Abstract
Rhabdoviruses are important pathogens of humans, livestock, and plants that are often vectored by insects. Rhabdovirus particles have a characteristic bullet shape with a lipid envelope and surface-exposed transmembrane glycoproteins. Sigma virus (SIGMAV) is a member of the Rhabdoviridae and is a naturally occurring disease agent of Drosophila melanogaster. The infection is maintained in Drosophila populations through vertical transmission via germ cells. We report here the nature of the Drosophila innate immune response to SIGMAV infection as revealed by quantitative reverse transcription-PCR analysis of differentially expressed genes identified by microarray analysis. We have also compared and contrasted the immune response of the host with respect to two nonenveloped viruses, Drosophila C virus (DCV) and Drosophila X virus (DXV). We determined that SIGMAV infection upregulates expression of the peptidoglycan receptor protein genes PGRP-SB1 and PGRP-SD and the antimicrobial peptide (AMP) genes Diptericin-A, Attacin-A, Attacin-B, Cecropin-A1, and Drosocin. SIGMAV infection did not induce PGRP-SA and the AMP genes Drosomycin-B, Metchnikowin, and Defensin that are upregulated in DCV and/or DXV infections. Expression levels of the Toll and Imd signaling cascade genes are not significantly altered by SIGMAV infection. These results highlight shared and unique aspects of the Drosophila immune response to the three viruses and may shed light on the nature of the interaction with the host and the evolution of these associations.
Sigma virus (SIGMAV; family Rhabdoviridae) occurs naturally in Drosophila melanogaster and is maintained in fly populations through vertical transmission via germ cells (31). Other viruses in this family are known pathogens of humans, livestock, fish, and plants (33). Insects commonly serve as vectors and replication hosts for many livestock and all well-characterized plant rhabdoviruses. Black flies, sand flies, and mosquitoes, for example, transmit vertebrate-infecting rhabdoviruses, e.g., Vesicular stomatitis virus and Bovine ephemeral fever virus (12, 27), whereas aphids, leafhoppers, and planthoppers vector plant rhabdoviruses (17, 19).
While rhabdoviruses can infect a variety of tissues in their invertebrate hosts, they appear to predominantly invade the central nervous system. In humans and other vertebrates, Rabies virus spreads throughout the body, including the central nervous system, and most importantly for transmission, the salivary glands (12). SIGMAV and some plant rhabdoviruses have been shown to replicate in neural and other tissues of Drosophila and their insect vectors (1, 2, 17, 31). SIGMAV does not appear to adversely affect Drosophila in their natural environment; however, SIGMAV-infected flies remain irreversibly paralyzed and die after CO2 anesthetization (7, 31). Vesiculoviruses also confer similar CO2 sensitivity to their black fly hosts (7).
Drosophila immune responses to various bacterial and fungal pathogens are well characterized at the molecular level. The elucidation of Drosophila antiviral immune responses began only recently and has focused on two other naturally occurring viruses, Drosophila C virus (DCV; family Dicistroviridae) (13, 28) and Drosophila X virus (DXV; family Birnaviridae) (35). SIGMAV differs from these two viruses in its mode of transmission, morphology, tissue tropism, and virulence (8, 16, 21, 31, 32, 35) (Table 1). Given SIGMAV's unique biology, we predicted that the Drosophila immune response might also differ with respect to this virus. Using quantitative reverse transcription-PCR (qRT-PCR) approaches, we have examined the expression of a number of innate immune genes in SIGMAV-infected Drosophila insects relative to uninfected flies. We have compared these patterns of transcription to those in response to DCV and DXV with the aim of shedding some light on how Drosophila responds to diverse viral infections.
TABLE 1.
Characteristics of DCV (Dicistroviridae), DXV (Birnaviridae), and SIGMAV (Rhabdoviridae)
| Virus | Characteristic (reference)
|
|||
|---|---|---|---|---|
| Structure | Mode of transmission | Tissue tropism | Effect(s)/virulence | |
| DCV | Nonenveloped, isometric, positive-sense single-stranded RNA genome (16) | Horizontal, ingestion (9, 15) | Reproductive tissue, fat body, thoracic muscle, tracheal cells, digestive tract (21) | Mortality, faster developmental time, increased daily fecundity (20) |
| DXV | Nonenveloped, icosahedral nucleocapsid, double-stranded RNA genome (35) | Horizontal, contact (32) | Brain, thorax, reproductive tissue, malpighian tubules, trachea, muscle sheath (32) | Anoxia sensitivity, mortality (32) |
| SIGMAV | Enveloped bullet-shaped, transmembrane glycoprotein protruding from lipid envelope, negative sense single stranded RNA genome (27) | Vertical, via germ cells (31) | All tissues except muscle, especially thoracic and cephalic ganglia (31) | CO2 sensitivity, reduced egg viability (31) |
MATERIALS AND METHODS
Drosophila stocks.
The D. melanogaster Fe strain (SIGMAV infected) and Canton-S strain (SIGMAV free) were used as starting stocks. All Drosophila stocks were maintained at 25°C in 70% humidity with a 12-h light-dark cycle on standard cornmeal-yeast medium. To minimize genetic background effects, Canton-S females were crossed with Fe males, and then the progeny females of each generation were backcrossed against Fe males for four generations to create a BC4 strain with 97% Fe background. A small portion of BC4 flies remained infected with SIGMAV because paternal transmission is possible although it is less efficient than maternal transmission (31). SIGMAV-infected BC4 flies were removed from the population using a CO2 sensitivity assay (see below). SIGMAV-negative samples were also screened via qRT-PCR using SIGMAV-specific primers (see below).
Confirming SIGMAV infection in flies by immunofluorescence microscopy.
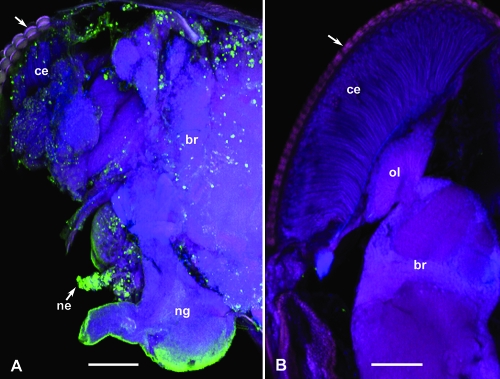
Three-day-old adult Drosophila SIGMAV-infected (Fe) or virus-free (BC4) flies were treated with CO2 gas and kept on ice for 10 min. The SIGMAV-infected flies remained irreversibly paralyzed whereas the virus-free flies recovered from anesthetization after returning to fresh air and room temperature. Subsequently, the heads of tested flies were separated from the bodies (thorax and abdomen) under a stereomicroscope. The bodies of tested flies were stored in RNAlater (Ambion, Austin, TX) at 4°C for subsequent RNA isolation and qRT-PCR whereas the heads were used to confirm the presence of SIGMAV in the brain and other tissues by immunofluorescence confocal laser scanning microscopy (iCLSM) (Fig. 1). To achieve this goal, each Drosophila head was split transversely with a sharp razor blade (to expose internal structures), kept in fixative (4% paraformaldehyde in 0.1 M phosphate buffer [pH 7.4], 0.1% Triton X-100) at 4°C overnight, and subsequently processed and examined by iCLSM as described previously (1). Briefly, the split heads were incubated with anti-SIGMAV antiserum (diluted 1/200) for 3 h and then incubated in a 1/600 dilution of the secondary antibody (goat anti-rabbit Alexa Fluor 488; Invitrogen Corp., Carlsbad, CA) for 1 h; samples were immersed first in the nuclear stain propidium iodide (Invitrogen Corp.) for 5 min and then in the actin stain phalloidin for 1 h before being examined by CLSM (Leica TCS SP). A subset of samples exhibiting substantial levels of infection (Fig. 1A) was then selected for downstream analysis. The status of SIGMAV-negative samples (Fig. 1B) was further confirmed with qRT-PCR (see below) using primers designed to amplify a fragment spanning the SIGMAV N and P genes (Table 2). Relative SIGMAV abundance per sample was compared following normalization against the host gene, Actin 88F (Table 2).
FIG. 1.
SIGMAV infection (green fluorescence) in the compound eye (ce), brain (br), other nerve ganglia (ng), and nerves (ne) in the head of an infected Drosophila (A) compared to that of a noninfected fly (B). In each case, the head was split transversely to expose internal structures and immediately fixed and processed for iCLSM using SIGMAV antiserum as a primary antibody, Alexa Fluor 488 as a secondary antibody, the nuclear stain propidium iodide (red), and the actin stain phalloidin (blue or purple). Arrows indicate compound eye lenses. ol, optic lobe. Scale bar, 50 μm.
TABLE 2.
Oligonucleotide primers for qRT-PCR
| Gene(s) | GeneIDa | Primer name | Nucleotide sequence (5′ to 3′) | Amplicon size (bp) |
|---|---|---|---|---|
| Actin88F | CG5178 | Actin88 305F | ATCGAGCACGGCATCATCAC | 78 |
| Actin88 349R | CACGCGCAGCTCGTTGTA | |||
| Attacin-A, Attacin-B | CG10146, CG18372 | AttB 362F | GGCCCATGCCAATTTATTCA | 101 |
| AttB 435R | CATTGCGCTGGAACTCGAA | |||
| Cecropin-A | CG1365 | CecA 91F | TCTTCGTTTTCGTCGCTCTC | 144 |
| CecA 234R | CTTGTTGAGCGATTCCCAGT | |||
| Defensin | CG1385 | Def 146F | GCCAGAAGCGAGCCACAT | 54 |
| Def 181R | CGGTGTGGTTCCAGTTCCA | |||
| Diptericin-A | CG12763 | Dipt 226F | AGGTGTGGACCAGCGACAA | 61 |
| Dipt 265R | TGCTGTCCATATCCTCCATTCA | |||
| Drosocin | CG10816 | Dro 30F | GCACAATGAAGTTCACCATCGT | 60 |
| Dro 71R | CCACACCCATGGCAAAAAC | |||
| Drosomycin-B | CG10810 | Dros 29F | CTCCGTGAGAACCTTTTCCA | 120 |
| Dros 149R | GTATCTTCCGGACAGGCAGT | |||
| Metchnikowin | CG8175 | Mtk 85F | GCTACATCAGTGCTGGCAGA | 102 |
| Mtk 187R | AATAAATTGGACCCGGTCT | |||
| Toll | CG5490 | Toll 2201F | AACTTGGGCAACCTTGTGAC | 180 |
| Toll 2380R | GTAACCAAACGGGGAGTTGA | |||
| PGRP-SA | CG11709 | PGRPSA 30F | CTGCGGCTGTTATCAGTGAA | 144 |
| PGRPSA 155R | TGATGGAATTTCCGCTTTTC | |||
| PGRP-SB1 | CG9681 | PGRPSB 27F | TGTGGCCGCTTTAGTGCTT | 57 |
| PGRPSB 65R | TCAATCTGCAGGGCATTGG | |||
| PGRP-SC1a, PGRP-SC1b | CG14746, CG8577 | SC1 330F | CGAGTGGAACCCCTACAGCAT | 65 |
| SC1 408R | GCTCCAGGGTGTCCCAGTT | |||
| PGRP-SD | CG7496 | PGRPSD 128F | CCTTGCCACGTGCTGTGA | 59 |
| PGRPSD 165R | TGTAACATCATCCGCACAAGCT | |||
| PGRP-LC | CG4432 | PGRPLC 211F | ACGGAATCCAAGCGTATCAG | 165 |
| PGRPLC 356R | GGCCTCCGAATCACTATCAA | |||
| Relish | CG11992 | Rel 2916F | TCCTTAATGGAGTGCCAACC | 181 |
| Rel 3097R | TGCCATGTGGAGTG ATTAT | |||
| SIGMAV N and P genes | NA | SIGMAV 1343F | ATGTAACTCGGGTGTGACAG | 154 |
| SIGMAV 1496R | CCTTCGTTCATCCTCCTGAG | |||
| vir-1 | CG31764 | vir-1 1361F | TGTGCCCATTGACCTATCCA | 109 |
| vir-1 1450R | GATTACAGCTGGGTGCACAA |
NA, not applicable.
Sample preparation and qRT-PCR.
A total of six samples were prepared for analysis for each SIGMAV-infected and uninfected Drosophila line. Each sample was comprised of paired male and female flies. Total RNA was extracted using Trizol (Invitrogen Corp.) according to the manufacturer's instructions. Initial homogenization was carried out using a Mini BeadBeater (BioSpec Products, Inc. Bartlesville, OK). The integrity and concentration of the RNA were determined spectrophotometrically using a NanoDrop and associated software, version 1000, (NanoDrop Technologies, Wilmington, DE). Extractions were treated with Turbo DNA-free (Ambion), and concentration was determined using a Quant-iT RiboGreen RNA reagent kit (Molecular Probes, Eugene, OR).
A SuperScript III Platinum Two-Step qRT-PCR kit with Sybr Green (Invitrogen Corp.) was used according to the manufacturer's protocol. cDNA was generated for each sample using random primers. Gene-specific primers were subsequently utilized for qRT-PCRs in a Rotor-Gene 3000 thermal cycler (Corbett Research, Brisbane, Australia). Real-time PCR primers (Table 2) were designed using Primer Express, version 1.5, software (Applied Biosystems, Foster City, CA) to yield 100- to 200-bp amplicons with a thermal denaturation midpoint temperature of ≥80°C. Threshold cycle values were normalized against Actin 88F as an internal control, and the ΔΔCT (where CT threshold cycle) method was used to calculate relative concentrations of target mRNA using Rest 2005, version 1.9.12, software (Corbett Research) (26). Two assay replicates and five to six biological replicates were compiled and averaged for each treatment.
RESULTS
Relative SIGMAV abundance.
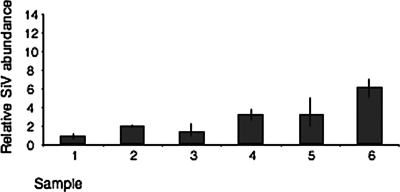
The mean relative values of SIGMAV infection, as revealed by qRT-PCR across the six samples that were found positive in the iCLSM study (Fig. 1A), was 2.3 ± 0.76 (mean ± standard error of the mean) with a range of 0.8 to 6.0 (Fig. 2). Of the six putative SIGMAV-negative samples based on the iCLSM study (Fig. 1B), one sample was apparently infected with SIGMAV, as revealed by qRT-PCR using SIGMAV primers (data not shown), and therefore was excluded for further analyses. Thus, iCLSM detected only relatively high levels of SIGMAV infection, which is reflected in the narrow range of qRT-PCR numbers for the six samples found positive in iCLSM (Fig. 2).
FIG. 2.
Relative abundance of SIGMAV (SiV) per sample based on the expression of the SIGMAV N and P genes normalized against host Actin 88F expression. Error bars represent the range from assay replicates.
In pilot experiments prior to employing selection of SIGMAV-positive samples by iCLSM, extremely variable results were obtained with respect to the transcriptional profiles of various immune genes. This variation can be explained by a polymorphism for both infection status and viral titer in laboratory stocks. The CO2 sensitivity assays are also not 100% accurate in identifying SIGMAV-free flies. Hence, we decided to focus on comparing the transcription profiles of highly infected flies and SIGMAV-negative flies as determined by iCLSM and qRT-PCR.
Expression of innate immunity-associated genes.
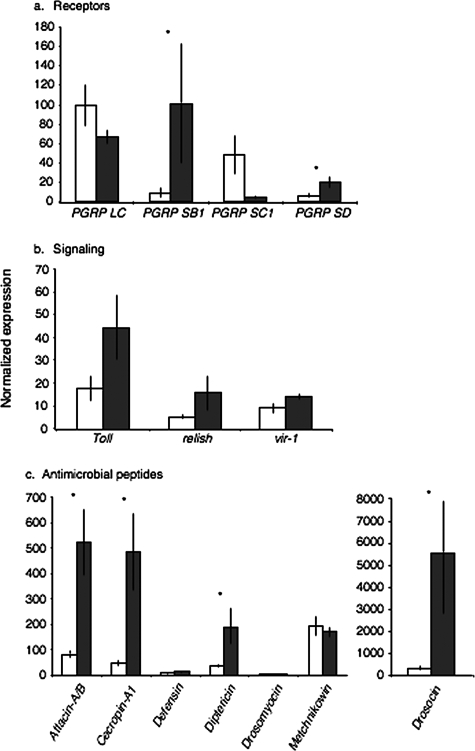
We tested the transcription levels of 15 immunity-related genes relative to the internal control gene Actin 88F by qRT-PCR. This indicated that six of the immune genes showed a consistent and statistically significant upregulation in the six SIGMAV-infected samples versus the five samples of uninfected flies. For the upstream genes involved in receptor activity and signaling, the peptidoglycan recognition protein (PGRP) genes PGRP-SB1 and PGRP-SD showed clear upregulation in infected flies (Fig. 3A and Table 3), whereas PGRP-LC, PGRP-SC1, and PGRP-SA were not upregulated (Fig. 3A and Table 3). Expression levels of PGRP-SB1 were particularly high (23.3-fold uninfected) whereas the expression level of PGRP-SD was only slightly higher (3.5-fold uninfected) (Table 3). Expression of Toll, Relish, and vir-1 showed increases in expression in SIGMAV-infected flies, but these increases were not statistically significant (Fig. 3B). For the genes encoding antimicrobial peptides (AMPs), significant upregulation was found for Attacin-A, Attacin-B, Cecropin-A1, Diptericin-A, and Drosocin in SIGMAV-infected flies but not for Defensin, Drosomycin-B, and Metchnikowin (Fig. 3C). PGRP-SB1, PGRP-SD, and Diptericin-A are primarily regulated by Relish of the Imd pathway, whereas Attacin-A, Attacin-B, Cecropin-A1, and Drosocin are regulated by Relish of the Imd pathway and Spaetzle of the Toll pathway (11). However, we find no evidence that SIGMAV infection induces expression of Toll and Relish (Table 3). SIGMAV infection also does not activate vir-1 of the Jak-STAT pathway (Table 3).
FIG. 3.
Innate immune gene expression normalized against host Actin 88F by functional group. Values are mean ± standard error of the mean. SIGMAV-negative (white) and SIGMAV-positive (gray) bars represent results of five and six samples, respectively. *, P <0.05 for the difference between SIGMAV-negative and SIGMAV-positive flies.
TABLE 3.
Mean ratios of expression for virus-infected relative to virus-free Drosophila
| Gene function and name | GeneID | Relative expression level (virus-infected flies/virus-free flies)a
|
|||
|---|---|---|---|---|---|
| SIGMAVb | DCV
|
DXVe | |||
| Oral infection routec | Intrathoracic infection routed | ||||
| Upstream genes and receptors | |||||
| PGRP-SA | CG11709 | x | − | 3.2 | ND |
| PGRP-SB1 | CG9681 | 23.5 | − | − | ND |
| PGRP-SC1a, PGRP-SC1b | CG14746, CG8577 | − | − | − | ND |
| PGRP-SD | CG7496 | 3.5 | − | − | ND |
| PGRP-LC | CG4432 | − | − | − | ND |
| Toll | CG5490 | − | − | − | ND |
| Spaetzle | CG6134 | ND | − | 3.0 | ND |
| Signaling cascade | |||||
| Relish | CG11992 | − | − | 3.5 | ND |
| vir-1 | CG31764 | − | − | 6.4 | ND |
| Antimicrobial peptides | |||||
| Drosomycin-B | CG10810 | − | 3.0 | 2.1 | 70 |
| Defensin | CG1385 | − | − | + | 4.8 |
| Metchnikowin | CG8175 | − | − | 3.0 | 60 |
| Drosocin | CG10816 | 10.3 | − | − | 3.2 |
| Diptericin-A | CG12763 | 5.6 | − | − | 3.2 |
| Diptericin-B | CG10794 | ND | − | 5.0 | ND |
| Attacin-A | CG10146 | 7.2 | 6.3 | 8.7 | 2.5 |
| Attacin-B | CG18372 | ND | − | 4.7 | ND |
| Attacin-C | CG4740 | ND | − | 2.7 | ND |
| Attacin-D | CG7629 | ND | − | + | ND |
| Cecropin-A1 | CG1365 | 8.8 | 2.6 | − | 1.8 |
| Cecropin-A2 | CG1367 | ND | 3.3 | − | ND |
| Cecropin-B | CG1878 | ND | − | + | ND |
| Cecropin-C | CG1373 | ND | − | + | ND |
−, no difference in expression levels; x, no detectable expression; ND, not determined; +, induced expression relatively to virus-free flies.
qRT-PCR data from this study; data reported for P values <0.05.
Microarray data from reference 28.
Microarray data from reference 13.
qRT-PCR data from reference 35.
Comparison of Drosophila immune responses toward SIGMAV, DCV, and DXV.
Signaling pathways controlling the Drosophila humoral defense have been well described (6, 11, 22), but an understanding of the antiviral response is less complete (9). There have been several transcriptional profiles generated of Drosophila in response to the viruses DCV (13, 28) and DXV (35) that we have attempted to summarize (Table 3). The platforms/methods utilized vary across these studies as do the genes compared. Additionally, the fly response to DCV differs whether the virus is administered orally (28) or by intrathoracic injection (13). In general, the transcriptional responses of flies infected with DCV and DXV appear to be more similar to each other than to flies infected with SIGMAV. SIGMAV heavily induces PGRP-SB1 and PGRP-SD while DCV increases transcription of only PGRP-SA (13). DCV also induces expression of Relish and other signaling cascade genes (13), which is not the case for SIGMAV (Table 3). Lastly, SIGMAV infection did not upregulate the AMP-encoding genes Drosomycin-B, Defensin, and Metchnikowin (Table 3) whereas DCV and DXV infections did (Table 3) (13, 28, 35). SIGMAV- and DCV-infected flies (13, 28) share upregulated expression of the Attacin-A, Attacin-B, and Cecropin A genes. SIGMAV- and DXV-infected flies (35) share upregulated expression of Drosocin, Diptericin-A, Attacin-A, and Attacin-B.
DISCUSSION
The pattern of induced PGRP gene expression by SIGMAV is distinct from that of other viruses, where only PGRP-SA shows induced transcription by DCV. SIGMAV induces both PGRP-SD and, more notably in terms of the magnitude of expression, PGRP-SB1. These two members of the short class of PGRP genes share a number of characteristics. Both genes exhibit low-level constitutive expression in adult Drosophila insects, are highly inducible in response to bacterial infection, are expressed mainly in the fat body, and encode proteins that are likely exported from the cell (34). PGRP-SB1 has also been shown to have amidase activity and bactericidal properties (25). Unlike DCV and DXV that have proteinaceous capsids, SIGMAV particles are surrounded by a lipid bilayer with glycoprotein spikes. PGRPs are the first receptors that recognize, bind, or catalytically cleave specific surface components of bacterial cell membranes (22, 29). Thus, the differential induction of the PGRPs among the viruses may be an indication of the different virus surface properties.
Unlike the case with DCV (13), we found little evidence of increased transcription in the signaling cascade genes of the Imd, Toll, and Jak-STAT pathways. However, one would expect a chance in expression of these signaling genes, because PGRP-SB1 and PGRP-SD expressions are primarily regulated by Relish of the Imd pathway (11), and PGRP-SD function is required for activation of the Toll pathway (4, 30). Also, Diptericin-A is primarily regulated by Relish of the Imd pathway, whereas Attacin-A, Attacin-B, Cecropin-A1, and Drosocin are regulated by Relish of the Imd pathway and Spaetzle of the Toll pathway (11). On the other hand, we did not find upregulation of Metchnikowin, which is also induced by both the Toll and Imd pathways (23). The type of infection dictates how the Toll and Imd pathways contribute to the expression of each AMP gene (11). For the AMP gene expression levels, the SIGMAV infection appears to be most similar to that of the gram-negative bacteria that also induce Diptericin, Attacin, Cecropin, and Drosocin but not Drosomycin and Metchnikowin (18). The outcomes of the Drosophila immune response to SIGMAV and gram-negative bacteria may be similar because both microbes have outer lipid bilayers and glucose.
Since SIGMAV is a vertically transmitted parasite, there would be substantial selection pressure for reduced virulence and for evasion of the host immune response in the virus (14). Like SIGMAV, DCV has an old and established relationship with Drosophila. Laboratory experiments comparing responses of flies following infection by intrathoracic injection with the more natural route of feeding indicate a weaker Toll response in the latter case (9, 13, 28). This may be due entirely to differences in the mode of immune system activation via the gut but could also reveal a history of adaptation. The constitutive upregulation of immunity genes in SIGMAV-infected flies nonetheless indicates evidence of host recognition and energetic investment in fighting the SIGMAV infection. Extreme overactivation of the Imd pathway has been associated with developmental defects and larval death (5), and a number of published works reveal tradeoffs between immune function and fitness in insects (3, 10, 24). Not surprisingly, SIGMAV has been shown to cause mild reductions in host egg viability; however, the 10 to 20% SIGMAV infection frequency in natural Drosophila populations (31) suggests that infected flies can compete in terms of fitness to some degree with virus-free flies.
Acknowledgments
This work was supported by the Australian Research Council Linkages International grant LX0452397.
We thank Scott O'Neill for his support and advice, Roger Mitchell for expert technical assistance, and Rebecca Elkington and Jennifer McMahon for qRT-PCR advice.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Ammar, E. D., and S. A. Hogenhout. 2008. A neurotropic route for Maize mosaic virus (Rhabdoviridae) in its planthopper vector Peregrinus maidis. Virus Res. 131:77-85. [DOI] [PubMed] [Google Scholar]
- 2.Ammar, E. D., T. Meulia, E. Özbek, and S. A. Hogenhout. 2004. Assembly and accumulation sites of Maize mosaic virus (Rhabdoviridae) in plant host and insect vector using transmission electron and confocal laser scanning microscopy, p. 56-64. In A. Mendez-Vilas and L. Labajos-Broncano (ed.), Current issues on multidisciplinary microscopy research and education. Formatex, Badajoz, Spain.
- 3.Armitage, S. A. O., J. J. W. Thompson, J. Rolff, and M. T. Siva-Jothy. 2003. Examining costs of induced and constitutive immune investment in Tenebrio molitor. J. Evol. Biol. 16:1038-1044. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff, V., C. Vignal, I. G. Boneca, T. Michel, J. A. Hoffmann, and J. Royet. 2004. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of gram-positive bacteria. Nat. Immunol. 5:1175-1180. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, V., C. Vignal, B. Duvic, I. G. Boneca, J. A. Hoffmann, and J. Royet. 2006. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, C. A., and K. V. Anderson. 2004. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22:457-483. [DOI] [PubMed] [Google Scholar]
- 7.Brun, G. 1991. Rhabdoviridae, p. 443-460. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, FL.
- 8.Cherry, S., and N. Perrimon. 2004. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat. Immunol. 5:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherry, S., and N. Silverman. 2006. Host-pathogen interactions in Drosophila: new tricks from an old friend. Nat. Immunol. 7:911-917. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, S. C., L. E. B. Kruuk, and K. Wilson. 2004. Costs of resistance: genetic correlations and potential trade-offs in an insect immune system. J. Evol Biol. 17:421-429. [DOI] [PubMed] [Google Scholar]
- 11.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Mattos, C. A., C. C. de Mattos, and C. E. Rupprecht. 2001. Rhabdoviruses, p. 1245-1277. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Dostert, C., E. Jouanguy, P. Irving, L. Troxler, D. Galiana-Arnoux, C. Hetru, J. A. Hoffmann, and J. L. Imler. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol. 6:946-953. [DOI] [PubMed] [Google Scholar]
- 14.Ewald, P. 1994. Evolution of infectious diseases. Oxford University Press, Oxford, United Kingdom.
- 15.Gomariz-Zilber, E., M. Poras, and M. Thomas-Orillard. 1995. Drosophila C virus: experimental study of infectious yields and underlying pathology in Drosophila melanogaster laboratory populations. J. Invertebr. Pathol. 65:243-247. [DOI] [PubMed] [Google Scholar]
- 16.Gravot, E., M. Thomas-Orillard, and B. Jeune. 2000. Virulence variability of the Drosophila C virus and effects of the microparasite on demographic parameters of the host (Drosophila melanogaster). J. Invertebr. Pathol. 75:144-151. [DOI] [PubMed] [Google Scholar]
- 17.Hogenhout, S. A., M. G. Redinbaugh, and E. D. Ammar. 2003. Plant and animal rhabdovirus host range: a bug's view. Trends Microbiol. 11:264-271. [DOI] [PubMed] [Google Scholar]
- 18.Imler, J. L., and P. Bulet. 2005. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem. Immunol. Allergy 86:1-21. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, A. O., R. G. Dietzgen, M. M. Goodin, J. N. Bragg, and M. Deng. 2005. Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43:623-660. [DOI] [PubMed] [Google Scholar]
- 20.Jousset, F. X., M. Bergoin, and B. Revet. 1977. Characterization of the Drosophila C virus. J. Gen. Virol. 34:269-283. [DOI] [PubMed] [Google Scholar]
- 21.Lautie-Harivel, N., and M. Thomas-Orillard. 1990. Location of Drosophila C virus target organs in Drosophila host population by an immunofluorescence technique. Biol. Cell 69:35-39. [DOI] [PubMed] [Google Scholar]
- 22.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 23.Levashina, E. A., S. Ohresser, B. Lemaitre, and J. L. Imler. 1998. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J. Mol. Biol. 278:515-527. [DOI] [PubMed] [Google Scholar]
- 24.McKean, K. A., and L. Nunney. 2001. Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98:7904-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellroth, P., and H. Steiner. 2006. PGRP-SB1: An N-acetylmuramoyl l-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Commun. 350:994-999. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the virus and their replication, p. 1221-1244. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 28.Roxstrom-Lindquist, K., O. Terenius, and I. Faye. 2004. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 5:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiner, H. 2004. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 198:83-96. [DOI] [PubMed] [Google Scholar]
- 30.Tanji, T., and Y. T. Ip. 2005. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 26:193-198. [DOI] [PubMed] [Google Scholar]
- 31.Teninges, D. 1999. Sigma rhabdoviruses (Rhabdoviridae), p. 1635-1639. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, vol. 2. Academic Press, San Diego, CA. [Google Scholar]
- 32.Teninges, D., A. Ohanessian, C. Richard-Molard, and D. Contamine. 1979. Isolation and biological properties of Drosophila X virus. J. Gen. Virol. 42:241-254. [Google Scholar]
- 33.Tordo, N., A. Benmansour, C. Calisher, R. G. Dietzgen, R. X. Fang, A. O. Jackson, G. Kurath, S. Nadin-Davis, R. B. Tesh, and P. J. Walker. 2005. Family Rhabdoviridae, p. 623-644. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Dessenberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 34.Werner, T., G. Liu, D. Kang, S. Ekengren, H. Steiner, and D. Hultmark. 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97:13772-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zambon, R. A., M. Nandakumar, V. N. Vakharia, and L. P. Wu. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 102:7257-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]