2007. Electron flow in acidic subsurface sediments co-contaminated with nitrate and uranium. Geochim. Cosmochim. Acta 71:643-654. [Google Scholar]
- 14.Eschbach, M., H. Mobitz, A. Rompf, and D. Jahn. 2003. Members of the genus Arthrobacter grow anaerobically using nitrate ammonification and fermentative processes: anaerobic adaptation of aerobic bacteria abundant in soil. FEMS Microbiol. Lett. 223:227-230. [DOI] [PubMed] [Google Scholar]
- 15.Fields, M. W., T. F. Yan, S. K. Rhee, S. L. Carroll, P. M. Jardine, D. B. Watson, C. S. Criddle, and J. Z. Zhou. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417-428. [DOI] [PubMed] [Google Scholar]
- 16.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 17.Fischer-Romero, C., B. J. Tindall, and F. Juttner. 1996. Tolumonas auensis gen. nov., sp. nov., a toluene-producing bacterium from anoxic sediments of a freshwater lake. Int. J. Syst. Bacteriol. 46:183-188. [DOI] [PubMed] [Google Scholar]
- 18.Francis, C. A., A. Y. Obraztsova, and B. M. Tebo. 2000. Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl. Environ. Microbiol. 66:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredrickson, J. K., and T. C. Onstott. 2001. Biogeochemical and geological significance of subsurface microbiology, p. 341. In J. K. Fredrickson and M. Fletcher (ed.), Subsurface microbiology and biogeochemistry. Wiley-Liss, New York, NY.
- 20.Heylen, K., B. Vanparys, L. Wittebolle, W. Verstraete, N. Boon, and P. De Vos. 2006. Cultivation of denitrifying bacteria: optimization of isolation conditions and diversity study. Appl. Environ. Microbiol. 72:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, D. E., K. P. Nevin, R. A. O'Neil, J. E. Ward, L. A. Adams, T. L. Woodard, H. A. Vrionis, and D. R. Lovley. 2005. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71:6870-6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn, M. A., J. Ihssen, C. Matthies, A. Schramm, G. Acker, and H. L. Drake. 2005. Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov. and Paenibacillus terrae strain MH72, N2O-producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int. J. Syst. Evol. Microbiol. 55:1255-1265. [DOI] [PubMed] [Google Scholar]
- 23.Hsi, C. K. D., and D. Langmuir. 1985. Adsorption of uranyl onto ferric oxyhydroxides: application of the surface complexation site-binding model. Geochim. Cosmochim. Acta 49:1931-1941. [Google Scholar]
- 24.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 25.Jardine, P. M., T. L. Melhorn, Y. Roh, and W. E. Sanford. 2003. Hydrological and geochemical processes controlling the fate and transport of contaminants in fractured bedrock, p. 1-24. In H. M. Selim and W. L. Kingery (ed.), Geochemical and hydrological reactivity of heavy metals in soils. Lewis Publishers, New York, NY.
- 26.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society of Microbiology, Washington, DC.
- 27.Reference deleted.
- 28.Khan, S. T., Y. Horiba, M. Yamamoto, and A. Hiraishi. 2002. Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl. Environ. Microbiol. 68:3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khardenavis, A., A. Kapley, and H. Purohit. 2007. Simultaneous nitrification and denitrification by diverse Diaphorobacter sp. Appl. Microbiol. Biotechnol. 77:403. [DOI] [PubMed] [Google Scholar]
- 30.Kostka, J. E., D. D. Dalton, H. Skelton, S. Dollhopf, and J. W. Stucki. 2002. Growth of iron(III)-reducing bacteria on clay minerals as the sole electron acceptor and comparison of growth yields on a variety of oxidized iron forms. Appl. Environ. Microbiol. 68:6256-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostka, J. E., and G. W. Luther. 1994. Partitioning and speciation of solid-phase iron in salt-marsh sediments. Geochim. Cosmochim. Acta 58:1701-1710. [Google Scholar]
- 32.Kusel, K., and H. L. Drake. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovell, C. R., and Y. Piceno. 1994. Purification of DNA from estuarine sediments. J. Microbiol. Methods 20:161. [Google Scholar]
- 34.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 36.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov sp. nov, a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 37.Lovley, D. R., and E. J. P. Phillips. 1992. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26:2228-2234. [Google Scholar]
- 38.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo, J., O. A. Cirpka, W. M. Wu, M. N. Fienen, P. M. Jardine, T. L. Mehlhorn, D. B. Watson, C. S. Criddle, and P. K. Kitanidis. 2005. Mass-transfer limitations for nitrate removal in a uranium-contaminated aquifer. Environ. Sci. Technol. 39:8453-8459. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Martinez, R. J., M. J. Beazley, M. Taillefert, A. K. Arakaki, J. Skolnick, and P. A. Sobecky. 2007. Aerobic uranium (VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ. Microbiol. 9:3122-3133. [DOI] [PubMed] [Google Scholar]
- 43.McGuinness, L. M., M. Salganik, L. Vega, K. D. Pickering, and L. J. Kerkhof. 2006. Replicability of bacterial communities in denitrifying bioreactors as measured by PCR/T-RFLP analysis. Environ. Sci. Technol. 40:509-515. [DOI] [PubMed] [Google Scholar]
- 44.Michalsen, M. M., B. A. Goodman, S. D. Kelly, K. M. Kemner, J. P. McKinley, J. W. Stucki, and J. D. Istok. 2006. Uranium and technetium bio-immobilization in intermediate-scale physical models of an in situ bio-barrier. Environ. Sci. Technol. 40:7048-7053. [DOI] [PubMed] [Google Scholar]
- 45.Moon, J. W., Y. Roh, T. J. Phelps, D. H. Phillips, D. B. Watson, Y. J. Kim, and S. C. Brooks. 2006. Physicochemical and mineralogical characterization of soil-saprolite cores from a field research site, Tennessee. J. Environ. Qual. 35:1731-1741. [DOI] [PubMed] [Google Scholar]
- 46.Natural and Accelerated Bioremediation Research program (NABIR). 2003. Bioremediation of metals and radionuclides…what it is and how it works. In T. C. Hazen, S. M. Benson, F. B. Metting, B. Faison, A. C. Palmisano, and J. McCullough (ed.), NABIR primer, 2nd ed. Lawrence Berkeley National Laboratory, Berkeley, CA.
- 47.Nevin, K. P., D. E. Holmes, T. L. Woodard, E. S. Hinlein, D. W. Ostendorf, and D. R. Lovley. 2005. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int. J. Syst. Evol. Microbiol. 55:1667-1674. [DOI] [PubMed] [Google Scholar]
- 48.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nyman, J. L., T. L. Marsh, M. A. Ginder-Vogel, M. Gentile, S. Fendorf, and C. Criddle. 2006. Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation 17:303-316. [DOI] [PubMed] [Google Scholar]
- 50.Nyman, J. L., H. I. Wu, M. E. Gentile, P. K. Kitanidis, and C. S. Criddle. 2007. Inhibition of a U(VI)- and sulfate-reducing consortia by U(VI). Environ. Sci. Technol. 41:6528-6533. [DOI] [PubMed] [Google Scholar]
- 51.Peacock, A. D., Y. J. Chang, J. D. Istok, L. Krumholz, R. Geyer, B. Kinsall, D. Watson, K. L. Sublette, and D. C. White. 2004. Utilization of microbial biofilms as monitors of bioremediation. Microb. Ecol. 47:284-292. [DOI] [PubMed] [Google Scholar]
- 52.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Read, D., T. A. Lawless, R. J. Sims, and K. R. Butter. 1993. Uranium migration through intact sandstone cores. J. Contam. Hydrol. 13:277-289. [Google Scholar]
- 54.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sani, R. K., B. M. Peyton, J. E. Amonette, and G. G. Geesey. 2004. Reduction of uranium(VI) under sulfate-reducing conditions in the presence of Fe(III)-(hydr)oxides. Geochim. Cosmochim. Acta 68:2639-2648. [Google Scholar]
- 55a.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senko, J. M., T. A. Dewers, and L. R. Krumholz. 2005. Effect of oxidation rate and Fe(II) state on microbial nitrate-dependent Fe(III) mineral formation. Appl. Environ. Microbiol. 71:7172-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senko, J. M., J. D. Istok, J. M. Suflita, and L. R. Krumholz. 2002. In-situ evidence for uranium immobilization and remobilization. Environ. Sci. Technol. 36:1491-1496. [DOI] [PubMed] [Google Scholar]
- 58.Senko, J. M., Y. Mohamed, T. A. Dewers, and L. R. Krumholz. 2005. Role for Fe(III) minerals in nitrate-dependent microbial U(IV) oxidation. Environ. Sci. Technol. 39:2529-2536. [DOI] [PubMed] [Google Scholar]
- 59.Senko, J. M., J. M. Suflita, and L. R. Krumholz. 2005. Geochemical controls on microbial nitrate-dependent U(IV) oxidation. Geomicrobiol. J. 22:371-378. [Google Scholar]
- 60.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153. [DOI] [PubMed] [Google Scholar]
- 61.Spain, A. M., A. D. Peacock, J. D. Istok, M. S. Elshahed, F. Z. Najar, B. A. Roe, D. C. White, and L. R. Krumholz. 2007. Identification and isolation of a Castellaniella species important during biostimulation of an acidic nitrate- and uranium-contaminated aquifer. Appl. Environ. Microbiol. 73:4892-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spear, J. R., L. A. Figueroa, and B. D. Honeyman. 2000. Modeling reduction of uranium U(VI) under variable sulfate concentrations by sulfate-reducing bacteria. Appl. Environ. Microbiol. 66:3711-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stucki, J. W., K. Lee, B. A. Goodman, and J. Kostka. 2007. Effects of in situ biostimulation on iron mineral speciation in sub-surface soil. Geochim. Cosmochim. Acta 71:835-843. [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vrionis, H. A., R. T. Anderson, I. Ortiz-Bernad, K. R. O'Neill, C. T. Resch, A. D. Peacock, R. Dayvault, D. C. White, P. E. Long, and D. R. Lovley. 2005. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71:6308-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wall, J. D., and L. R. Krumholz. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149-166. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, W. M., J. Carley, M. Fienen, T. Mehlhorn, K. Lowe, J. Nyman, J. Luo, M. E. Gentile, R. Rajan, D. Wagner, R. F. Hickey, B. H. Gu, D. Watson, O. A. Cirpka, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ. Sci. Technol. 40:3978-3985. [DOI] [PubMed] [Google Scholar]
- 69.Wu, W. M., J. Carley, T. Gentry, M. A. Ginder-Vogel, M. Fienen, T. Mehlhorn, H. Yan, S. Caroll, M. N. Pace, J. Nyman, J. Luo, M. E. Gentile, M. W. Fields, R. F. Hickey, B. H. Gu, D. Watson, O. A. Cirpka, J. Z. Zhou, S. Fendorf, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986-3995. [DOI] [PubMed] [Google Scholar]
- 70.Yan, T. F., M. W. Fields, L. Y. Wu, Y. G. Zu, J. M. Tiedje, and J. Z. Zhou. 2003. Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 5:13-24. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating microorganism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]