Abstract
Brachyspira hyodysenteriae is an anaerobic spirochete and the etiologic agent of swine dysentery. The genome of this spirochete contains a mitomycin C-inducible, prophage-like gene transfer agent designated VSH-1. VSH-1 particles package random 7.5-kb fragments of the B. hyodysenteriae genome and transfer genes between B. hyodysenteriae cells. The chemicals and conditions inducing VSH-1 production are largely unknown. Antibiotics used in swine management and stressors inducing traditional prophages might induce VSH-1 and thereby stimulate lateral gene transfer between B. hyodysenteriae cells. In these studies, VSH-1 induction was initially detected by a quantitative real-time reverse transcriptase PCR assay evaluating increased transcription of hvp38 (VSH-1 head protein gene). VSH-1 induction was confirmed by detecting VSH-1-associated 7.5-kb DNA and VSH-1 particles in B. hyodysenteriae cultures. Nine antibiotics (chlortetracycline, lincomycin, tylosin, tiamulin, virginiamycin, ampicillin, ceftriaxone, vancomycin, and florfenicol) at concentrations affecting B. hyodysenteriae growth did not induce VSH-1 production. By contrast, VSH-1 was detected in B. hyodysenteriae cultures treated with mitomycin C (10 μg/ml), carbadox (0.5 μg/ml), metronidazole (0.5 μg/ml), and H2O2 (300 μM). Carbadox- and metronidazole-induced VSH-1 particles transmitted tylosin and chloramphenicol resistance determinants between B. hyodysenteriae strains. The results of these studies suggest that certain antibiotics may induce the production of prophage or prophage-like elements by intestinal bacteria and thereby impact intestinal microbial ecology.
In the United States, various antimicrobials are added to feed to prevent diseases and to promote growth or to enhance the feeding efficiency of swine (14, 28). Antibiotics commonly used in feed for swine include tetracyclines, carbadox, macrolides, and lincosamides (19). At higher concentrations, carbadox, lincomycin, tylosin, and tiamulin are added to feed or drinking water for the treatment of swine intestinal diseases, notably swine dysentery (25, 28). In Australia and some European countries, nitroimidazole antibiotics, such as metronidazole, ronidazole, and dimetridazole, have been used to treat swine dysentery (22, 25; D. Trott, personal communication), although legislation in several countries has restricted the use of these antibiotics in food animals (2, 44).
The etiologic agent of swine dysentery is the anaerobic spirochete Brachyspira hyodysenteriae. Within their genome, B. hyodysenteriae cells carry a mitomycin C-inducible prophage-like element, designated VSH-1 (30, 31, 63). Unlike traditional prophages, VSH-1 particles contain random 7.5-kb fragments of the B. hyodysenteriae genome. VSH-1 head, tail, and lysis genes total at least 16.3 kb of DNA (38). Consequently, an individual VSH-1 particle is incapable of lytic growth, and there are no bioassays (i.e., plaque formation) for measuring VSH-1 production. Although VSH-1 particles do not self-propagate, they transfer genes between B. hyodysenteriae cells (31, 54). These unusual properties of VSH-1 are shared by similar elements in species of Rhodobacter, Methanococcus, and Desulfovibrio (37, 52). VSH-1 and the other elements have collectively been designated gene transfer agents (GTAs) (15).
One goal of our research is to identify environmental inducers of VSH-1 production. B. hyodysenteriae cells are undoubtedly exposed to antimicrobials in the swine intestinal tract or in the farm environment. In these investigations, different antibiotics were tested as inducers of VSH-1. Carbadox and metronidazole were potent VSH-1 inducers. VSH-1 virions induced by these antibiotics transferred chloramphenicol resistance and tylosin resistance between B. hyodysenteriae cells.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. hyodysenteriae strains B78T (type strain), B204, and A203 were used in these studies. Strain B78T is sensitive (MIC = 4 to 8 μg/ml) to the macrolide antibiotic tylosin (33). Strain B204 is naturally resistant to tylosin (Tyr) (MIC > 256 μg/ml) due to a 23S rRNA mutation, T in place of A2058 (33). Strain A203 is chloramphenicol resistant (Cmr) (MIC > 10 μg/ml) and was derived from strain B204 by inserting a cat cassette into the flaA1 gene (31, 46).
B. hyodysenteriae cells were routinely grown at 38°C in stirred 10-ml BHIS broth cultures beneath an initial N2-O2 (99:1) culture atmosphere (53). BHIS broth is anaerobically prepared brain heart infusion broth supplemented with 10% (vol/vol) heat-treated calf serum. (For unknown reasons, the BD Bacto brand of BHI was superior to BHI obtained from BBL for optimum induction of VSH-1.) All cultures were in the exponential phase of growth at the time of use (optical density at 620 nm [OD620] = 0.5 to 1.0; 18-mm path length). VSH-1 particles for antibiotic resistance transfer experiments were obtained from B. hyodysenteriae A203 cells cultured in NT broth, a serum-free, low-protein-content medium essential for VSH-1 purification (31). Culture media for strain A203 cells contained chloramphenicol (10-μg/ml final concentration).
Identification of VSH-1 inducers and their effects on cell growth.
B. hyodysenteriae B204 cultures in early exponential growth phase (OD620 = 0.5) in BHIS broth were treated with potential VSH-1 inducers. The culture ODs were monitored over time to detect differences in cell growth between treated and untreated (control) cultures. Specific treatments impairing or inhibiting growth (defined below) were subsequently tested for the ability to stimulate VSH-1 hvp38 transcription.
Antibiotics tested for their growth effects were as follows (range of final concentrations in μg/ml of BHIS culture): carbadox (0.005 to 0.2), metronidazole (0.005 to 2), chlortetracycline (5 to 50), lincomycin (10 to 100), tylosin (500 to 1,000), tiamulin (0.02 to 0.2), virginiamycin (0.1 to 15), ampicillin (0.5 to 30), ceftriaxone (5 to 100), vancomycin (500 to 1,000), and florfenicol (2 to 20). Within the concentration ranges, two- or fourfold dilutions of antibiotics were tested. Stock solutions or suspensions (50 to 100×) were prepared in sterile water, because ethanol was a weak inducer of VSH-1. In each experiment, parallel control cultures were either untreated or treated with 10 μg mitomycin C/ml (30).
RNA purification.
Six hours after treatment with potential inducers of VSH-1, samples (2 to 5 ml) of B. hyodysenteriae cultures were diluted 1:3 in RNA Protect (Qiagen, Valencia, CA). After 5 min at room temperature, spirochete cells were harvested by centrifugation (5,000 × g; 5 min), and the cell pellets were stored frozen (−85°C) for up to 2 weeks. Total RNA was extracted from the cell pellets by using RNAeasy minicolumn kits following the manufacturer's instructions (Qiagen). DNA was removed by treating the RNA preparations with Turbo DNase following the manufacturer's instructions (Applied Biosystems/Ambion, Foster City, CA). RNA concentrations were estimated spectrophotometrically (OD260) by using microcapillary cuvettes and a Beckmann DU-650 spectrophotometer and were based on standard conversion values (49). This protocol yielded 1 to 2 μg of RNA/ml of culture.
The purity and quality of RNA preparations were assessed by examining the banding patterns after gel electrophoresis of 300 ng of RNA (4% Nu-Sieve agarose; 1× Tris-borate-EDTA). RNA solutions were diluted in nuclease-free water (Integrated DNA Technologies, Coralville, IA) to a final concentration of 5 ng/ml, dispensed in aliquots, and frozen (−85°C) until they were used.
Identification of VSH-1 inducers by QRT-PCR of hvp38 transcription.
Quantitative real-time reverse transcriptase PCR (QRT-PCR) was used to detect increases in mRNA transcribed from the VSH-1 hvp38 gene. The hvp38 gene encodes the VSH-1 major capsid protein, and increased hvp38 transcription is an early event in VSH-1 induction by mitomycin C (38, 39). QRT-PCR was also used to determine the 16S rRNA content of each RNA preparation, and this value was used as an internal reference standard against which relative changes in hvp38 mRNAs were calculated.
Based on the hvp38 sequence (GenBank accession no. AY971355), the primers for amplifying hvp38 mRNA were 38F (5′-TTCAAGACTTGGGCTTTTAAGAG) and 38R (5′-TTGGTTTGGCACTTAAATCAAC), and the probe was 5′-GTGTTTGTGCTTCCATAAAGTTCTGCATCTGT. Based on the B. hyodysenteriae rrs sequence (GenBank accession no. M57741), the primers for amplifying 16S rRNA were 16F (5′-TCATGGCCCTTATGTCCAG) and 16R (5′-CGAACTGAGGCAACTTTTTTG), and the probe was 5′-CACGTGCTACAATGGCAAGTACAAAGAGA. Probes were labeled at the 3′ end with the quencher dye TAMRA (6-carboxytetramethylrhodamine). The hvp38 probe was labeled at the 5′ end with VIC (PE Biosystems, Foster City, CA), and the 16S rRNA probe was labeled with TET (6-carboxy-2′,4,7,7′-tetracholorofluorescein). Primers and probes were designed using Primer Express software (PE Applied Biosystems, Foster City, CA) and were synthesized by Integrated DNA Technologies.
For QRT-PCR analysis, 1 pg of total RNA was used for 16S rRNA amplification and 1 ng for hvp38 mRNA amplification. A reaction mixture contained 2 μl of target RNA, 15 pmol of each primer, and 0.126 pmol of probe added to a TaqMan One Step Master Mix reagent kit (Applied Bioystems) in a final volume of 25 μl.
Amplification and detection were carried out in optical-grade 96-well plates in an ABI Prism 7700 sequence detection system (PE Biosystems). Following an initial RT-PCR consisting of 30 min of incubation at 48°C for converting mRNA to cDNA, amplification of cDNA was carried out with one cycle of 95°C for 10 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s. The final extension was carried out at 72°C for 2 min, followed by cooling of samples to 25°C.
Copies of the B. hyodysenteriae rrs (16S rRNA) and hvp38 genes cloned in pBlueScript II SK(−) (Stratagene, La Jolla, CA) were used as templates to generate RNA gene transcripts using the RNAMaxx high-yield transcription kit (Stratagene). Standard curves were generated for estimating 16S rRNA and hvp38 mRNA levels in cells by linear regression analysis.
In each experiment, RNA levels were assayed in triplicate for at least two cultures. Control amplifications containing known quantities (copy numbers) of rrs and hvp38 RNAs were included. Control amplifications lacking reverse transcriptase were included for each RNA preparation to confirm that B. hyodysenteriae DNA was absent from the sample.
Identification of VSH-1 inducers and detection of VSH-1 7.5-kb DNA.
B. hyodysenteriae cells in cultures (5 ml) exposed to potential VSH-1 inducers were harvested by centrifugation and resuspended in 0.35 ml of 50 mM Tris, pH 8.0, containing 0.05 mM EDTA. Lysozyme (0.1 ml of a 10-mg/ml solution in 0.25 mM Tris, pH 8) was added, and the cell suspension was placed on ice for 45 min. Proteinase K (0.1 ml of a solution containing 0.1 mg proteinase K, 0.5% sodium dodecyl sulfate, 50 mM Tris, pH 7.5, 10 mM EDTA, 50 μM CaCl2) was then added.
After incubation at 50°C for 1 h, the solution was extracted twice, first with an equal volume of Tris-buffered phenol-chloroform-isoamyl alcohol (Sigma-Aldrich, St. Louis, MO) and then with CHCl3. Phase Lock 1 tubes (Eppendorf) were used to separate the extraction phases. DNA in the final aqueous phase was precipitated with 3 M sodium acetate-ethanol (49), air dried, and dissolved in 0.5 ml of 50 mM Tris, pH 7.4, containing 1 mM EDTA. After 1 h of RNase (RNAce it; Stratagene) digestion, the DNA was again solvent extracted, precipitated with acetate-ethanol, and dissolved in 50 μl of Tris-EDTA buffer, as described above. This method yielded 5 to 25 μg of DNA, as determined spectrophotometrically using microcapillary cuvettes (Beckman DU650 spectrophotometer).
Identification of VSH-1 inducers by electron microscopy.
B. hyodysenteriae cells exposed to potential inducers of VSH-1 were examined by electron microscopy to detect VSH-1 particles as described previously (30). Up to 50 microscope fields containing well-stained spirochete cells were examined. Negative control cultures (untreated) and positive control cultures (treated with mitomycin C or carbadox) were examined in parallel.
VSH-1 transfer of tylosin and chloramphenicol resistances.
NT broth cultures of B. hyodysenteriae strain A203 (Cmr Tyr; OD620 = 0.5) were treated with mitomycin C (20 μg/ml), metronidazole (1 μg/ml), or carbadox (1 μg/ml) to obtain VSH-1 particles. Five hours after induction, VSH-1 virions were harvested from 400-ml cultures and resuspended in SM buffer (31). Purified virions (10 μl containing 1 to 2.5 μg of DNA) were added to 10-ml BHIS broth cultures of B. hyodysenteriae B204 (Cms Tyr) and strain B78T (Cms Tys) in exponential growth phase (∼5 × 107 cells/ml). After overnight incubation at 39°C, the cultures (OD620 = 1.0; 3 × 108 to 5 × 108 CFU/ml) were transferred into a Coy anaerobic chamber inflated with a gas mixture of 85% N2-10% CO2-5% H2. The cultures were serially diluted 10-fold, and 0.1 ml of the dilutions was plated onto Trypticase soy blood (TSB), TSB plus tylosin (10-μg/ml final concentration), and TSB plus chloramphenicol (10 μg/ml) agar. TSB agar plates (54) had been placed into the Coy chamber 18 to 24 h before they were used. After 4 days of incubation at 39°C in the chamber, B. hyodysenteriae antibiotic-resistant colonies (hemolytic zones) were counted. Cmr genotypes were confirmed by PCR detection of the cat gene (54). Tyr genotypes were confirmed by sequence analysis of a PCR-amplified region of the 23S rRNA gene containing the A2058→T mutation associated with resistance to macrolides (33). Control B. hyodysenteriae cultures for assessing spontaneously resistant mutants did not receive VSH-1.
RESULTS
Antibiotic concentrations affecting B. hyodysenteriae growth.
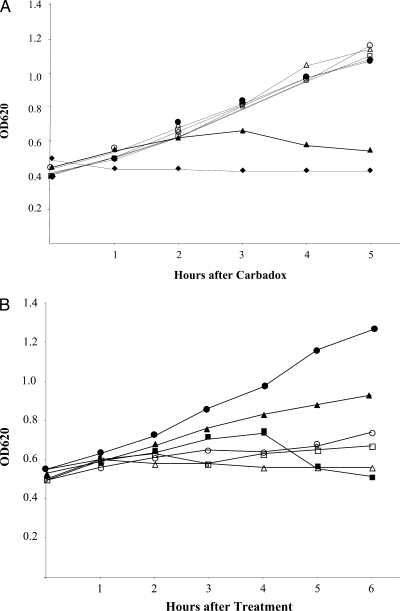
B. hyodysenteriae B204 growth was inhibited (no increase in the culture OD) by 2 μg carbadox/ml (Fig. 1A). At 0.5 μg carbadox/ml, bacterial growth was first impaired (a slow increase in the OD), and then the culture OD declined between 3 and 5 h after exposure to the antibiotic. This decline in the OD620 in cultures treated with carbadox resembled that of a mitomycin C-treated culture during VSH-1 induction (30). Concentrations of 0.2 μg carbadox/ml or lower were subinhibitory for B. hyodysenteriae growth in BHIS broth (Fig. 1A).
FIG. 1.
(A) Carbadox effects on B. hyodysenteriae B204 growth. Carbadox was added (0 h) to cultures at final concentrations (μg/ml) of 0 (open circles), 0.005 (open triangles), 0.02 (open diamonds), 0.05 (open squares), 0.2 (closed circles), 0.5 (closed triangles), and 2.0 (closed diamonds). (B) Effects on B. hyodysenteriae B204 growth of no addition (control; closed circles), mitomycin C (10 μg/ml; closed squares), metronidazole (0.5 μg/ml; closed triangles), virginiamycin (0.5 μg/ml; open circles), tiamulin (0.1 μg/ml; open squares), and H2O2 (300-μM final concentration; open triangles).
In similar growth studies, different concentrations of antibiotics and 300 μM H2O2 were found to impair or inhibit B. hyodysenteriae growth in BHIS broth (Fig. 1B). Among these, only mitomycin C consistently produced a decline in the culture OD620. B. hyodysenteriae cultures exposed to these and other concentrations were examined for induction of hvp38 transcription.
Antibiotic induction of the GTA VSH-1.
Chlortetracycline, lincomycin, tylosin, tiamulin, virginiamycin, ampicillin, ceftriaxone, vancomycin, and florfenicol, at the tested concentrations, did not stimulate hvp38 transcription (Table 1) and thus were not inducers of VSH-1. Chlortetracycline, lincomycin, tylosin, tiamulin, and virginiamycin are approved medicated-feed additives for treating swine dysentery, for promoting growth/feeding efficiency, or for both applications (28). Florfenicol and ampicillin are approved for the treatment of swine diseases (14).
TABLE 1.
Antimicrobial compounds and conditions inducing VSH-1
| Treatment | Antimicrobial concn
|
hvp38 transcription (fold increase)a | VSH-1 productionb
|
||
|---|---|---|---|---|---|
| μg/ml | μM | DNA | EM | ||
| Control (untreated) | 1 | − | − | ||
| Mitomycin C | 10 | 30 | 260 | + | + |
| Metronidazole | 0.5 | 2.5 | 720 | + | + |
| 0.05 | 0.25 | 50 | + | − | |
| 0.005 | 0.025 | 7 | − | − | |
| Carbadox | 0.5 | 1.9 | 290 | + | + |
| 0.05 | 0.19 | 30 | + | + | |
| 0.005 | 0.019 | 14 | − | − | |
| H2O2 | 300 | 100 | + | + | |
Increase in hvp38 transcription over untreated (control) cultures. Levels of hvp38 mRNA in control cultures were arbitrarily standardized to a value of 1. An increase of 5-fold or greater in each of triplicate cultures undergoing a particular treatment was considered significant. The values are mean values for at least two experiments. The following antimicrobials at the indicated concentrations (μg/ml) were not found to stimulate hvp38 transcription: chlortetracycline (5, 10, 50), lincomycin (10, 50, 100), tylosin (500, 1,000), tiamulin (0.02, 0.1), virginiamycin (3), ampicillin (3, 15), ceftriaxone (30), vancomycin (500, 1,000), and florfenicol (2, 10, 20).
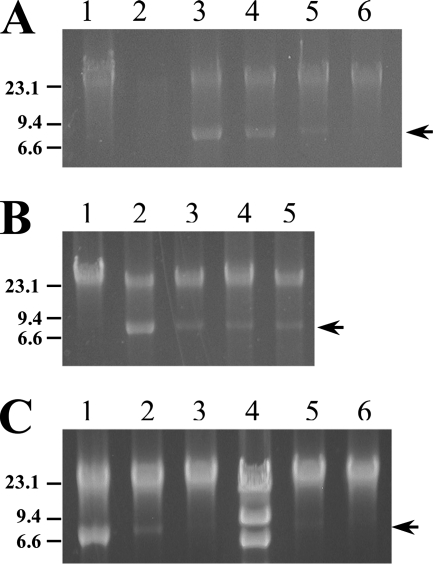
Transcription of hvp38 in B. hyodysenteriae cultures treated with carbadox (0.5 μg/ml) or metronidazole (0.5 μg/ml) increased 290- and 720-fold, respectively, over that in untreated cultures (Table 1). VSH-1 production in treated cultures was confirmed by detecting both VSH-1-associated 7.5-kb DNA fragments (Table 1 and Fig. 2) and VSH-1 particles (Table 1 and Fig. 3). Carbadox at 2 μg/ml inhibited growth (Fig. 1A), and intact DNA could not be obtained from the cultures, presumably an indication of DNA degradation (Fig. 2).
FIG. 2.
Induction of VSH-1 7.5-kb DNA in B. hyodysenteriae B204 cultures. (A) DNAs from carbadox-treated cultures. Lane 1, control (untreated) culture; lane 2, carbadox (2 μg/ml)-treated culture; lane 3, carbadox (0.5 μg/ml); lane 4, carbadox (0.1 μg/ml); lane 5, carbadox (0.05 μg/ml); lane 6, carbadox (0.01 μg/ml). The absence of DNA in lane 2 is likely due to DNA degradation in spirochete cells. (B) DNAs from cultures treated with various antimicrobials. Lane 1, control (untreated) culture; lane 2, carbadox (0.5 μg/ml); lane 3, carbadox (0.1 μg/ml); lane 4, H2O2 (200 μM); lane 5, H2O2 (300 μM). (C) DNAs from B. hyodysenteriae cultures treated with carbadox or metronidazole. Lane 1, carbadox (0.5 μg/ml); lane 2, carbadox (0.05 μg/ml); lane 3, carbadox (0.005 μg/ml); lane 4, HindIII-digested lambda size markers; lane 5, metronidazole (0.05 μg/ml); lane 6, metronidazole (0.005 μg/ml). DNA sizes estimated from markers (kb) are indicated to the left of the panels. The arrows on the right of the panels indicate the positions of VSH-1-associated DNA (7.5 kb). Each lane contained 0.5 μg of DNA extracted from a B. hyodysenteriae culture.
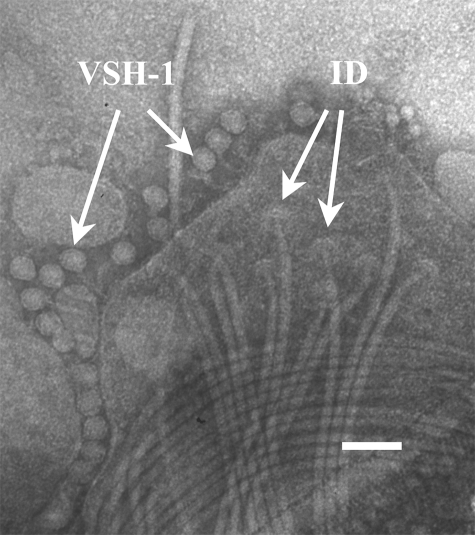
FIG. 3.
Transmission electron micrograph of VSH-1 particles (VSH-1) surrounding a disrupted B. hyodysenteriae B204 cell from a culture treated with 0.5 μg carbadox/ml. Flagella and flagellum insertion disks (ID) are visible. Phosphotungstic acid stain (2%; pH 6.5). Marker bar = 0.1 μm.
Transcription of hvp38 was induced by carbadox or metronidazole at concentrations ranging between 0.005 and 0.5 μg/ml (Table 1). VSH-1-associated DNA was detected in B. hyodysenteriae cultures treated with either antibiotic at concentrations between 0.05 and 0.5 μg/ml (Fig. 1 and 2), and particles were detected in cultures treated with 0.05 μg carbadox/ml. In cultures treated with 0.005 μg/ml of either carbadox or metronidazole, VSH-1 production was not directly detected, and if it occurred, was likely below the detection limits of the two assays.
Mitomycin C is commonly used to induce prophages and was used in experiments leading to the discovery of VSH-1 (30). Hydrogen peroxide can also induce bacteriophage production (20, 56, 58) and induces VSH-1 in vitro and possibly in the swine intestinal tract (39). Transcription of hvp38 in B. hyodysenteriae cells treated with mitomycin C (10 μg/ml) or H2O2 (300 μM) increased 260-fold and 100-fold, respectively, over that in untreated cultures (Table 1). VSH-1-associated 7.5-kb DNA and particles were detected in H2O2-treated cultures (Table 1 and Fig. 2B), although VSH-1 particles were produced at much lower levels in these cultures than in carbadox-treated cultures. A previous study used semiquantitative RT-PCR and Northern slot blot techniques and demonstrated that H2O2 and mitomycin C induce transcription of hvp38 and VSH-1 production (39). The present results support the findings of that study. The QRT-PCR method used in this study, however, is a more sensitive assay of hvp38 transcription than the Northern slot blot technique and provides quantitative measurements useful for comparing inducing treatments. While electron microscopy is the most reliable method for detecting VSH-1 production, it is, unfortunately, the least sensitive and most labor-intensive. Attempts to develop a VSH-1 bioassay (by measuring gene transfer frequencies) in our laboratory have been unsuccessful.
Gene transfer by metronidazole- or carbadox-induced VSH-1.
To determine whether antimicrobial-induced VSH-1 particles were capable of transferring genes between B. hyodysenteriae strains, VSH-1 particles were purified from B. hyodysenteriae A203 (Tyr Cmr) cultures treated with carbadox or metronidazole. The VSH-1 particles were added to cultures of B. hyodysenteriae B78T (Tys Cms). After overnight incubation, strains resistant to either tylosin or chloramphenicol were present in cultures that received VSH-1 particles and were not detected or were beneath the limit of detection in control cultures (Table 2). Transductant cells doubly resistant to both antibiotics were not recovered.
TABLE 2.
VSH-1-mediated transduction of chloramphenicol and tylosin resistances between B. hyodysenteriae strains A203 and B78
| VSH-1 virion | No. of transductants/108 CFUa
|
|
|---|---|---|
| Tylosin resistant (10 μg/ml) | Chloramphenicol resistant (10 μg/ml) | |
| None (control) | <LODb | <LOD |
| Mitomycin C induced | 470 | 800 |
| Metronidazole induced | 405 | 390 |
| Carbadox induced | 250 | 820 |
VSH-1 virions were induced by treating NT cultures of B. hyodysenteriae strain A203 (Tyr Cmr) with mitomycin C (20 μg/ml), metronidazole (1 μg/ml), or carbadox (1 μg/ml). Purified virions (containing 1 to 2.5 μg DNA) were added to B. hyodysenteriae B78 (Tys Cms) cultures. The values represent average numbers of resistant CFU per total numbers of CFU as determined in two experiments using two VSH-1 preparations for each treatment.
LOD, limit of detection. Spontaneously occurring tylosin-resistant cells were too few for accurate estimates (i.e., 2 to 4 CFU/108 CFU). Spontaneously occurring chloramphenicol-resistant cells were not detected.
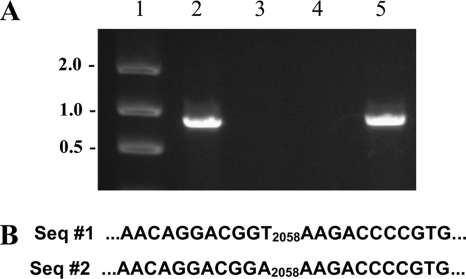
Genotype analysis of six randomly selected chloramphenicol-resistant transductants from two experiments were examined, and all contained the cat gene of strain A203 (Fig. 4A). Eight randomly selected tylosin-resistant transductants had the same A2058→T 23S rRNA gene base modification as the B. hyodysenteriae A203 strain from which VSH-1 had been induced (Fig. 4B).
FIG. 4.
Genotype analyses of antibiotic resistance determinants of B. hyodysenteriae strains. (A) PCR amplification of cat gene. Lane 1, molecular size markers (kb); lane 2, strain A203 (Cmr Tyr); lane 3, strain B204 (Cms Tyr); lane 4, strain B78 (Cms Tys); lane 5, transductant strain CM-1 (Cmr Tys). Seven other randomly selected transductant strains gave the same-size amplicon as CM-1 in the PCR assay. (B) DNA sequence analysis to detect rrl (23S rRNA gene) nucleotide change associated with tylosin (macrolide) resistance. Sequence 1 (Seq #1), strain B78 (Cms Tys); sequence 2, strain B204 (Cms Tyr) (strain A203, strain Ty1, and five other randomly selected transductant strains had the same rrl sequence).
DISCUSSION
Antimicrobials can have effects on target bacteria, in addition to their bactericidal or bacteriostatic effects. Different antibiotics have been found to modulate gene transcription in Salmonella enterica (23) and Escherichia coli (27), to induce Pseudomonas aeruginosa biofilm formation (29), and to stimulate toxin production by Staphylococcus aureus (42) and E. coli (24, 59, 61). These “collateral” effects are manifested at antibiotic concentrations subinhibitory for bacterial growth.
Although from a clinical perspective bacterial exposure to subinhibitory levels of antibiotics seems incongruous, it does occur. Antibiotic concentrations considered therapeutic for bacterial infections are subinhibitory for pathogenic bacteria with acquired resistance and for commensal species with acquired resistance or natural insensitivity to the antibiotic. Doxycycline is used at subantibacterial levels to reduce tissue destruction in periodontal disease (47). Animal feeds promoting weight gain/feeding efficiency contain antibiotic levels lower than those of feeds for disease therapy (14, 28). Explanations for the effectiveness of performance-enhancing medicated feeds are that they favor the growth/metabolism of insensitive, beneficial commensal species in the animal intestinal tract or prevent growth of subclinical sensitive pathogenic species (41).
From a genetic and evolutionary perspective, a noteworthy bacterial response to subinhibitory antibiotic exposure is the activation of mobile genetic elements carried by bacteria. These mobile elements include both chromosomally integrated conjugative elements and traditional prophages. The excision and transfer of SXT, a 100-kb chromosomally integrated conjugative element of Vibrio cholerae and a carrier of multiple-drug resistance and virulence-encoding genes, is activated by low levels of ciprofloxacin (5, 6). Tetracycline stimulates 100- to 1,000-fold the transfer of Bacteroides conjugative transposons carrying resistance to tetracycline and other antibiotics (48, 60).
Prophages of bacteria colonizing animals and humans often carry genes that are nonessential for the bacteriophage but that provide selective advantages for their bacterial host. Examples include genes encoding toxins and proteins affecting immune responses and antibiotic resistance (9, 26, 50). Stx (shiga toxin)-transducing prophages of E. coli have been induced by norfloxacin (40). Ciprofloxacin given in subtherapeutic doses increases gene transduction by E. coli Stx2 phages in the mouse gastrointestinal tract (61), although, interestingly, not in sheep rumens (16).
VSH-1 is the only known gene transfer mechanism of B. hyodysenteriae. The involvement of this GTA in the evolution of B. hyodysenteriae as a recombinant population has been proposed (57). VSH-1-like elements appear to be widely distributed among Brachyspira species, including strains in the human intestinal tract (11, 12, 30, 55). The evolutionary origins of VSH-1 are unclear, but the current and simplest explanation is that its ancestor was a fully functioning prophage (52).
Carbadox, a quinoxaline-di-N-oxide compound, and metronidazole, a 5-nitro-imidazole compound, are potent inducers of VSH-1. On a molar basis, they are 12 to 15 times more effective than mitomycin C at inducing hvp38 transcription (Table 1).
Carbadox is an effective antimicrobial for preventing and treating intestinal diseases of postweaning swine, notably swine dysentery caused by B. hyodysenteriae. Carbadox is also used as a feed additive to promote swine growth (28, 43), although to our knowledge the basis for growth promotion is unknown. Although metronidazole and related 5-nitroimidazoles are no longer used for swine applications, they are used for treating anaerobic infections of nonfood animals (44). Nitroimidazole antibiotics are commonly used to treat human intestinal diseases caused by anaerobic bacteria and parasitic protozoa (4, 21, 45).
Under anaerobic conditions, carbadox and metronidazole are chemically reduced by bacterial metabolism to products that directly interact with bacterial DNA, causing mutations and DNA strand breaks (7, 51). Thus, an early event for VSH-1 induction by carbadox and metronidazole, and by mitomycin C and H2O2, is presumed to be DNA damage leading to a RecA-centered SOS response, as reported for other bacterial species (3, 5, 6, 10, 18, 32, 62).
Carbadox seems to be generally useful for inducing prophages from various bacterial species. Carbadox induces lambda-like prophages carrying shiga toxin (stx) genes in E. coli cultures (36) and is a component of a commercial test, EMD Duopath Verotoxins (Merck catalog no 1.04144.0001), to stimulate phage-associated Stx production (1). Recently, we and a colleague (S. Casjens, personal communication) have used 0.5 μg carbadox/ml of culture to induce prophages from Shigella and Salmonella cultures. In view of its low cost (approximately 10−6 the cost of a prophage-inducing amount of mitomycin C), carbadox is a more economical alternative to mitomycin C for bacteriophage or GTA induction.
Does carbadox in medicated feeds induce production of VSH-1, other GTAs, or traditional prophages by bacteria in the intestinal tracts of swine? The answer is not known. This is a question that is worth pursuing, in view of growing awareness that prophages play significant roles in bacterial evolution and ecology (8, 13). VSH-1 involvement in B. hyodysenteriae evolution has been proposed (57), and carbadox-induced VSH-1 particles transfer tylosin resistance between B. hyodysenteriae cells in culture (Table 2). Swine dysentery, or bloody scours, is associated with passage of blood through the intestinal mucosa at sites of B. hyodysenteriae colonization (34, 35). Swine feed contains 10 to 25 g carbadox/ton (11 to 28 mg/kg) for growth promotion and 50 g/ton (55 mg/kg) for disease prophylaxis (28). In a study of carbadox pharmacokinetics, swine fed 30 mg carbadox/kg of feed had 0.03 μg carbadox/ml blood (17). In B. hyodysenteriae cultures, VSH-1 is induced by carbadox at similar concentrations (Table 1 and Fig. 1).
The findings of these studies suggest a need to evaluate VSH-1 induction and lateral gene transfer in vivo as possible collateral effects of carbadox medication. Additionally, GTA or prophage induction is a lethal event for the host bacterium and might contribute to the therapeutic or performance-enhancing properties of the antibiotic. Thus, a potentially broader impact of carbadox (and metronidazole) as a prophage inducer on intestinal microbial ecology in animals and humans deserves consideration.
Acknowledgments
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Akkaya, L., H. Atabay, B. Kenar, and M. Alisarli. 2006. Prevalence of verocytotoxigenic Escherichia coli O157:H7 on chicken carcasses sold in Turkey. Bull. Vet. Inst. Pulawy. 50:513-516. [Google Scholar]
- 2.Anonymous. 1995. EC bans use of dimetridazole in food animals. Vet. Rec. 137:230. [DOI] [PubMed] [Google Scholar]
- 3.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 102:12554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagopal, A., and C. L. Sears. 2007. Clostridium difficile: new therapeutic options. Curr. Opin. Pharmacol. 7:455-458. [DOI] [PubMed] [Google Scholar]
- 5.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 6.Beaber, J. W., and M. K. Waldor. 2004. Identification of operators and promoters that control SXT conjugative transfer. J. Bacteriol. 186:5945-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin, L., E. Preller, and B. Kowalski. 1981. Mutagenicity of quindoxin, its metabolites, and two substituted quinoxaline-di-N-oxides. Antimicrob. Agents Chemother. 20:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brüssow, H. 2007. Impact of phages on evolution of bacterial pathogenicity, p. 267-300. In M. J. Pallen, K. E. Nelson, and G. M. Preston (ed.), Bacterial pathogenomics. ASM Press, Washington, DC.
- 9.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman, F. 2002. Lateral DNA transfer. Mechanisms and consequences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Calderaro, A., G. Dettori, L. Collini, P. Ragni, R. Grillo, P. Cattani, G. Fadda, and C. Chezzi. 1998. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. J. Basic Microbiol. 38:323-335. [PubMed] [Google Scholar]
- 12.Calderaro, A., G. Dettori, R. Grillo, P. Plaisant, G. Amalfitano, and C. Chezzi. 1998. Search for bacteriophages spontaneously occurring in cultures of haemolytic intestinal spirochaetes of human and animal origin. J. Basic Microbiol. 38:313-322. [PubMed] [Google Scholar]
- 13.Canchaya, C., G. Fournous, and H. Brüssow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 14.Carlson, M., and T. Fangman. 2000. G2353, antibiotics and other additives for swine: food safety considerations. Columbia Extension Service, University of Missouri, Columbia.
- 15.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 16.Cornick, N. A., A. F. Helgerson, V. Mai, J. M. Ritchie, and D. W. Acheson. 2006. In vivo transduction of an Stx-encoding phage in ruminants. Appl. Environ. Microbiol. 72:5086-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Graaf, G. J., L. P. Jager, A. J. Baars, and T. J. Spierenburg. 1988. Some pharmacokinetic observations of carbadox medication in pigs. Vet. Q. 10:34-41. [DOI] [PubMed] [Google Scholar]
- 18.DeMarini, D. M., and B. K. Lawrence. 1992. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat. Res. 267:1-17. [DOI] [PubMed] [Google Scholar]
- 19.Dewey, C. E., B. D. Cox, B. E. Straw, E. J. Bush, and S. Hurd. 1999. Use of antimicrobials in swine feeds in the United States. Swine Health Prod. 7:19-25. [Google Scholar]
- 20.Frye, J. G., S. Porwollik, F. Blackmer, P. Cheng, and M. McClelland. 2005. Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 187:1485-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung, H. B., and T. L. Doan. 2005. Tinidazole: a nitroimidazole antiprotozoal agent. Clin. Ther. 27:1859-1884. [DOI] [PubMed] [Google Scholar]
- 22.Galvin, J. E., D. L. Harris, and M. J. Wannemuehler. 1997. Prevention and control of intestinal spirochaetal disease: immunological and pharmacological mechanisms, p. 343-374. In D. J. Hampson and T. B. Stanton (ed.), Intestinal spirochaetes in domestic animals and humans. CAB International, New York, NY.
- 23.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grif, K., M. P. Dierich, H. Karch, and F. Allerberger. 1998. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 17:761-766. [DOI] [PubMed] [Google Scholar]
- 25.Harris, D. L., D. J. Hampson, and R. D. Glock. 1999. Swine dysentery, p. 579-600. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. E. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 26.Hens, D. K., N. C. Chatterjee, and R. Kumar. 2006. New temperate DNA phage BcP15 acts as a drug resistance vector. Arch. Virol. 151:1345-1353. [DOI] [PubMed] [Google Scholar]
- 27.Herold, S., J. Siebert, A. Huber, and H. Schmidt. 2005. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob. Agents Chemother. 49:931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrman, T., and P. Sundberg. 2002. MF-2042 Medicated feed additives for swine. Kansas State University, Manhattan.
- 29.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 30.Humphrey, S. B., T. B. Stanton, and N. S. Jensen. 1995. Mitomycin C induction of bacteriophages from Serpulina hyodysenteriae and Serpulina innocens. FEMS Microbiol. Lett. 134:97-101. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey, S. B., T. B. Stanton, N. S. Jensen, and R. L. Zuerner. 1997. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J. Bacteriol. 179:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jerman, B., M. Butala, and D. Zgur-Bertok. 2005. Sublethal concentrations of ciprofloxacin induce bacteriocin synthesis in Escherichia coli. Antimicrob. Agents Chemother. 49:3087-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson, M., C. Fellström, M. U. Heldtander, K. E. Johansson, and A. Franklin. 1999. Genetic basis of macrolide and lincosamide resistance in Brachyspira (Serpulina) hyodysenteriae. FEMS Microbiol. Lett. 172:255-260. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy, G. A., and A. C. Strafuss. 1976. Scanning electron microscopy of the lesions of swine dysentery. Am. J. Vet. Res. 37:395-401. [PubMed] [Google Scholar]
- 35.Kennedy, G. A., A. C. Strafuss, and D. A. Schoneweis. 1973. Scanning electron microscopic observations on swine dysentery. J. Am. Vet. Med. Assoc. 163:53-55. [PubMed] [Google Scholar]
- 36.Köhler, B., H. Karch, and H. Schmidt. 2000. Antibacterials that are used as growth promoters in animal husbandry can affect the release of Shiga-toxin-2-converting bacteriophages and Shiga toxin 2 from Escherichia coli strains. Microbiology 146:1085-1090. [DOI] [PubMed] [Google Scholar]
- 37.Lang, A. S., and J. T. Beatty. 2007. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 15:54-62. [DOI] [PubMed] [Google Scholar]
- 38.Matson, E. G., M. G. Thompson, S. B. Humphrey, R. L. Zuerner, and T. B. Stanton. 2005. Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J. Bacteriol. 187:5885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson, E. G., R. L. Zuerner, and T. B. Stanton. 2007. Induction and transcription of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. Anaerobe 13:89-97. [DOI] [PubMed] [Google Scholar]
- 40.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Research Council. 1999. The use of drugs in food animals: benefits and risks. National Academy Press, Washington, DC. [PubMed]
- 42.Ohlsen, K., W. Ziebuhr, K. P. Koller, W. Hell, T. A. Wichelhaus, and J. Hacker. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 42:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phibro Animal Health. 2004. NADA 141-211 combined use of mecadox 10 (carbadox) and terramycin 50, terramycin 100, and terramycin 200 (oxytetracycline) in swine feeds. Phibro Animal Health, Fairfield, NJ.
- 44.Polzer, J., C. Stachel, and P. Gowik. 2004. Treatment of turkeys with nitroimidazoles: impact of the selection of target analytes and matrices on an effective residue control. Anal. Chim. Acta 521:189-200. [Google Scholar]
- 45.Raether, W., and H. Hanel. 2003. Nitroheterocyclic drugs with broad spectrum activity. Parasitol. Res. 90(Suppl. 1):S19-S39. [DOI] [PubMed] [Google Scholar]
- 46.Rosey, E. L., M. J. Kennedy, D. K. Petrella, R. G. Ulrich, and R. J. Yancey. 1995. Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J. Bacteriol. 177:5959-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan, M. E., and D. M. Baker. 2001. Tetracycline treatment of periodontal disease: antimicrobial and non-antimicrobial mechanisms, p. 237-265. In M. Nelson, W. Hillen, and R. A. Greenwald (ed.), Tetracyclines in biology, chemistry, and medicine. Birkhauser Verlag, Berlin, Germany.
- 48.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1 to 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Schaefler, S. 1982. Bacteriophage-mediated acquisition of antibiotic resistance by Staphylococcus aureus type 88. Antimicrob. Agents Chemother. 21:460-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sisson, G., J. Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanton, T. B. 2007. Prophage-like gene transfer agents—novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 13:43-49. [DOI] [PubMed] [Google Scholar]
- 53.Stanton, T. B., and C. P. Cornell. 1987. Erythrocytes as a source of essential lipids for Treponema hyodysenteriae. Infect. Immun. 55:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanton, T. B., E. G. Matson, and S. B. Humphrey. 2001. Brachyspira (Serpulina) hyodysenteriae gyrB mutants and interstrain transfer of coumermycin A1 resistance. Appl. Environ. Microbiol. 67:2037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanton, T. B., M. G. Thompson, S. B. Humphrey, and R. L. Zuerner. 2003. Detection of bacteriophage VSH-1 svp38 gene in Brachyspira spirochetes. FEMS Microbiol. Lett. 224:225-229. [DOI] [PubMed] [Google Scholar]
- 56.Stickler, D. J., R. G. Tucker, and D. Kay. 1965. Bacteriophage-like particles released from Bacillus subtilis after induction with hydrogen peroxide. Virology 26:142-145. [DOI] [PubMed] [Google Scholar]
- 57.Trott, D. J., S. L. Oxberry, and D. J. Hampson. 1997. Evidence for Serpulina hyodysenteriae being recombinant, with an epidemic population structure. Microbiology 143:3357-3365. [DOI] [PubMed] [Google Scholar]
- 58.Ventura, M., J. H. Lee, C. Canchaya, R. Zink, S. Leahy, J. A. Moreno-Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, D. J. O'Sullivan, and D. van Sinderen. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 71:8692-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walterspiel, J. N., S. Ashkenazi, A. L. Morrow, and T. G. Cleary. 1992. Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I. Infection 20:25-29. [DOI] [PubMed] [Google Scholar]
- 60.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 62.Zirkle, R. E., and N. R. Krieg. 1996. Development of a method based on alkaline gel electrophoresis for estimation of oxidative damage to DNA in Escherichia coli. J. Appl. Bacteriol. 81:133-138. [DOI] [PubMed] [Google Scholar]
- 63.Zuerner, R. L., T. B. Stanton, F. C. Minion, C. Li, N. W. Charon, D. J. Trott, and D. J. Hampson. 2004. Genetic variation in Brachyspira: chromosomal rearrangements and sequence drift distinguish B. pilosicoli from B. hyodysenteriae. Anaerobe 10:229-237. [DOI] [PubMed] [Google Scholar]