Abstract
The dynamics of Legionella spp. and of dominant bacteria were investigated in water from a cooling tower plant over a 9-month period which included several weeks when Legionella pneumophila proliferated. The structural diversity of both the bacteria and the Legionella spp. was monitored by a fingerprint technique, single-strand conformation polymorphism, and Legionella spp. and L. pneumophila were quantified by real-time quantitative PCR. The structure of the bacterial community did not change over time, but it was perturbed periodically by chemical treatment or biofilm detachment. In contrast, the structure of the Legionella sp. population changed in different periods, its dynamics at times showing stability but also a rapid major shift during the proliferation of L. pneumophila in July. The dynamics of the Legionella spp. and of dominant bacteria were not correlated. In particular, no change in the bacterial community structure was observed during the proliferation of L. pneumophila. Legionella spp. present in the cooling tower system were identified by cloning and sequencing of 16S rRNA genes. A high diversity of Legionella spp. was observed before proliferation, including L. lytica, L. fallonii, and other Legionella-like amoebal pathogen types, along with as-yet-undescribed species. During the proliferation of L. pneumophila, Legionella sp. diversity decreased significantly, L. fallonii and L. pneumophila being the main species recovered.
Legionella species are relatively slow-growing, ubiquitous, aquatic bacteria (3). Natural freshwater environments are the major reservoirs. However, various human-made systems such as heated water in spas, showerheads, sanitary hot water networks, or cooling towers provide ideal habitats for Legionella species (30). In such water systems, pathogenic Legionella spp. responsible for acute respiratory infections can proliferate. Most cases of legionellosis can be traced to human-made aquatic environments where the water temperature is higher than the ambient temperature (17). In particular, cooling towers have been implicated in major outbreaks of legionellosis caused by Legionella pneumophila (7, 10, 19, 22). Legionella species are facultative intracellular gram-negative bacilli which multiply in protozoan hosts and can also survive within microbial biofilm communities (17). Amoebal cysts provide a protective environment for Legionella species, which can then withstand treatments with biocides such as chlorine (21, 34). Keeping Legionella under control in cooling towers is the necessary condition for reducing legionellosis outbreaks. However, due to the ecology of this bacterium, reducing the risk related to Legionella remains a challenge. Indeed, not only physical parameters (temperature, fouling of the network, etc.) induce a risk of proliferation, but biotic parameters such as the presence of a biofilm or amoebae also increase the risk (33). Understanding of the factors that contribute to the survival or active growth of L. pneumophila in the environment is still very limited. In particular, very little information is available on microbial diversity in systems contaminated with L. pneumophila. Clearly, controlling the Legionella risk in cooling towers requires a better understanding of the dynamics of the different microbial constituents during the proliferation of pathogenic species.
In this study, the ecology of Legionella spp. in relation to the bacterial community was investigated in a cooling tower plant. The dynamics of the bacteria's structural diversity during the proliferation of L. pneumophila were analyzed. The dynamics of Legionella spp. were also investigated to determine the changes that arose in the Legionella population during the proliferation of the pathogenic species L. pneumophila. In fact, little is known regarding the Legionella species present in cooling tower systems or their dynamics within the same cooling tower (28). The growth requirements of Legionella, its ability to enter a viable-but-nonculturable state, the association of Legionella with protozoa, and the occurrence of Legionella in biofilms all tend to complicate its detection (7), and cultures on selective media have sometimes failed to isolate Legionella species from environmental samples (13). Cultivation-independent methods have thus been used to characterize the Legionella population in aquatic environments (12, 13, 30, 35). In the present study, PCR methods were used to quantify Legionella spp. and L. pneumophila and to analyze Legionella sp. diversity. The dynamics of Legionella spp., in relationship to those of the dominant bacteria, were followed by single-strand conformation polymorphism (SSCP) in a cooling tower plant over a 9-month period which included several weeks when L. pneumophila proliferated.
MATERIALS AND METHODS
Cooling tower facility.
The cooling tower facility studied, located in France, had the following characteristics: a total volume of 250 m3, 16 MW of power, and 100 m3 of water used per day with a flux rate of 300 to 1,800 m3/h in the cooling tower circuit. The municipal water (tap water quality) was treated in the circuit by biocide, descaling, antialgal, anticorrosion, and UV treatments. Hot water was cooled down by air after spraying at the top of the eight cooling towers. It was retrieved in a 150-m3 basin. The temperature of the incoming water was around 15 to 16°C. From May to September 2005, the average temperature of the water before release at the top of the towers (sampling point) was 27°C, including an increase to 29 to 30°C in July and August.
Chemical treatment.
Isothiazolone was used as a biocide. Volumes of 5 to 65 liters were applied weekly from 26 May 2005 to 28 September 2005 in order to reach concentrations of 100 to 250 ppm. On three dates (28 July, 11 August, and 2 September), the biocide and biodispersant (detergent) were added simultaneously. On these occasions, isothiazolone volumes were 65, 62, and 60 liters, respectively, while the biodispersant volume added was 20 liters on each date.
Sampling.
Water was sampled at different dates at a point located on the supply pipe common to all eight cooling towers, prior to its release at the top of the towers. One liter of water was collected, filtered through 0.45-μm-pore-size filters (Millipore, Billerica, MA), and frozen while awaiting DNA extraction.
ATP measurements.
For ATP measurements, the WaterGiene ATP test was used (Charm Sciences Inc., Lawrence, KS).
DNA extraction.
DNA was extracted directly from filters with the Aquadien extraction kit (Bio-Rad, Hercules, CA). For each sample, all analyses were carried out with the same DNA extract.
Real-time quantitative PCR.
Quantitative PCRs were performed with IQ-Check Legionella and IQ-Check L. pneumophila kits from Bio-Rad by following the stipulations of the manufacturer and using the iCycler IQ apparatus (Bio-Rad). Standard DNA curves were generated by amplification of serial 10-fold dilutions of genomic DNA of L. pneumophila ATCC 33152 in sterilized water and designed by an iCycler IQ apparatus. The L. pneumophila genome mass used was 4.3 fg of DNA. A standard DNA curve was generated for each assay. The cycle threshold (CT) corresponding to the number of cycles at which the reaction becomes exponential, was compared to the standard curve in order to calculate the number of genomic units (GU) in the DNA extract of the samples. The PCR equation was CT = −1/[log (1 + E)] × (log X0 − log X), where E is the efficiency of the PCR, X is the concentration, and X0 is the initial concentration. The standard DNA curve was validated when the slope was between −3.9 and −3.1, corresponding to 80 and 110% PCR efficiency, respectively. The intercept varied from 37 to 40. The Bio-Rad kit contained an internal control present in each amplification mixture to check the presence of inhibitory factors. In the event of PCR inhibition, the sample was further diluted and reanalyzed. Two negative controls were performed for each assay, a negative control for PCR (obtained by replacing the DNA with water) and a negative control for DNA extraction. Both controls had to be negative to validate the assay. PCR results were converted to genomic units per liter. Average values were calculated from duplicates. The detection threshold for Legionella spp. was 30 GU/well, which corresponded to 960 GU liter−1. In the event of PCR inhibition, the dilution of the sample brought the detection threshold to 1,920 GU liter−1 (3.3 log GU liter−1). The detection threshold for L. pneumophila was 640 GU liter−1 (2.8 log GU liter−1). The protocol of quantification by PCR of Legionella spp. and L. pneumophila followed the XP T90-471 standard elaborated by the T90E WG AFNOR (Association Française de Normalization) (6).
PCR-SSCP.
To analyze the overall structure of the bacterial community as a whole, the V3 region of the 16S rRNA gene was amplified with primers W49 (5′-ACGGTCCAGACTCCTACGGG-3′, Escherichia coli position F331) and 5′-fluorescein phosphoramidite-labeled W104 (5′-TTACCGCGGCTGCTGGCAC-3′, E. coli position R533) (14). PCR amplifications were performed with a Mastercycler thermal cycler (Eppendorf, Hamburg, Germany). The reaction mixtures contained 1× polymerase buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 130 ng of each primer, 0.5 U of Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), 1 μl of genomic DNA, and water added to obtain a final volume of 50 μl. The PCR conditions were an initial denaturation step of 2 min at 94°C; 25 cycles of a three-stage program of 30 s at 94°C, 30 s at 61°C, and 30 s at 72°C; and a final elongation for 10 min at 72°C. The reactions were stopped by cooling the mixture to 4°C. The analysis of Legionella sp. diversity was performed after a nested PCR. First, primers specific to the 16S rRNA gene of Legionella spp. were used to amplify a 653-bp fragment including the V3 region, i.e., LEG-225 (5′-AAGATTAGCCTGCGTCCGAT-3′) and LEG-858 (5′-GTCAACTTATCGCGTTTGCT-3′) (27). The reaction mixtures contained 1× polymerase buffer, 0.2 mM dNTPs, 200 ng of each primer, 1 U of redTaq DNA polymerase (Sigma-Aldrich, St. Louis, MO), 1 μl of genomic DNA, and water added to obtain a final volume of 50 μl. The PCR conditions were an initial denaturation step of 90 s at 95°C; 30 cycles of 30 s at 95°C, 60 s at 64°C, and 60 s at 72°C; and a final elongation for 5 min at 72°C. Then, 1 μl of the PCR product was used to amplify the V3 16S rRNA gene bacterial region with primers W49 and W104 as described above. Amplification product sizes were confirmed by electrophoresis on a 2% (wt/vol) agarose gel.
SSCP analysis permits the separation of DNA fragments of the same size but with different compositions. One microliter of further diluted PCR products was added to 18 μl of formamide and 1 μl of internal size standard Rox 400 HD (Applied Biosystems, Foster City, CA) diluted 10 times. The sample was then denatured for 5 min at 95°C and placed directly on ice for 10 min. SSCP was performed with the ABI 3130 genetic analyzer (Applied Biosystems) equipped with four 50-cm capillary tubes filled with 5.6% conformation analysis polymer (Applied Biosystems) in corresponding buffer and 10% glycerol. The injection of DNA in capillaries required 5 kV for 3 s. Electrophoresis was carried out at 15 kV and 32°C for 30 min per sample. Raw SSCP data were exported into the easily handled csv format with the Chromagna shareware (developed by Mark J. Miller at the U.S. National Institutes of Health), and statistical analyses were performed with SAFUM (37) and the Matlab 6.5 software (MathWorks).
Identification of Legionella species and eukaryotic species by cloning and sequencing.
Two libraries of Legionella 16S rRNA genes were built, one with DNA from the 22 June sample and the other with DNA from the sample collected on 13 July. For the eukaryotic library, the DNA extract from water collected on 13 July was used. PCR mixtures contained 1× polymerase buffer, 0.2 mM dNTPs, 200 ng of each primer, 1 U of redTaq DNA polymerase, 1 μl of genomic DNA, and water added to obtain a final volume of 50 μl. A 653-bp fragment of Legionella 16S rRNA genes was amplified by PCR with primers LEG-225 and LEG-858 as described above, but with 35 cycles instead of 30. For amplification of a fragment of the eukaryotic 18S ribosomal DNA, primers W16 (5′-CTTAATTTGACTCAACACGG-3′) (20) and W176 (5′-GGGCATCACAGACCTGTT-3′) were used. The PCR conditions were an initial denaturation step of 2 min at 94°C; 35 cycles of 60 s at 94°C, 60 s at 51°C, and 60 s at 72°C; and a final elongation for 10 min at 72°C. All PCR products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions. The purified PCR products were cloned and transformed with the pCR4-TOPO plasmid and TOP10 E. coli competent cells, as indicated by the manufacturer (TOPO TA cloning kit; Invitrogen, Carlsbad, CA). Recombinant cells were selected by kanamycin resistance and ccd gene killer inactivation before cultivation at 37°C for 24 h in LB2× medium (tryptone at 20 g liter−1, yeast extract at 10 g liter−1, NaCl at 10 g liter−1). Sequences were obtained from clone culture (Millegen, Toulouse, France). The primer sequences were removed, and the presence of chimerical sequences was checked for with the CHECK-CHIMERA tool available at the Ribosomal Data Project (26) and the Pintail program (5). Sequences were compared with GenBank databases (www.ncbi.nlm.nih.gov/BLAST) by using the BLASTN program (4). They were imported and aligned into the January 2004 ARB database (25). The aligned sequences were added to the ARB tree by using the parsimony tool. A tree gathering sequences from environmental clones and from described Legionella species (613 bp) was then built by neighbor joining (29). The tree was rooted with the sequence from Coxiella burnetii (D89798). Bootstrap analyses (1,000 replicates) were used to assess the robustness of inferred monophyletic groups.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of Legionella spp. determined in this study were deposited in the GenBank database under accession numbers EU309480 to EU309490.
RESULTS
Water from the cooling tower facility was collected at different dates between April 2005 and January 2006. Water was sampled since it contains the microflora which will be discharged from the towers after aerosolization. Water was sampled at a point located just before its dispersal into the air inside the eight cooling towers. This enabled us to analyze the microbiological constituents in contact with the outside and the potential microbial contamination by the plant. It is also the sampling point generally used to monitor the Legionella risk in cooling towers.
Abundance of Legionella spp. and L. pneumophila and effect of chemical treatments.
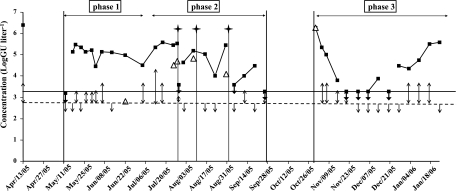
Concentrations of Legionella spp. and L. pneumophila were determined by real-time quantitative PCR from April 2005 to January 2006. Results are gathered in Fig. 1. The threshold of detection by quantitative PCR was 3.3 log GU liter−1 for Legionella spp. and 2.8 log GU liter−1 for L. pneumophila. In April 2005, the concentration of Legionella spp. was high (6.4 log GU liter−1) and the plant was shut down and cleaned. At the subsequent startup of the cooling tower in May, the Legionella sp. concentration was below 3.3 log GU liter−1 and L. pneumophila was not detected. However, after only 5 days, the concentration of Legionella spp. increased to 5.1 log GU liter−1 and then remained between 4.4 and 5.6 log GU liter−1 for several weeks. L. pneumophila was detected in May and June, but its concentration remained under the quantification threshold until the end of June (phase 1). It started to rise in July, and the highest concentrations of L. pneumophila were obtained in July and August, at up to 4.7 log GU liter−1 (phase 2). In September, concentrations of L. pneumophila remained low. The plant was shut down in October, and from the end of October to January (phase 3), L. pneumophila was quantified at a high concentration only once, when the plant was restarted.
FIG. 1.
Concentrations of Legionella spp. (▪) and L. pneumophila (▵) in a cooling tower facility as determined by quantitative PCR. When detection of L. pneumophila occurred but at concentrations below the quantification threshold, results are represented by double arrows. When the concentration was below the detection threshold, results are represented by single arrows. The detection thresholds of Legionella spp. and L. pneumophila are indicated, respectively, by solid and dotted lines (Legionella detection threshold = 3.3 log GU liter−1, L. pneumophila detection threshold = 2.8 log GU liter−1). Stars indicate combined biodispersant and biocide treatments.
From May, biocides were regularly injected into the system without any effect on Legionella sp. concentrations. After the proliferation of L. pneumophila, a combined biocide and biodispersant treatment was carried out on 28 July, 8 August, and 2 September. This treatment reduced Legionella sp. and L. pneumophila concentrations significantly, below the detection or quantification threshold, even when the system was highly contaminated (Fig. 1). Simultaneous decreases in ATP content and biofilm size were also observed after the treatment (data not shown). The dispersal agent was effective in biofilm removal, and dispersal and biocide treatment decreased not only the total microbial population size (based on ATP levels) but also the Legionella sp. concentration. However, a few days after treatment, high concentrations of Legionella and L. pneumophila were observed once again, showing that although the treatment effect was real, it was only transient. L. pneumophila was quantified in five samples, with values ranging from 2.8 to 4.9 log GU liter−1. In these samples, the concentration of Legionella spp. was around 5 log GU liter−1. The corresponding percentage of L. pneumophila among the total Legionella population varied from less than 1% to up to 50%. Legionella spp. and L. pneumophila did not follow the same dynamic; the increase in the L. pneumophila concentration, concomitant with a constant concentration of Legionella spp., may indicate that the concentration of other Legionella spp. decreased.
Dynamics of the microbial community. (i) Bacterial population.
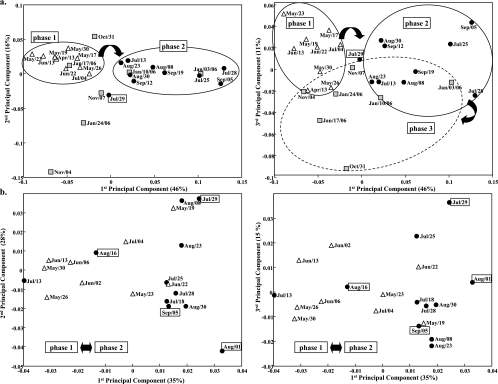
The structure of the bacterial community was examined from May to September by using SSCP fingerprints. SSCP fingerprints were compared by principal-component analysis (PCA). Close SSCP fingerprints gather together on the PCA plots. The comparison is based on the totality of the SSCP signal. The first two dimensions of the PCA plots represent the two components which best highlight the differences between the SSCP fingerprints. Figure 2 presents two PCA plots, one for components 1 and 2 and one for components 1 and 3.
FIG. 2.
PCA of SSCP fingerprints obtained for Legionella spp. (a) and bacteria (b) during phase 1 (▵), phase 2 (•), and phase 3 (▪). Samples in boxes were collected after chemical biodispersant and biocide treatment.
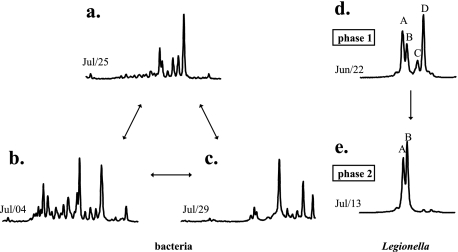
Six bacterial species were continuously present during the 3.5 months of the study in various amounts. In some samples, great diversity was observed, 5 to 15 peaks appearing on top of the six species. The samples displaying great diversity harbored the same bacterial species in various proportions. The increase in diversity did not seem to occur after chemical biocide and biodispersant treatment. Rather, PCA analysis of the SSCP data showed that this treatment led, in some cases, to disrupted fingerprints (samples in boxes, Fig. 2b) located away from the main groupings on the PCA plot. In these samples, the six most common species were not retrieved. Figure 3 gathers three SSCP fingerprints showing the six resilient species (Fig. 3a), one high-diversity SSCP fingerprint (Fig. 3b), and one fingerprint obtained after the chemical treatment of 28 July (Fig. 3c). Overall, the system was colonized by few dominant and resilient bacterial species, an observation that held even after the community structure was strongly disrupted, and over and above this resilient flora, some more species were sometimes present. There was no variation with the passage of time in the dynamics of the bacterial diversity. In particular, no significant change in the bacterial population was observed in July during the increase in the L. pneumophila concentration.
FIG. 3.
SSCP fingerprints showing different bacterial community structures (resilient species [a], high-diversity SSCP fingerprint [b], and after chemical treatment on 28 July [c]) and Legionella sp. diversity during phases 1 (d) and 2 (e).
(ii) Legionella sp. population.
The structure of the Legionella sp. population was determined from April 2005 to January 2006. The PCA plot of SSCP fingerprints (Fig. 2a) showed two main groupings, i.e., samples from April to July (phase 1) on the one hand and samples from mid-July to September (phase 2) on the other. The same four dominant species were observed from the beginning of April to the beginning of July (peaks A, B, C, and D in Fig. 3d). After this date, the diversity decreased and SSCP fingerprints then harbored one or two peaks (peaks A and B in Fig. 3e). Peak B, observed from May to September, was dominant during the summer, although not always recovered in October and November. SSCP fingerprints obtained during phase 3 formed another grouping on the PCA plot (Fig. 2a). Peaks A and B were present and even dominant in some of the fingerprints obtained in January. The effect of chemical treatment (biocide and biodispersant) on the diversity of Legionella spp. could not be analyzed since the water collected just after treatment had insufficient concentrations for PCR amplification. The beginning of the proliferation of the system by L. pneumophila was simultaneous with a major decrease in Legionella sp. diversity. Indeed, changes in Legionella sp. community structure appeared mainly between the end of June and the beginning of July, which was precisely the same period when high concentrations of L. pneumophila were first observed.
Overall, the community structure of Legionella spp. varied depending on the time of year, whereas bacterial community structure was composed largely by the same group of a few dominant species and was reestablished within a few days after any disruption. Legionella spp. changed throughout the time period studied, with no reoccurrence of the initial population, whereas the same bacterial flora was observed during different periods of the year. Therefore, there could be no correlation between the dynamics of Legionella spp. and those of the dominant bacteria.
Phylogenetic positioning of Legionella species.
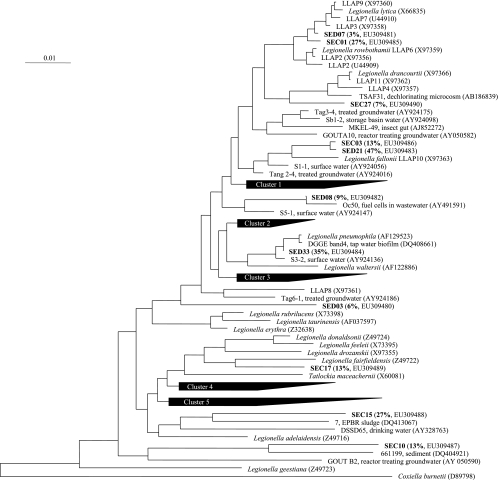
The Legionella spp. present before the proliferation of L. pneumophila (sample collected on 22 June during phase 1) and during the proliferation (sample collected on 13 July during phase 2) were identified after cloning and sequencing of 16S rRNA genes. The 16 sequences obtained from the first sample were coded SEC, and the 34 sequences obtained during phase 2 were coded SED. All 16S rRNA gene sequences had the greatest similarity to 16S rRNA gene sequences of Legionella spp., confirming the specificity of the primers used. The percentages of similarity with the closest match in the GenBank database, as given by the BLASTN program, ranged from 94 to 99%; low similarity percentages were obtained for both libraries. A phylogenetic tree was built with 16S rRNA gene sequences from described Legionella species and from the closest environmental clones (Fig. 4). The diversity of Legionella spp. was high before the proliferation of L. pneumophila, with phylotypes differently positioned on the phylogenetic tree and distantly related. The dominant species were close to L. fallonii (renamed after Legionella-like amoebal pathogen [LLAP] 10), L. fairfieldensis, L. lytica, and other LLAPs. Several sequences (SEC10, SEC15, and SED03) were only distantly related to described species and were associated with uncultured Legionella spp. from drinking water, groundwater, sludge, or sediment. In the sample collected in July, L. fallonii and L. pneumophila were the main species recovered, with, respectively, 47 and 33% of the sequences. Positioning on the phylogenetic tree was not directly related to positioning on the SSCP fingerprints, since SSCP analysis was based on a smaller fragment of the 16S rRNA gene (around 200 bp). However, peak A gathered L. lytica and other LLAPs, as well as L. pneumophila, and peak B could be identified as L. fallonii. The other peaks corresponded mainly to environmental clones. Peak B dominated in July and August and was present from April to September.
FIG. 4.
Phylogenetic tree showing positioning of Legionella spp. present in a cooling tower facility prior to L. pneumophila proliferation (phase 1, SEC clones) and during its proliferation (phase 2, SED clones). The percentage of each phylotype in the corresponding library is shown in parentheses. Cluster 1 includes L. moravica (Z49729), L. quateirensis (Z49732), L. worsleiensis (Z49739), and L. shakespearei (Z49736). Cluster 2 includes L. anisa (X73394), L. parisiensis (Z49731), L. dumoffii (Z32637), L. gormanii (Z32639), L. cherrii (X73404), L. wadsworthii (Z49738), L. steigerwaltii (Z49737), L. tucsonensis (Z32644), and L. bozemanae (Z49719). Cluster 3 includes L. cincinnatiensis (Z49721), L. santicrucis (Z49735), L. longbeachae (AY444741), and L. sainthelensi (X73399). Cluster 4 includes L. jamestowniensis (X73409), L. jordanis (X73396), L. brunensis (X73403), L. birminghamensis (Z49717), and L. quinlivanii (Z49733). Cluster 5 includes L. israelensis (Z32640), L. nautarum (Z49730), L. oakridgensis (X73397), L. impletisoli (AB233209), L. yabuuchiae (AB233212), L. busanensis (AF424887), L. gresilensis (AF122883), and L. beliardensis (AF122884).
Identification of amoebae.
Eukaryotic diversity was analyzed in the sample from July with universal primers for the domain Eukarya. Of the 14 sequences analyzed, 6 were close to sequences from Ochromonas, a golden brown alga found mostly in freshwater, and two were associated with amoebae. For the first amoebal sequence, the closest relatives belonged to the genera Plantyamoeba and Vannella. The second sequence corresponded to Acanthamoeba.
DISCUSSION
This study gathers qualitative and quantitative data on the dynamics of Legionella species and of dominant bacteria in a cooling tower facility over a 9-month period which included several weeks when L. pneumophila proliferated. Previous studies of Legionella in cooling towers reported quantification of Legionella spp. and L. pneumophila, but to our knowledge, none of them followed the dynamics of the Legionella population in relation to the bacterial microflora in the same cooling tower system.
During the spring and summer, Legionella sp. concentrations varied between 4.5 and 5.5 log GU liter−1. The maximal concentrations of L. pneumophila were obtained in July and August and were between 4.5 and 5 log GU liter−1. These concentrations are within the range of values previously reported for cooling towers (36). The high level of L. pneumophila obtained when the plant was restarted in October may be due to fouling of the network and considerable biofilm development during the shutdown period. This result fits in with the fact that cooling towers are implicated in outbreaks of legionellosis, particularly at startup or during construction (7, 10). It has been suggested that there may be a relationship between high Legionella counts in cooling towers and the occurrence of outbreaks of legionellosis (31). In this study, for similar Legionella sp. concentrations, some samples were highly contaminated with L. pneumophila, while in others the pathogenic bacterium was not detected. These results reinforce the existing assumption that the assessment of health risks from cooling towers cannot be reliably based upon single and infrequent Legionella tests (9, 24).
Combining biocides and biodispersants did reduce the concentration of Legionella spp. and of L. pneumophila. However, this effect was transient. Chemical treatment used to control Legionella in human-made water systems does not lead to total eradication of the bacterium, and recolonization occurs as soon as the treatment is interrupted (34). Legionellae are, in fact, protected inside amoebae and in biofilms and can proliferate again, recontaminating the water once the biocides lose their effect (21, 34). Amoebae present during the proliferation of L. pneumophila were identified after amplification of the 18S ribosomal DNA with primers universal for the domain Eukarya. Sequences close to those of Plantyamoeba and Vannella were obtained. These flagellate amoebae are frequently found in freshwater or seawater and are affiliated with Legionella (32). Acanthamoeba was also present. This amoeba is commonly isolated from Legionella-contaminated plumbing systems (32) and is an important host of L. pneumophila in water (7). The survival of L. pneumophila within Acanthamoeba cells during biocide treatment has been reported (21), and the presence of this amoeba could explain why the effect of biocides on Legionella abundance was only transient.
The treatment reduced the concentration of Legionella spp. to such a low level that it was not possible to analyze their diversity. However, when the population grew back to its initial level, the Legionella spp. were the same as prior to treatment. This shows clearly that even if the treatment induced a change in the structure of the Legionella population, its effect was only transient. Concerning dominant bacteria, a major disruption of the community structure due to biocide and biodispersant treatment was observed (samples collected on 29 July and 1 August, i.e., 1 and 4 days after treatment). The differences observed between the other SSCP bacterial fingerprints may relate to the origin of the cells. The similar fingerprints showing a low diversity may correspond to planktonic bacteria present in all samples, and the increase in bacterial diversity may originate from pieces of biofilm detached from the surfaces. This is congruent with the fact that when the biofilm was greater in size, the diversity increased (data not shown). It would be interesting to prove this in a subsequent experiment by comparing the bacterial diversity in water filtered through 10-μm filters with that obtained with 0.45-μm filters. Overall, it appears that the modification of the bacterial flora was probably due, in some cases, to circulating pieces of biofilm, resulting in a greater diversity in the SSCP fingerprints, while in other cases it resulted from chemical treatments, which led to very disrupted fingerprints (Fig. 3).
The suitability of primers LEG-225 and LEG-858 for detecting Legionella spp. in water has been reported previously (12, 13, 27, 35). The present study has shown that these primers can also be used to analyze Legionella diversity by SSCP. Concerning the phylogenetic positioning of Legionella spp., some relationships in the phylogenetic tree were supported moderately by bootstrap values, as previously observed by Carvalho et al. (13).
The high diversity of Legionella spp. in cooling towers has been clearly demonstrated (Fig. 4). This is an important result because the diversity of Legionella spp. in the environment, and especially in cooling tower systems, is poorly documented (28). Several sequences were close to those retrieved from treated surface water supplies and treated groundwater supplies in The Netherlands (clones Tag and S in Fig. 4) (35). Furthermore, LLAPs were particularly well represented in the cooling tower network studied. L. fallonii, which was constantly present in the system and dominated during the summer, was only described fairly recently (1) and has as its type strain LLAP-10T. A recent study of Dutch tap water installations also reported a large proportion of LLAPs (16). Those protozoonotic bacilli, which initially were isolated in coculture with protozoa, were named LLAPs because of their ability to infect and multiply intracellularly within amoebae in the same way that legionellae do (1, 2). Overall, a large proportion of sequences determined in the present study belong to as-yet-uncultured legionellae, a result which highlights the discrepancy between described Legionella species and the Legionella species present in the environment, including human-made systems.
L. pneumophila was one of the two dominant Legionella species during the summer (13 July). However, it was not detected by sequencing at the end of June (22 June). This rapid proliferation of L. pneumophila, in less than 3 weeks, may correspond to an increase in the “background” concentration of L. pneumophila in the environment during the summer, leading to the contamination of the system through water or air. A summer and autumn peak in incidence has been described as a characteristic epidemiological feature of Legionnaires' disease in Europe (11). Furthermore, a seasonal pattern of infection has been reported by Fliermans et al. (18), who injected guinea pigs with sample water collected monthly from a thermally altered lake, the highest frequency of infection by L. pneumophila occurring during the summer months. Such a seasonal variation may well be due to the facts that Legionella spp. multiply faster in the warmer waters of summer and that the greater use of cooling towers in summer provides opportunities for dissemination (11). During the investigation of Legionella colonization in 31 cooling towers in South Australia, it was found that between 60 and 75% of the cooling towers studied were colonized by Legionella during the summer months but only 20 to 30% were colonized during the winter (8). In the present study, the increase in L. pneumophila concentrations, concomitant with stability in the Legionella concentration, indicates that there must have been a decrease in the concentrations of other Legionella species during the same period. Furthermore, peak B dominated the SSCP fingerprints in phase 2, whereas it was nondominant in phase 1. This means that not only did L. pneumophila become one of the dominant species during phase 2, but the relative proportions of other Legionella species also changed significantly. In particular, peaks C and D disappeared from the SSCP fingerprints and the apparent diversity decreased. Factors which might have led to these important changes in the structure of the Legionella population still remain to be identified, i.e., the antagonistic relationships between certain Legionella species and L. pneumophila, differences in temperature growth ranges, changes in amoebal hosts, etc. The results obtained thus open new perspectives for future research. It would also be interesting to analyze Legionella diversity during the proliferation of L. pneumophila in other cooling towers in order to ascertain whether the dynamics observed in the present study also occur in other systems.
The objective of this study was to determine whether the different dynamics of bacteria and of Legionella spp. were correlated in cooling towers. A quantitative correlation was found in some cases (a decrease in ATP levels simultaneous with a decrease in the Legionella sp. concentration after chemical treatment), but it was not qualitative. In particular, no change in the structure of the bacterial community was observed during the proliferation of L. pneumophila. Bacterial diversity was modified by the chemical treatment or the development of biofilms but did not depend on the time period. In contrast, variations in the Legionella population structure were observed over time, with significant changes occurring at the end of June, following a stable phase from mid-May to mid-June. In a previous study of microbial communities in a wide range of aquatic samples containing L. pneumophila, no relationship was found between the occurrence of L. pneumophila and the associated microbiota (15). It was observed, however, that the occurrence of L. pneumophila was possible within a certain range of species richness and diversity. The authors concluded that the relationship between the occurrence of L. pneumophila and the bacteriological characteristics of water is complex and that it may therefore be interesting to concentrate on specific groups of microorganisms. In this study, we used a different approach, focusing our investigation on one cooling tower facility over several months, but we also found no relationship between the dynamics of the Legionella population during the proliferation of L. pneumophila and the dynamics of the dominant bacteria in the system. It may be that the factors that regulate the occurrence of Legionella are different from those that govern overall bacterial populations in cooling tower systems (23).
This study has provided new input on the diversity of Legionella spp. in cooling towers, showing that structural changes in the Legionella population are linked to the time of year, displaying stable periods but also a rapid major shift during the proliferation of L. pneumophila. This study has also demonstrated the application of molecular techniques which enabled us to conduct a comprehensive study of the dynamics of Legionella in relation to the bacterial community as a whole, gathering data on abundance and population structures which cannot easily be obtained by culture-dependent approaches. Controlling the Legionella risk in cooling towers requires exact knowledge of which constituents of the microbial community play a role in the proliferation of the pathogenic species, as well as the importance of these microorganisms compared to abiotic parameters such as the temperature, the mineral composition of the water, or the structure of the network. In this perspective, this study has demonstrated the value of studying the dynamics of the microflora rather than using one-off analyses.
Acknowledgments
This work was supported by the Languedoc-Roussillon Regional Government Council (France) and the L.-R. Service for Industry, Research and the Environment (DRIRE). Bouisson Bertrand Laboratories was the project leader, its coordination ensured jointly by Transfert LR and ARIA.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Adeleke, A. A., B. S. Fields, R. F. Benson, M. I. Daneshvar, J. M. Pruckler, R. M. Ratcliff, T. G. Harrison, R. S. Weyant, R. J. Birtles, D. Raoult, and M. A. Halablab. 2001. Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov., and Legionella fallonii sp. nov.: three unusual new Legionella species. Int. J. Syst. Evol. Microbiol. 51:1151-1160. [DOI] [PubMed] [Google Scholar]
- 2.Adeleke, A. A., J. Pruckler, R. Benson, T. Rowbotham, M. A. Halablab, and B. Fields. 1996. Legionella-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert-Weissenberger, C., C. Cazalet, and C. Buchrieser. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 64:432-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Association Française de Normalization. 2006. Qualité de l'eau—détection et quantification des Legionella et/ou Legionella pneumophila par concentration et amplification génique par réaction de polymérisation en chaîne (PCR). XP T90-471. http://www.boutique.afnor.org.
- 7.Atlas, R. M. 1999. Legionella: from environmental habitats to disease pathology, detection and control. Environ. Microbiol. 1:283-293. [DOI] [PubMed] [Google Scholar]
- 8.Bentham, R. H. 1993. Environmental factors affecting the colonization of cooling towers by Legionella spp. in South Australia. Int. Biodeterior. Biodegrad. 31:55-63. [Google Scholar]
- 9.Bentham, R. H. 2000. Routine sampling and the control of Legionella spp. in cooling tower water systems. Curr. Microbiol. 41:271-275. [DOI] [PubMed] [Google Scholar]
- 10.Bentham, R. H., and C. R. Broadbent. 1995. Field trial of biocides for control of Legionella in cooling towers. Curr. Microbiol. 30:167-172. [DOI] [PubMed] [Google Scholar]
- 11.Bhopal, R. S., and R. J. Fallon. 1991. Seasonal variation of Legionnaires' disease in Scotland. J. Infect. 22:153-160. [DOI] [PubMed] [Google Scholar]
- 12.Calvo-Bado, L. A., J. A. W. Morgan, M. Sergeant, T. R. Pettitt, and J. M. Whipps. 2003. Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops. Appl. Environ. Microbiol. 69:533-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho, F. R. S., R. F. Vazoller, A. S. Foronda, and V. H. Pellizari. 2007. Phylogenetic study of Legionella species in pristine and polluted aquatic samples from a tropical Atlantic forest ecosystem. Curr. Microbiol. 55:288-293. [DOI] [PubMed] [Google Scholar]
- 14.Delbès, C., R. Moletta, and J.-J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction—single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 15.Devos, L., K. Clymans, N. Boon, and W. Verstraete. 2005. Evaluation of nested PCR assays for the detection of Legionella pneumophila in a wide range of aquatic samples. J. Appl. Microbiol. 99:916-925. [DOI] [PubMed] [Google Scholar]
- 16.Diederen, B. M. W., C. M. A. de Jong, I. Aarts, M. F. Peeters, and A. van der Zee. 2007. Molecular evidence for the ubiquitous presence of Legionella species in Dutch tap water installations. J. Water Health 5:375-383. [DOI] [PubMed] [Google Scholar]
- 17.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Fulgueiras, A., C. Navarro, D. Fenoll, J. Garcia, P. Gonzalez-Diego, T. Jimenez-Bunuales, M. Rodriguez, R. Lopez, F. Pacheco, J. Ruiz, M. Segovia, B. Baladron, and C. Pelaz. 2003. Legionnaires' disease outbreak in Murcia, Spain. Emerg. Infect. Dis. 9:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godon, J.-J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilvington, S., and J. Price. 1990. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 22.Kirrage, D., G. Reynolds, G. E. Smith, and B. Olowokure. 2007. Investigation of an outbreak of Legionnaires' disease: Hereford, UK 2003. Respir. Med. 101:1639-1644. [DOI] [PubMed] [Google Scholar]
- 23.Kusnetsov, J. M., P. J. Martikainen, H. R. Jousimies-Somer, M.-L. Vaisanen, A. I. Tulkki, H. E. Ahonen, and A. I. Nevalainen. 1993. Physical, chemical and microbiological water characteristics associated with the occurrence of Legionella in cooling tower systems. Water Res. 27:85-90. [Google Scholar]
- 24.Leoni, E., G. De Luca, P. P. Legnani, R. Sacchetti, S. Stampi, and F. Zanetti. 2005. Legionella waterline colonization: detection of Legionella species in domestic, hotel and hospital hot water systems. J. Appl. Microbiol. 98:373-379. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak, B., J. Cole, T. Lilburn, C. J. Parker, P. Saxman, J. Stredwick, G. M. Garrity, B. Li, G. Olsen, S. Pramanik, T. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragull, S., M. Garcia-Nunez, M. L. Pedro-Botet, N. Sopena, M. Esteve, R. Montenegro, and M. Sabria. 2007. Legionella pneumophila in cooling towers: fluctuations in counts, determination of genetic variability by pulsed-field gel electrophoresis (PFGE), and persistence of PFGE patterns. Appl. Environ. Microbiol. 73:5382-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelton, B. G., W. D. Flanders, and G. K. Morris. 1994. Legionnaire's disease outbreaks and cooling towers with amplified Legionella concentrations. Curr. Microbiol. 28:359-363. [Google Scholar]
- 32.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 33.Storey, M. V., N. Ashbolt, and T. A. Stenstrom. 2004. Biofilms, thermophilic amoebae and Legionella pneumophila—a quantitative risk assessment for distributed water. Water Sci. Technol. 50:77-82. [PubMed] [Google Scholar]
- 34.Thomas, V., T. Bouchez, V. Nicolas, S. Robert, J. F. Loret, and Y. Lévi. 2004. Amoebae in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence. J. Appl. Microbiol. 97:950-963. [DOI] [PubMed] [Google Scholar]
- 35.Wullings, B. A., and D. van der Kooij. 2006. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15°C. Appl. Environ. Microbiol. 72:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaradou, D. F., S. Hallier-Soulier, S. Moreau, F. Poty, Y. Hillion, M. Reyrolle, J. Andre, G. Festoc, K. Delabre, F. Vandenesch, J. Etienne, and S. Jarraud. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zemb, O., B. Haegeman, J. P. Delgenes, P. Lebaron, and J.-J. Godon. 2007. SAFUM: statistical analysis of SSCP fingerprints using PCA projections, dendrograms and diversity estimators. Mol. Ecol. Notes 7:767-770. [Google Scholar]