Abstract
The mitogen-activated protein kinase Sty1 is essential for the regulation of transcriptional responses that promote cell survival in response to different types of environmental stimuli in Schizosaccharomyces pombe. Upon stress activation, Sty1 reversibly accumulates in the nucleus, where it stimulates gene expression via the Atf1 transcription factor. The Atf1 protein forms a heterodimer with Pcr1, but the specific role of this association is controversial. We have carried out a comparative analysis of strains lacking these proteins individually. We demonstrate that Atf1 and Pcr1 have similar but not identical roles in S. pombe, since cells lacking Pcr1 do not share all the phenotypes reported for Δatf1 cells. Northern blot and microarray analyses demonstrate that the responses to specific stresses of cells lacking either Pcr1 or Atf1 do not fully overlap, and even though most Atf1-dependent genes induced by osmotic stress are also Pcr1 dependent, a subset of genes require only the presence of Atf1 for their induction. Whereas binding of Atf1 to most stress-dependent genes requires the presence of Pcr1, we demonstrate here that Atf1 can bind to the Pcr1-independent promoters in a Δpcr1 strain in vivo. Furthermore, these analyses show that both proteins have a global repressive effect on stress-dependent and stress-independent genes.
Eukaryotic cells respond to environmental stimuli by activating a number of signaling cascades. Prominent among these are the mitogen-activated protein kinase (MAPK) cascades, which mediate the response to extracellular signals. MAPKs phosphorylate and activate a variety of target substrates, including a number of transcription factors. Once activated, these factors mediate the specific transcriptional responses to the environmental signals. Activation of these pathways is frequently accomplished by many different stimuli.
The stress-induced Schizosaccharomyces pombe MAPK Sty1 protein (also known as Spc1 or Phh1), which is the homologue to the Saccharomyces cerevisiae HOG1 protein (5), can be activated by heat shock, high-osmolarity stress, nutrient depletion, oxidative stress, and arsenic (for reviews, see references 14 and 40). The cascade begins upstream with activation of the histidine kinases Mak1, -2 and -3 (3, 6, 24). These kinases activate the phosphotransmitter Spy1/Mpr1 (4, 25), which in turn controls the response regulator Mcs4 (32, 34). Mcs4 then activates the upstream components of the MAPK module, the MAPK kinase kinases Wak1/Wis4 and Win1. These kinases are functionally redundant, and their only reported substrate is the MAPK kinase Wis1. Activated Wis1 dually phosphorylates Sty1 on neighboring threonine and tyrosine residues (30, 33, 34, 43). The basal activity of the pathway and its inactivation after stress are regulated by tyrosine and serine-threonine phosphatases, such as Pyp1 and Pyp2 and Ptc1 to -4 (13, 22, 26). Which pathway components are required to sense and transduce each distinct signal has not yet been established at the molecular level.
Upon stress-mediated phosphorylation, Sty1 accumulates in the nucleus (11, 12), where it activates a complex transcriptional program of stress defense mechanisms (8). Thus, in the response to four out of five types of stress stimuli, Sty1 is required for the transcriptional regulation of a large set of genes that constitute the core environmental stress response (CESR) (8). For the majority of these genes, regulation is also dependent on the transcription factor Atf1; and although several Sty1 substrates have been identified, the main one seems to be Atf1 (16, 33, 37, 43).
Atf1 contains a basic leucine zipper (bZIP) DNA-interacting domain, in common with five other S. pombe transcription factors: Pap1, Pcr1, Atf21, Atf31, and Zip1 (10, 28, 37, 42). Phosphorylation of Atf1 by Sty1 upon stress activation has been demonstrated both in vitro (43) and in vivo (33). Atf1 has nuclear localization prior to stress (11). It has been long believed that Atf1 phosphorylation by Sty1 is essential for gene activation. However, it has been recently reported that an Atf1 mutant protein lacking all 11 MAPK phosphorylation sites has an activity similar to that of the wild-type protein when expressed at wild-type levels (20). The same report also shows that the nonphosphorylatable mutant is barely expressed in cells because it is very unstable, and the authors suggest that the main role of Sty1-dependent phosphorylation is to stabilize Atf1, rather than to activate it.
Soon after it was isolated, Atf1 was shown to activate transcription of some target genes by forming a heterodimer with a small bZIP transcription factor, Pcr1 (16, 41, 42). In mammalian cells, formation of heterodimers by distinct bZIP transcription factors has been suggested to control a large number of transcriptional regulatory systems, thereby increasing and diversifying gene responses (for a review, see reference 44). In yeast, it has been reported that while cells lacking Pcr1 share many stress-related phenotypes with Sty1- and Atf1-deficient cells, they also show some distinct features. Both Δpcr1 and Δatf1 cells are cold sensitive, are inefficient in mating and spore formation upon nitrogen starvation, and show no transcriptional activation of nutrient depletion-induced genes, such as fbp1 and ste11 (16, 42). The single mutants are also deficient in meiotic recombination, because the Atf1/Pcr1 heterodimer (also known as Mts1/Mts2) binds to hot spots more efficiently than the individual transcription factors (18, 41). The participation of the Atf1/Pcr1 transcription factor in hot spot recognition explains the observation that most recombination hot spots are located in promoter regions (45). Once bound to hot spots, Atf1/Pcr1 triggers chromatin remodeling to activate meiotic recombination (46). However, a role for Atf1/Pcr1 heterodimers is not so clear in the case of stress responses. Although Atf1 and Pcr1 have both been reported to bind to stress-regulated promoters (16, 42), it has not yet been demonstrated that the heterodimer binds to cis elements with greater affinity than the individual transcription factors. Furthermore, Wahls and coworkers reported that Atf1 but not Pcr1 is required for resistance to extracellular NaCl (18, 19) and suggested that the heterodimer plays a key role in mating, meiosis, and hot spot-mediated recombination but not in global stress resistance.
Here we further investigate the participation of Pcr1 in the fission yeast transcriptional program in response to stress. We show that Pcr1 is essential for some but not all extracellular stress responses and that even though it forms in vivo a heterodimer with Atf1, it does not regulate Atf1 stability. The transcriptional response of wild-type cells to KCl stress is more extensive than that of cells lacking Pcr1, which in turn is larger than that of Δatf1 cells. It is remarkable that cells lacking Pcr1 and, to a lesser extent, cells lacking Atf1 show a general derepression of stress-related and stress-unrelated genes under basal conditions, which suggests a major role of these proteins, and especially of Pcr1, in the regulation of basal expression levels. Finally, we have identified a subset of stress-dependent genes whose activation is Pcr1 independent, suggesting that Atf1 can function as a transcription factor in the absence of Pcr1.
MATERIALS AND METHODS
Yeast strains and growth conditions.
We used the wild-type S. pombe strains 972 (h−) and JA366 (h− leu1), as well as other strains, such as JM1066 (h+ leu1 atf1::ura4+) (31), NT224 (h− leu1 ura4 sty1-1) (22), JM1821 (h− leu1 ura4 atf1-HA6His::ura4+) (stock of Jonathan Millar), and JX26 (h90 ade6 leu1 ura4 pcr1::ura4+) (42). To construct an S. pombe strain expressing a hemagglutinin (HA)-tagged version of Pcr1, we designed specific primers to PCR amplify a fragment containing the carboxyl-terminal pcr1-coding sequence fused in-frame to an HA-coding sequence followed by the selectable marker kanMX6 and a 3′, noncoding pcr1 sequence using as the template plasmid pFA6a-3HA-kanMX6. Wild-type strain 972 was transformed with the linear fragment, and recombinants were selected for their ability to grow on G418/Geneticin (Invitrogen), as described previously (7). The resulting strain was named MS65 (h− pcr1-HA::kanMX6). To construct S. pombe strains with specific loci deleted, we transformed wild-type strains with linear fragments containing open reading frame (ORF)::kanMX6 or ORF::ura4+, obtained by PCR amplification using ORF-specific primer and plasmid pFA6a-kanMX6 or KS-ura4 as the templates as described previously (7). We obtained AV15 (h− atf1::kanMX6), AV18 (h− sty1::kanMX6), MS5 (h− pcr1::kanMX6), MS7 (h+ leu1 pcr1::kanMX6), and MS38 (h+ leu1 ura4 pcr1::ura4+). To construct a strain deleted in both atf1 and pcr1, we crossed strains JX26 and AV15, yielding strain MS48 (h+ leu1 ura4 pcr1::ura4+ atf1::kanMX6). Cells were grown in rich medium or in synthetic minimal medium as described previously (2, 23).
Plasmids.
The atf1 and pcr1 coding sequences were PCR amplified from an S. pombe cDNA library using primers specific for the Atf1- and Pcr1-coding genes. Plasmid p151.41x (pHA-atf1.41x) was obtained by digestion of p123.41x (pHA-tpx1.41x) (38) with BamHI and SmaI to eliminate tpx1 ORF and ligation with atf1 ORF flanked with BglII and SmaI sites. The same strategy was used to generate the integrative plasmid p137.41x′ (pHA-atf1.41x′) from plasmid p136.41x′ (pHA.41x′; from our laboratory collection). For plasmid p138.41x (pHA-pcr1.41x), the pcr1 ORF flanked with BamHI and SmaI restriction sites was cloned into p123.41x (pHA-tpx1.41x) (38), previously digested with BamHI-SmaI to eliminate the tpx1 ORF. Plasmid AY025 is a pJK148 derivative (17) containing a PstI-SacI fragment with the nmt promoter, multiple cloning site, and terminator from pREP.81x (21). A PstI-SmaI fragment of p138.81x (pHA-pcr1.81x), containing the nmt promoter linked to HA-pcr1, was cloned into the vector AY025 to yield the integrative plasmid p139.81x′ (pHA-pcr1.81x′). To trigger the expression of green fluorescent protein (GFP)-tagged Atf1 and Pcr1 fusion proteins in S. pombe, we obtained plasmids p179.41x′ (pGFP-HA-atf1.41x′) and p171.41x′ (pGFP-HA-pcr1.41x′) by cloning a PCR-amplified atf1 ORF digested with BglII and SmaI or a pcr1 ORF digested with BamHI and SmaI into p85.41x′ (pGFP-HA-pap1.41x′) (7) digested with BglII-SmaI to eliminate most pap1 ORFs. All the new clones obtained from PCR-amplified DNA fragments were confirmed by sequencing.
Construction of yeast strains with integrated versions of the atf1 and pcr1 genes.
The plasmid p139.81x′ (pHA-pcr1.81x′) was linearized with NruI and integrated at the leu1 locus of MS38, yielding strain MS10 (h+ leu1 ura4 pcr1::ura4+ nmt81x::HA-pcr1::leu1). Strain MS9 was obtained linearizing p137.41x′ (pHA-atf1.41x′) with NruI and integrating it into the leu1 locus of JM1066. To construct S. pombe strains expressing GFP-tagged Atf1 or Pcr1, we transformed the MS7 or JM1066 strain with NruI-linearized plasmids p179.41x′ (pGFP-HA-atf1.41x′) and p171.41x′ (pGFP-HA-pcr1.41x′), yielding strains MS13 (h+ leu1 atf1::ura4+ nmt41x::GFP-HA-atf1::leu1+), MS14 (h+ leu1 pcr1::kanMX6 nmt41x::GFP-HA-atf1::leu1+), MS15 (h+ leu1 atf1::ura4+ nmt41x::GFP-HA-pcr1::leu1+), and MS16 (h+ leu1 pcr1::kanMX6 nmt41x::GFP-HA-pcr1::leu1+).
Preparation of formaldehyde-cross-linked extracts for immunoprecipitation analysis.
Formaldehyde cross-linking was performed as described previously (27) with some modifications. Pelleted cells were resuspended in buffer B (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2) and lysed with three 30-s pulses in a bead beater (FastPrep; Bio 101) at 4°C. Lysates were centrifuged for 5 min at 1,600 × g and supernatants transferred to fresh microtubes. HA-Atf1 was immunoprecipitated from cleared supernatants by adding 150 μl of monoclonal anti-HA antibody (12CA5) for 1 h at 4°C and then 25 μl of protein A-Sepharose beads (Amersham Biosciences) for 30 min. Immunoprecipitates were washed three times with buffer B, and proteins were released from cross-linked immunocomplexes by boiling for 10 min in sodium dodecyl sulfate (SDS) loading buffer. Samples were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and detected by immunoblotting with polyclonal anti-Atf1 or anti-Pcr1 antiserum raised against bacterial fusion proteins of glutathione-S-transferase (GST)-Atf1 (amino acids 71 to 291) or GST-Pcr1, following standard rabbit immunization procedures.
Fluorescence microscopy.
Fluorescence microscopy and capture imaging were performed as described previously (39), with one modification. When preparing the samples in the slides, the three microliters of 50% glycerol (mounting medium) contained 4′,6′-diamidino-2-phenylindole at a concentration of 0.5 μg/ml.
Preparation of extracts to detect phosphorylated and nonphosphorylated Pcr1.
S. pombe cells were grown in minimal medium to an optical density at 600 nm (OD600) of 0.5 and harvested by centrifugation. For detection of plasmid-derived HA-Pcr1, native extracts were obtained. Pelleted cells were washed once with distilled water and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM KCl, 5 mM EDTA, 0.1% NP-40, 10% glycerol). Cell suspensions were disrupted by adding glass beads and lysing with three 30-s pulses in a bead beater (Fast Prep; Bio 101) at setting 4.5 and at 4°C. Lysates were then centrifuged to remove cell debris. The protein concentration was determined by using the Bradford protein assay (Bio-Rad). Thirty micrograms of protein extracts were separated electrophoretically by 12% SDS-PAGE and transferred to Protran nitrocellulose membranes (Whatman). Pcr1 was immunodetected with polyclonal anti-Pcr1 antiserum. For dephosphorylation of HA-Pcr1 protein, lysates were incubated with 15 U/μg protein of lambda phosphatase (New England Biolabs) for 60 min at 30°C before being subjected to SDS-PAGE. When indicated, phosphatase inhibitors were added at the following final concentrations: 1 mM sodium fluoride, 1 mM β-glycerolphosphate, 1 mM sodium pyrophosphate, and 0.2 mM activated sodium orthovanadate. For detection of endogenous Pcr1, it was necessary to obtain boiled extracts. Pelleted cells were washed once with distilled water and resuspended in HB buffer (25 mM morpholinepropanesulfonic acid [pH 7.2], 60 mM β-glycerolphosphate, 15 mM p-nitrophenyl phosphate, 15 mM MgCl2, 15 mM EGTA, 1% Triton X-100, 1 mM dithiothreitol, and 170 mg/liter phenylmethylsulfonyl fluoride [PMSF]). The cell suspension was boiled for 6 min at 100°C and then disrupted by glass beads and lysed with two 30-s pulses in a bead beater (Fast Prep; Bio 101) at setting 5.5 and at 4°C. The protein concentration was determined by using the Bradford protein assay (Bio-Rad). One hundred micrograms of protein extracts were loaded onto gels.
Solid and liquid sensitivity assay.
For survival on solid plates, S. pombe strains were grown in liquid minimal medium to an OD600 of 0.5. Cells were then diluted in water, and the indicated number of cells in 5 μl was spotted onto minimal medium plates containing (or not) 1 M KCl, 1 M NaCl, 2.4 M sorbitol, or 1 mM hydrogen peroxide (H2O2). The spots were allowed to dry, and the plates were incubated at 30°C for 3 to 4 days. To determine survival in liquid cultures, cells were grown in rich medium to an OD600 of 0.5. Acute doses of 1 or 2 mM H2O2, as indicated, were then added for 60 min. Similarly, other flasks were shifted to 45°C for 15 or 30 min, as appropriate. Cells were then washed, diluted, and plated on rich medium agar plates to determine survival.
RNA analysis.
Total RNA from S. pombe cultures was obtained, processed, and transferred to membranes as described previously (39). Membranes were hybridized with the [α-32P]dCTP-labeled gpx1, ctt1, zym1, cta3, hsp9, gpd1, srx1, hsp16, ntp1, or cdc2 probe, containing the complete ORFs of the glutathione peroxidase-, catalase-, metallothionein-, calcium ATPase transporter-, heat shock protein 9-, glycerol-3-phosphate dehydrogenase-, sulfiredoxin-, heat shock protein 16-, neutral trehalase-, and cyclin-dependent kinase 2-coding genes.
Preparation of S. pombe trichloroacetic acid (TCA) extracts to measure Atf1 concentration.
S. pombe cultures (5 ml) at an OD600 of 0.5 were pelleted just after the addition of 10% TCA (from a 100% stock) and washed in 20% TCA. The pellets were lysed by vortexing for 5 min, following the addition of glass beads and 100 μl 12.5% TCA. Cell lysates were pelleted, washed in iced acetone, and dried at 55°C for 15 min. Pellets were resuspended in 50 μl of a solution containing 1% SDS, 100 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Samples were electrophoretically separated by 8% SDS-PAGE and immunodetected with anti-Atf1. As a loading control, we used monoclonal antitubulin antibodies in our blots (Tub2; Sigma).
Microarray experiments and data evaluation.
Global expression analysis utilized custom-designed S. pombe microarrays. Array construction, sample labeling, and hybridization were carried out as described previously (35). The arrays consisted of 8,785 70-mer oligonucleotides designed to measure gene expression (http://research.stowers-institute.org/microarray/S_pombe/). Briefly, total RNA was prepared from strains 972 (wild type), AV15 (Δatf1), and MS5 (Δpcr1), treated (or not) for 20 min with 0.4 M KCl. Polyadenylated RNA was extracted from total RNA by purification with an oligo(dT) cellulose column. RNA quality was assessed on a Bioanalyzer 2100 machine (Agilent). RNA (2 μg per sample) was converted to cDNA by priming with oligo(dT18) and poly(dN9) in the presence of aminoallyl-dUTP (Ambion), followed by conjugation to Cy5 or Cy3 fluorescent dye. Samples were prepared from three biological replicates, and all samples were dye swapped for further technical replication. Labeled samples were mixed for comparative hybridization on poly-l-lysine-coated microarrays printed with 70-mers representing all known S. pombe reading frames. The microarrays were scanned with a GenePix 4000B scanner and the images analyzed using the GenePix Pro 6.0 software program (Molecular Devices, Union City, CA). Data analysis was performed with the R programming language. Data were normalized via the print-tip loess method, and differential expression was assessed using the LIMMA software package (36). Genes exhibiting at least a twofold difference between the wild-type and mutant or treated versus untreated, with unadjusted P values of less than 0.05, were considered differentially expressed.
Chromatin immunoprecipitation.
For immunoprecipitation of HA-tagged Atf1 and Pcr1 proteins linked to DNA promoter regions, cells were grown in liquid minimal medium to an OD600 of 0.5 and formaldehyde (1.5% vol/vol) was added for 30 min at 25°C. Cross-linking was stopped by adding 187.5 mM glycine. After 5 min, cells were collected by centrifugation and washed twice with cold distilled water. Pellets were resuspended in 250 μl breaking buffer (0.1 M Tris-HCl [pH 8.0], 20% glycerol, and 1 mM PMSF) and lysed with glass beads in a bead beater (Biospect Products). Pellets were collected, washed twice with lysis buffer (50 mM HEPES-NaOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF), and resuspended in 250 μl lysis buffer. Lysates were then sonicated in a Bioruptor (Diagenode) sonicator with eight 30-s high-sonication pulses at 4°C and 1-min pauses between pulses, yielding chromatin fragments with an average size of 500 bp. Lysis buffer was added up to 1 ml, and samples were centrifuged at 16,000 × g for 30 min at 4°C. Fifty microliters from the soluble chromatin samples were kept as inputs, and the remaining was immunoprecipitated with monoclonal anti-HA antiserum overnight at 4°C, followed by addition of protein A-Sepharose beads and incubation for 4 h at 4°C. Immunocomplexes were washed once in lysis buffer, twice in lysis buffer containing 0.5 M NaCl, twice in washing buffer (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 1 mM PMSF), and finally once in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Beads were pelleted, and DNA was eluted in 100 μl elution buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA [pH 8.0], 1% SDS) during 20 min at 65°C. Beads were repelleted, the supernatants were transferred to fresh tubes, and any remaining DNA was eluted from the beads by washing it once in 150 μl TE-0.67% SDS. Corresponding elution supernatants were pooled, and formaldehyde cross-linking of both the 50 μl of soluble chromatin and immunoprecipitated chromatin was reversed by overnight incubation at 65°C. DNA was cleaned up by incubation for 2 h at 37°C with 0.3 mg/ml proteinase K and 0.04 mg/ml glycogen and was then purified by phenol-chloroform extraction and precipitated with ethanol and NaCl. DNA was resuspended in 100 μl TE. Recovered DNA was PCR amplified with specific primers, and products were resolved on ethidium bromide-containing 2% Tris-borate-EDTA agarose gels. The specific primers amplified promoter and ORF regions corresponding to the following sequences with respect to the translation initiation sites: −543 to −217 of the gpd1 gene; −385 to −129 of the hsp9 gene; −381 to −152 of the srx1 gene; −1425 to −1063 of the cta3 gene; and +1248 to +1561 of the cdc18 ORF.
Microarray data accession number.
Microarray data are available at ArrayExpress (www.ebi.ac.uk/arrayexpress/) under accession number E-TABM-447.
RESULTS
Atf1 and Pcr1 are nuclear phosphoproteins.
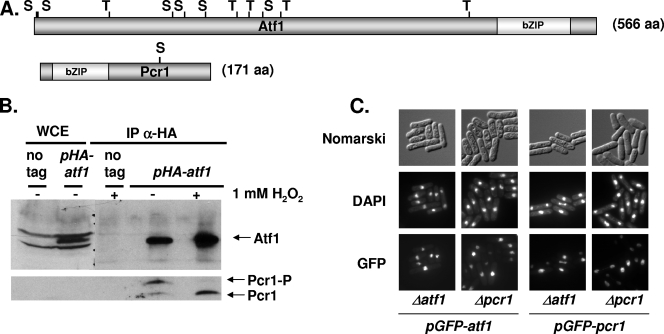
The bZIP-containing proteins Atf1 and Pcr1 (Fig. 1A) were isolated in several laboratories through different screening strategies (16, 18, 33, 37, 42). They were soon reported to coimmunoprecipitate in extracts, and we therefore decided to analyze whether they can form complexes in vivo. To preserve possible interactions between Atf1 and Pcr1, we used formaldehyde to cross-link all macromolecules in cultured cells. Before cross-linking, we exposed cell cultures to H2O2 stress. We took the soluble fractions of the cell extracts, reversed the cross-links, and analyzed by Western blotting whether Atf1 and Pcr1 interact with and/or without stress imposition (Fig. 1B). Even though the levels of Pcr1 were too low to be detected in whole-cell formaldehyde extracts (Fig. 1B, left two lanes, lower panel), two bands corresponding to Pcr1 could be detected in the immunoprecipitates of HA-Atf1 (Fig. 1B, right two lanes, lower panel). This interaction between Atf1 and Pcr1 was detected both before and after stress. Regarding the role of such interaction, a previous report showed by immunofluorescence analysis that both Atf1 and Pcr1 are nuclear proteins and that the nuclear localization of Atf1 was dependent on the presence of Pcr1 (11). However, we detected nuclear localization of both proteins, expressed from a heterologous promoter, independently of the presence of the other bZIP partner (Fig. 1C). It is worth noting that the amounts of the fusion proteins in the two strain backgrounds are very similar, as determined by Western blotting (data not shown).
FIG. 1.
Atf1 and Pcr1 interact in vivo and display nuclear localization. (A) Schematic representation of Atf1 and Pcr1 proteins. The bZIP domains and the potential MAPK sites (S or T) are indicated. (B) HA-Atf1 and Pcr1 interact in vivo before and after stress. Strain 972 (no tag) or MS9 (Δatf1 with integrated, nmt-driven pHA-atf1), were treated (+) or not (−) for 30 min with 1 mM H2O2, and formaldehyde extracts were obtained. Ten milligrams of total protein extracts were immunoprecipitated with anti-HA antibodies (IP α-HA), and the resulting immunoprecipitates were analyzed by SDS-PAGE and blotted with anti-HA or anti-Pcr1 antibodies. As a loading control, 150 μg of whole-cell extracts were loaded (WCE). (C) Nuclear localization of GFP-Atf1 and GFP-Pcr1 proteins in Δatf1 and Δpcr1 strains. We used strains MS13 (Δatf1 with integrated, nmt-driven pGFP-atf1), MS14 (Δpcr1 with integrated, nmt-driven pGFP-atf1), MS16 (Δpcr1 with integrated, nmt-driven pGFP-pcr1), and MS15 (Δatf1 with integrated, nmt-driven, pGFP-pcr1). Cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) to label DNA (center panels). The cellular distributions of the fusion proteins under nonstressed conditions were determined by fluorescence microscopy (GFP) (lower panels). The same cells under differential interference contrast (Nomarski) optics are shown in the upper panels.
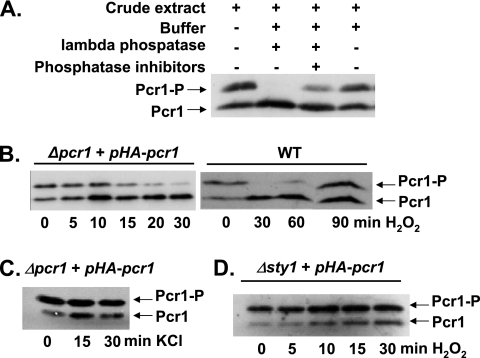
While analyzing the interaction between Atf1 and Pcr1, we noticed that endogenous Pcr1 appeared as a double band on Western blots probed with polyclonal anti-Pcr1 antibodies (Fig. 1B). Pretreatment of crude extracts from HA-Pcr1-expressing cells with lambda phosphatase prior to electrophoresis eliminated the slow-migrating band, confirming that Pcr1 is a phosphoprotein (Fig. 2A). The ratio of phosphorylated to nonphosphorylated Pcr1 was reversibly decreased by H2O2 treatment both in a wild-type strain (Fig. 2B, right panel) and in Δpcr1 cells expressing HA-Pcr1 from an inducible promoter (Fig. 2B, left panel). The H2O2-dependent dephosphorylation, which was not observed upon KCl treatment (Fig. 2C), was Sty1 dependent (Fig. 2D). The biological role of Pcr1 phosphorylation will require further study.
FIG. 2.
Pcr1 is a phosphoprotein. (A) Pcr1 is phosphorylated under unstressed conditions. Native extracts from MS7 cells (Δpcr1) transformed with p138.41x, expressing HA-Pcr1, were prepared and incubated with (+) or without (−) lambda phosphatase in the presence or absence of phosphatase inhibitors, as indicated. Pcr1 was detected after SDS-PAGE followed by Western blotting using polyclonal anti-Pcr1 antibodies. (B) Pcr1 is dephosphorylated upon oxidative stress. Strains MS7 with p138.41x (Δpcr1 + pHA-pcr1) or 972 (WT) were treated with 1 mM H2O2 stress for the times indicated. Native (to detect HA-Pcr1) or boiled (to detect endogenous Pcr1 in strain 972) protein extracts, obtained as described in Materials and Methods, were analyzed by Western blot analysis with anti-Pcr1 antibodies. Phosphorylated (Pcr1-P) and unphosphorylated (Prc1) protein forms, either HA tagged or untagged, are indicated with arrows. (C) Dephosphorylation of Pcr1 does not occur upon KCl stress. Strain MS7 with p138.41x (Δpcr1 + pHA-pcr1) was treated with 0.4 M KCl for 15 or 30 min or left untreated. The phosphorylation status of HA-Pcr1 was analyzed as panel B. (D) H2O2-dependent dephosphorylation of Pcr1 is Sty1 dependent. Strain NT224 transformed with p138.41x (Δsty1 + pHA-pcr1) was treated with 1 mM H2O2 stress for the times indicated. The phosphorylation status of HA-Pcr1 was analyzed as for panel B.
The Atf1/Pcr1 heterodimer is required for survival of some, but not all, stress situations.
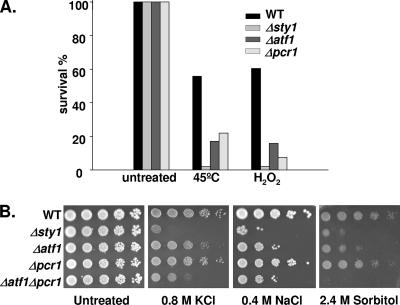
To test whether Atf1 and Pcr1 are both equally required for stress survival, we next compared the phenotypes of cells lacking (or not) each transcription factor under a variety of adverse stress conditions. In liquid cultures, both Atf1 and Pcr1 were required for full survival upon oxidative stress or heat shock (Fig. 3A). In contrast, when spread on solid plates containing NaCl, KCl, or sorbitol, although cells lacking Atf1 showed severely impaired survival, Δpcr1 cells showed an intermediate behavior, with survival efficiencies closer to those of the wild type than to those of Δatf1 cells (Fig. 3B). Pcr1 was similarly not essential for survival upon H2O2 stress on solid plates (data not shown).
FIG. 3.
Atf1 but not Pcr1 is essential for survival in front of all major types of stresses. (A) Survival of wild-type, Δsty1, Δatf1, and Δpcr1 strains in response to heat shock and oxidative stress. Strains 972 (WT), AV18 (Δsty1), AV15 (Δatf1), and MS5 (Δpcr1) were grown in rich medium to a final OD600 of 0.5. Cells were then incubated at 45°C for 30 min or with 2 mM H2O2 for 1 h before plating on rich-medium agar plates. Survival was measured as a percentage of the colony number at time zero. The experiments were repeated at least three times with very similar results. (B) Analysis of the osmotic stress resistance of wild-type, Δsty1, Δatf1, Δpcr1, and Δatf1 Δpcr1 strains. Strains 972 (WT), AV18 (Δsty1), AV15 (Δatf1), MS5 (Δpcr1), and MS48 (Δatf1 Δpcr1) were grown in liquid minimal medium to a final OD600 of 0.5, and the number of cells indicated at the top of the panels was spotted onto minimal medium plates containing KCl, NaCl, or sorbitol at the indicated concentrations and incubated at 30°C for 3 to 4 days.
Induction of most, but not all, stress-dependent genes requires Pcr1.
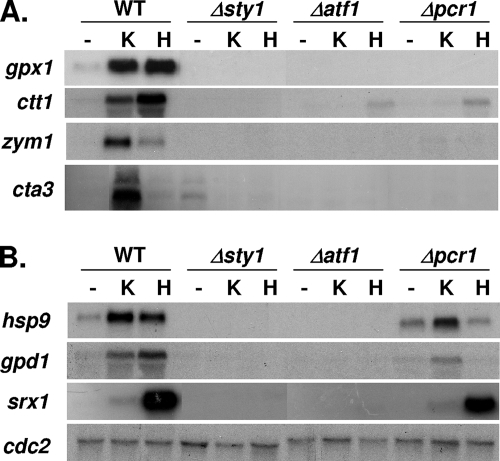
Since Atf1 but not Pcr1 was required for survival upon osmotic or oxidative stress on plates, we tested whether induction of some Sty1-dependent CESR genes would be impaired in Δatf1 cells but not in Δpcr1 cells. In wild-type cells, the induction of most stress-inducible genes by KCl or H2O2 was dependent on Sty1, Atf1, and Pcr1 (Fig. 4A). However, some target genes were induced upon some stress conditions in Δpcr1 cells, such as hsp9 (encoding a heat shock protein) (29), gpd1 (encoding glycerol-3P-dehydrogenase) (1), or srx1 (encoding the peroxiredoxin reductase Srx1) (38) (Fig. 4B). These results suggest that whereas the Atf1/Pcr1 dimer seems to be responsible for transcriptional activation of most CESR genes, Pcr1 is dispensable for the activation of gpd1 and hsp9 upon KCl stress and for activation of srx1 upon H2O2 stress.
FIG. 4.
Pcr1 is required for transcription of most but not all stress response genes. (A) Transcription of many stress response genes requires the presence of both Atf1 and Pcr1 transcription factors. Cultures of strains 972 (WT), AV18 (Δsty1), AV15 (Δatf1), and MS5 (Δpcr1) were left untreated (−) or were treated for 30 min with 0.4 M KCl (K) or 1 mM H2O2 (H). Total RNA was extracted and analyzed by Northern blotting with probes for gpx1, ctt1, zym1, or cta3. (B) Transcription of hsp9, gpd1, or srx1 does not require Pcr1 for all stress situations. Strains and conditions are as described for panel A. cdc2 was used as a loading control.
Atf1 protein stability is not directly regulated by Pcr1 or Sty1.
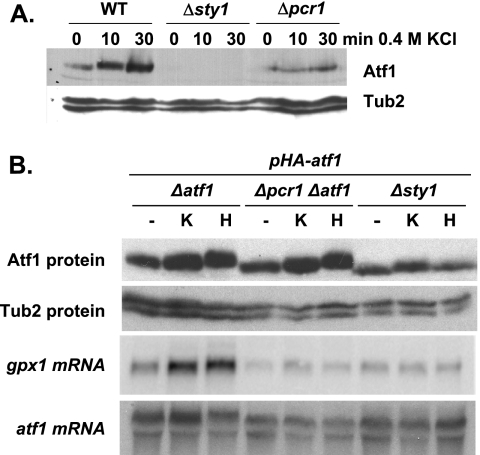
During preparation of extracts from wild-type and mutant strains, we observed that the amounts of Atf1 protein were significantly increased by exposure of cells to stress conditions (Fig. 5A, WT), as previously reported (8, 11). Atf1 basal levels were significantly lower in extracts from cells lacking Sty1 or Pcr1, even though some accumulation was observed following stress treatment of cells lacking Pcr1 (Fig. 5A). It has recently been suggested that Atf1 stability is regulated by its heterodimeric partner, Pcr1, and by the Sty1 kinase (20). We tested this hypothesis by integrating an nmt-driven atf1 gene into Δatf1, Δsty1, and Δpcr1 Δatf1 cells. In these engineered strains, the levels of Atf1 were almost identical to those of the wild-type strain (Fig. 5B, upper panel), which indicated that Atf1 stability does not require the Sty1 or Pcr1 protein. We confirmed this result by treating cells with the translation inhibitor cycloheximide and analyzing the stability of Atf1, both the native form expressed from its genomic locus and the nmt-driven form expressed from the integrated gene construct. The half-life of Atf1 was comparable in the presence or absence of Pcr1 (data not shown). To discard the possibility that the partial defects of cells lacking Pcr1 were due to the reduced concentration of Atf1 in these cells, we analyzed the expression of gpx1 in a Δpcr1 Δatf1 strain ectopically expressing wild-type levels of Atf1. As shown in Fig. 5B (gpx1 mRNA), the reduced levels of Atf1 observed in Δpcr1 or Δsty1 cells do not explain the lack of gpx1 induction in these strains, since increasing the amount of Atf1 does not restore gpx1 induction in the absence of either Pcr1 or Sty1.
FIG. 5.
Pcr1 does not regulate the levels of Atf1. (A) Atf1 protein levels are lower in Δsty1 and Δpcr1 mutants than in wild-type cells. TCA extracts from strains 972 (WT), AV18 (Δsty1), and MS5 (Δpcr1) were obtained from cultures grown in minimal medium before (0) or after a 10- or 30-min exposure to 0.4 M KCl. The relative amounts of Atf1 protein were determined by Western blotting with polyclonal anti-Atf1 antibodies (Atf1). Expression of tubulin was detected as a loading control (Tub2). (B) Atf1 protein levels are very similar in wild-type, Δsty1, and Δpcr1 cells when expressed from the Sty1-independent nmt promoter. Strains JM1066 (Δatf1), MS48 (Δpcr1 Δatf1), and NT224 (Δsty1) were transformed with plasmid p151.41x (containing the nmt-driven HA-atf1 gene, pHA-atf1), and TCA extracts were obtained before (−) or after 30 min with 0.4 M KCl (K) or 1 mM H2O2 (H). Cell lysates were analyzed by Western blotting with anti-Atf1 and anti-Tub2 antibodies as for panel A. RNA from the same strains was also isolated, and the expression of gpx1 was determined by Northern blotting (gpx1 mRNA). atf1 transcript levels are shown as a loading control (atf1 mRNA).
Atf1 can bind to some promoters in cells lacking Pcr1.
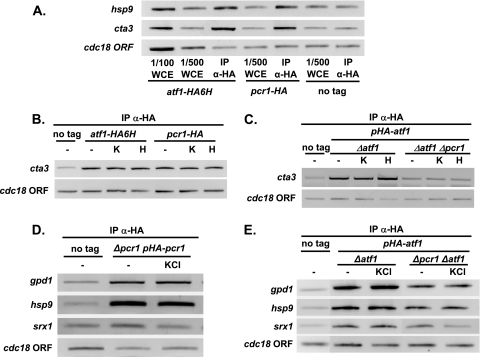
Most CESR genes, such as gpx1, ctt1, zym1, or cta3, require both Atf1 and Pcr1 for their stress-dependent induction (Fig. 4A). We used chromatin immunoprecipitation to examine the binding of Atf1 and Pcr1 to these stress-regulated promoters in vivo. Using strains carrying chromosomal HA-tagged versions of the atf1 or pcr1 genes, we observed that both Atf1 and Pcr1 are bound to the cta3 and hsp9 promoters before stress imposition (Fig. 6A). Formaldehyde cross-linking did not preactivate the Sty1 pathway, as demonstrated by Western blot analysis with anti-phosphorylated MAPK antibody (data not shown). KCl or H2O2 treatments did not significantly enhance Atf1 or Pcr1 binding to the cta3 promoter, a classical CESR gene (Fig. 6B). To determine whether the binding of Atf1 to the cta3 promoter requires the presence of Pcr1, we transformed a Δatf1 strain or a Δatf1 Δpcr1 strain with an episomal plasmid expressing HA-Atf1 so that they would express the same levels of the fusion protein (Fig. 5B). Binding of Atf1 to the cta3 promoter was detected only in the strain expressing Pcr1 (Fig. 6C).
FIG. 6.
Atf1 is bound to promoters before and after stress. (A) Chromatin immunoprecipitation analysis of Atf1-HA and Pcr1-HA bound to stress promoters. Strains JM1821 (atf1-HA6H), MS65 (pcr1-HA) (both strains tagged at the chromosomal loci), or wild-type 972 with no plasmid (no tag) were grown, formaldehyde extracts were obtained, and chromatin bound to Atf1-HA or Pcr1-HA was isolated using anti-HA monoclonal antibodies (IP α-HA). Recovered DNA was assayed by PCR amplification with primers encompassing the hsp9 and cta3 promoters or the cdc18 ORF as a negative control. Two different concentrations of whole-cell extracts (WCE) were also analyzed with the primer pairs to demonstrate that the quantity of amplified DNA is dependent on the amount of input (WCE) (1/100 or 1/500). (B) Atf1-Pcr1 heterodimer binds to standard stress-dependent promoters in vivo. Strains JM1821 (atf1-HA6H) and MS65 (pcr1-HA) or wild-type strain 972 (no tag) was grown in the absence of stress (−) or presence of 0.4 M KCl (KCl) or 1 mM H2O2 (H) for 15 min. We obtained formaldehyde extracts, and performed chromatin immunoprecipitation of Atf1-HA6H and Pcr1-HA with anti-HA monoclonal antibodies (IP α-HA) to assay Atf1 and Pcr1 binding to specific DNA sites. PCR was performed with primers encompassing the classical CESR promoter cta3 and the cdc18 ORF as a negative control. (C) Atf1 binding is Pcr1 dependent in the cta3 promoter. Strains JM1066 (Δatf1) and MS48 (Δatf1 Δpcr1) transformed with nmt-driven p151.41x (pHA-atf1) or wild-type 972 with no plasmid (no tag) were grown as described for panel B. We obtained formaldehyde extracts, and chromatin immunoprecipitation of HA-Atf1 (IP α-HA) was performed as for panel B. (D) Pcr1 binding to Pcr1-independent stress promoters in vivo. Strains MS10 (Δpcr1 pHA-pcr1) transformed with the integrated, nmt-driven p139.81x′ (pHA-pcr1) or wild-type 972 with no plasmid (no tag) were grown in the presence (KCl) or not (−) of 0.4 M KCl for 15 min. Formaldehyde extracts and chromatin immunoprecipitation of HA-Pcr1 were performed as for panel B. Specific primers were used for PCR amplification of the gpd1, hsp9, and srx1 promoters, as well as the cdc18 ORF as a negative control. (E) Atf1 binding to Pcr1-independent stress promoters in vivo. The same experiment as for panel D was performed with strains JM1066 (Δatf1) and MS48 (Δatf1 Δpcr1) transformed with nmt-driven p151.41x (pHA-atf1) or 972 (WT), with no plasmid (no tag).
As shown in Fig. 4B, the gpd1 and hsp9 genes require Atf1 and Pcr1 for induction upon H2O2 stress, whereas srx1 induction by peroxides is Pcr1 independent. However, the induction of gpd1 and hsp9 can be triggered in a Pcr1-independent manner upon KCl stress (Fig. 4B). We again used chromatin immunoprecipitation to examine the binding of Atf1 and Pcr1 to these stress-regulated promoters in vivo. As shown before for the cta3 promoter (Fig. 6C), we ensured similar levels of Pcr1 or Atf1 in the different strain backgrounds by examining HA-tagged proteins expressed from the nmt promoter. We determined that Pcr1 is bound to the gpd1 and hsp9 but not the srx1 promoters both before and after KCl stress (Fig. 6D). On the other hand, Atf1 was bound to all three promoters in the presence or absence of Pcr1 even prior to stress (Fig. 6E), indicating that Atf1 does not require its partner Pcr1 to bind to these particular cis elements in vivo.
The transcriptome of KCl-treated Δatf1 cells differs from that of Δpcr1 cells.
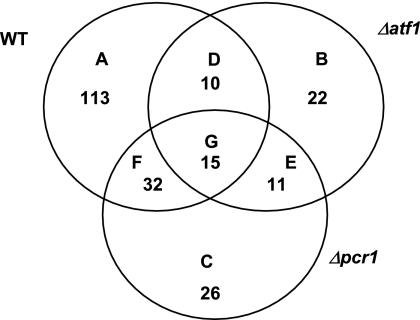
To globally identify new Atf1-dependent, Pcr1-independent stress genes, we performed genome-wide expression analyses of wild-type, Δatf1, and Δpcr1 cells upon exposure to 0.4 M KCl stress for 30 min. This stress condition was chosen because it caused a differential induction of the gpd1 and hsp9 genes in strains lacking Atf1 or Pcr1 (Fig. 4B). RNA samples, collected before and after KCl stress, were subjected to microarray analysis. Figure 7 shows the Venn diagram of genes up-regulated by at least twofold (in relation to the mRNA levels with the untreated condition in each strain) in wild-type, Δatf1, and Δpcr1 cells. A total of 170 genes were up-regulated by KCl in wild-type cells, whereas only 58 and 84 genes were induced by twofold or above in Δatf1 and Δpcr1 cells, respectively.
FIG. 7.
Microarray analysis of the responses of wild-type, Δatf1, and Δpcr1 cells to KCl stress. A Venn diagram of genes up-regulated more than twofold after 30 min of 0.4 M KCl stress is shown. mRNA from strains 972 (WT), AV15 (Δatf1), and MS5 (Δpcr1), grown in minimal medium, was analyzed by microarray hybridization. For the wild type, 170 genes were induced by KCl (A, D, F, and G); for Δatf1 cells, 58 genes were induced (B, D, E, and G); and for Δpcr1 cells, 84 genes were induced (C, E, F, and G). n-fold inductions upon stress are relative to mRNA levels for the untreated condition of each strain. The list of all A-to-G subsets of genes is provided in Table S1 in the supplemental material.
We carefully analyzed each subset of genes on the Venn diagram to determine the requirement of Atf1 and Pcr1 for their induction (grouped from A to G; see Table S1 in the supplemental material). From the total of 170 genes induced by KCl in the wild-type strain, 113 require the presence of both Atf1 and Pcr1 (group A). The genes in groups B, C, and E are those identified in the initial analysis as being induced by KCl more than twofold only in the mutant strains, not in the wild-type background. A closer look at their individual inductions showed that some of these genes either were induced just above twofold by KCl treatment (while in the wild-type cells they were barely reaching twofold) or their basal levels were already over twofold in the untreated mutant strains. Therefore, these genes were derepressed in Δatf1 and/or Δpcr1 cells (see Table S1 in the supplemental material), indicating that Atf1 and/or Pcr1 regulate the basal expression levels of some stress-dependent genes (see below). Regarding the genes in group D (induced by KCl in wild-type and Δatf1 cells but not in Δpcr1 cells), we observed that their expression levels were just bordering the twofold value; that is, none of those genes was dependent on Pcr1 without also showing some dependency on Atf1. With a few particular exceptions, very similar conclusions could be drawn from the analysis of group G: upon KCl treatment, there were no or very few genes whose expression was independent of both Pcr1 and Atf1.
In contrast, the genes in group F (those which were expressed more than twofold above the basal level upon KCl treatment in both wild-type and Δpcr1 cells) were all confirmed to be Pcr1 independent for their KCl induction (Table 1). Of these 32 genes, half behaved similarly to gpd1: their stress-induced up-regulation was dependent on Atf1 but not on Pcr1. On the other hand, for 15 of the 32 genes, basal levels were derepressed both in Δatf1 and Δpcr1 cells or only in Δpcr1 cells, but they showed KCl-dependent induction only in Δpcr1 cells. Therefore, KCl treatment induces most genes from group F more than twofold in an Atf1-dependent but Pcr1-independent manner.
TABLE 1.
Genes induced more than twofold 20 min after treatment with 0.4 M KCl in wild-type and Δpcr1 cells but not Δatf1 cells (Atf1-dependent but Pcr1-independent genes)
| Gene name or identifierd | Fold induction of gene for cell genotype, treatmenta
|
|||||
|---|---|---|---|---|---|---|
| WT, Unt | Δatf1, Untb | Δpcr1, Untb | WT, KClb | Δatf1, KClc | Δpcr1, KClc | |
| SPAC22A12.17c | 1.00 | 3.38 | 2.62 | 19.57 | 1.28 | 2.16 |
| SPAC22F8.05 | 1.00 | 1.13 | 1.78 | 11.28 | 1.33 | 2.50 |
| SPBC56F2.06 | 1.00 | 2.86 | 6.38 | 8.17 | 1.10 | 3.56 |
| SPAC4H3.03c | 1.00 | 3.70 | 3.91 | 7.15 | 1.37 | 2.57 |
| SPBC12C2.04 | 1.00 | 1.60 | 2.14 | 6.77 | 1.20 | 2.19 |
| SPBC11C11.06c | 1.00 | 0.38 | 1.16 | 6.15 | 1.22 | 3.34 |
| SPBC23G7.10c | 1.00 | 0.45 | 0.62 | 6.01 | 1.43 | 3.17 |
| SPAC3C7.05c | 1.00 | 1.75 | 1.89 | 5.71 | 1.66 | 2.58 |
| SPBC428.10 | 1.00 | 0.87 | 1.01 | 5.18 | 1.56 | 2.23 |
| SPBC16A3.02c | 1.00 | 1.25 | 4.18 | 4.81 | 1.57 | 4.23 |
| SPAC26F1.07 | 1.00 | 0.41 | 3.81 | 4.51 | 1.42 | 4.12 |
| tps1 | 1.00 | 1.49 | 2.53 | 4.25 | 1.96 | 2.81 |
| ntp1 | 1.00 | 2.56 | 4.07 | 4.07 | 1.92 | 2.74 |
| SPBC21C3.19 | 1.00 | 0.19 | 0.63 | 3.60 | 1.33 | 5.72 |
| gpd1 | 1.00 | 0.34 | 1.11 | 3.41 | 1.21 | 3.70 |
| SPCC965.06 | 1.00 | 0.31 | 0.48 | 3.39 | 1.25 | 2.26 |
| SPAPJ691.02 | 1.00 | 1.10 | 2.07 | 3.33 | 1.81 | 2.04 |
| git5 | 1.00 | 0.82 | 1.32 | 3.17 | 1.21 | 2.19 |
| SPCPB16A4.06c | 1.00 | 1.15 | 1.10 | 3.17 | 1.21 | 2.07 |
| SPBC23E6.03c | 1.00 | 1.01 | 1.41 | 3.06 | 1.26 | 2.02 |
| SPCC63.14 | 1.00 | 2.38 | 3.20 | 2.72 | 0.92 | 2.04 |
| SPAC9E9.04 | 1.00 | 0.32 | 1.01 | 2.54 | 1.09 | 2.46 |
| SPCC306.08c | 1.00 | 2.22 | 2.93 | 2.41 | 1.84 | 2.19 |
| SPACUNK4.15 | 1.00 | 2.17 | 4.07 | 2.30 | 0.88 | 2.69 |
| ght2 | 1.00 | 1.21 | 1.22 | 2.30 | 1.76 | 2.90 |
| SPAC26F1.04c | 1.00 | 0.65 | 0.83 | 2.27 | 1.70 | 2.94 |
| SPAC4H3.04c | 1.00 | 1.23 | 1.89 | 2.25 | 1.76 | 2.00 |
| SPAC3G9.11c | 1.00 | 1.58 | 6.70 | 2.20 | 1.14 | 4.05 |
| SPAC186.02c | 1.00 | 0.80 | 0.94 | 2.16 | 1.59 | 2.21 |
| SPAC16A10.01 | 1.00 | 1.27 | 1.47 | 2.05 | 1.14 | 2.74 |
| SPBC21H7.06c | 1.00 | 2.10 | 1.73 | 2.02 | 1.36 | 3.13 |
| psi1 | 1.00 | 1.55 | 2.65 | 2.02 | 1.98 | 2.27 |
WT, wild type; Unt, untreated.
n-fold induction is indicated using the wild-type strain as a reference (with an assigned value of 1).
n-fold induction for these KCl treatments is in reference to the respective untreated values for the same strain and not to the wild-type untreated values.
Those 15 genes whose untreated levels in Δatf1 and/or Δpcr1 strains are derepressed more than twofold with respect to levels for untreated wild-type cells are emphasized by boldface type.
An important outcome of the analysis of the transcriptome of Δatf1 and Δpcr1 strains is that these transcription factors modulate the basal expression level of a large number of stress-inducible genes. Of the 170 genes induced by KCl in wild-type cells, 20 are derepressed by more than twofold in the absence of stress in both Δatf1 and Δpcr1 cells, 20 are derepressed more than twofold only in Δpcr1 cells, 13 are repressed only in Δatf1 cells, and 7 are repressed in both Δatf1 and Δpcr1 cells (Table 2; see also Table S2 in the supplemental material). Furthermore, the basal expression levels of 4.3% and 6.5% of all the S. pombe genes were derepressed by more than 1.5-fold in Δatf1 and Δpcr1 strains, respectively (data not shown).
TABLE 2.
KCl-induced genes derepressed or repressed more than twofold under untreated conditions in Δatf1 and/or Δpcr1 cellsa
| Category or gene name or identifier | Fold induction of gene for cell genotype
|
||
|---|---|---|---|
| WT | Δatf1 | Δpcr1 | |
| KCl-dependent genes derepressed >2-fold in Δatf1 and Δpcr1 cells | |||
| SPBPB21E7.04c | 1.00 | 2.84 | 10.04 |
| SPAC6B12.03c | 1.00 | 9.06 | 8.81 |
| SPBC56F2.06 | 1.00 | 2.86 | 6.38 |
| isp6 | 1.00 | 2.55 | 4.31 |
| SPACUNK4.15 | 1.00 | 2.17 | 4.07 |
| ntp1 | 1.00 | 2.56 | 4.07 |
| SPAC4H3.03c | 1.00 | 3.70 | 3.91 |
| fbp1 | 1.00 | 2.87 | 3.25 |
| SPCC63.14 | 1.00 | 2.38 | 3.20 |
| SPBC660.06 | 1.00 | 2.29 | 3.17 |
| SPAC13F5.07c | 1.00 | 2.42 | 3.15 |
| SPCP31B10.06 | 1.00 | 2.12 | 3.02 |
| SPCC306.08c | 1.00 | 2.22 | 2.93 |
| SPAC29A4.17c | 1.00 | 2.28 | 2.71 |
| SPAC22A12.17c | 1.00 | 3.38 | 2.62 |
| mug66 | 1.00 | 2.70 | 2.41 |
| SPBC216.03 | 1.00 | 2.04 | 2.30 |
| SPAC13D6.01 | 1.00 | 2.87 | 2.25 |
| SPCC4G3.03 | 1.00 | 2.02 | 2.13 |
| SPBC24C6.09c | 1.00 | 2.70 | 2.09 |
| KCl-dependent genes derepressed >2-fold only in Δpcr1 cells | |||
| SPAC3G9.11c | 1.00 | 1.58 | 6.70 |
| SPAC29A4.12c | 1.00 | 1.25 | 4.73 |
| atf1 | 1.00 | 0.13 | 4.53 |
| SPBC16A3.02c | 1.00 | 1.25 | 4.18 |
| SPAC26F1.07 | 1.00 | 0.41 | 3.81 |
| SPAC27D7.11c | 1.00 | 1.50 | 3.53 |
| hsp16 | 1.00 | 0.58 | 3.22 |
| psi1 | 1.00 | 1.55 | 2.65 |
| SPACUNK4.16c | 1.00 | 1.48 | 2.61 |
| SPCC794.04c | 1.00 | 0.21 | 2.60 |
| tps1 | 1.00 | 1.49 | 2.53 |
| SPCPB1C11.02 | 1.00 | 1.52 | 2.50 |
| SPAC23G3.13c | 1.00 | 1.44 | 2.39 |
| SPAPB1A11.03 | 1.00 | 1.43 | 2.32 |
| SPCC417.13 | 1.00 | 1.57 | 2.30 |
| SPBC12C2.04 | 1.00 | 1.60 | 2.14 |
| SPAPJ691.02 | 1.00 | 1.10 | 2.07 |
| SPBC1711.11 | 1.00 | 1.89 | 2.06 |
| SPAC13F5.03c | 1.00 | 0.75 | 2.03 |
| SPCC1183.09c | 1.00 | 1.45 | 2.02 |
| KCl-dependent genes repressed >2-fold only in Δatf1 cells | |||
| hsp9 | 1.00 | 0.03 | 1.08 |
| SPBC21C3.19 | 1.00 | 0.19 | 0.63 |
| SPCC794.04c | 1.00 | 0.21 | 2.60 |
| SPAC9E9.04 | 1.00 | 0.32 | 1.01 |
| gpd1 | 1.00 | 0.34 | 1.11 |
| SPAC27D7.10c | 1.00 | 0.36 | 1.10 |
| SPBC11C11.06c | 1.00 | 0.38 | 1.16 |
| SPAC27D7.09c | 1.00 | 0.40 | 1.06 |
| SPAC19G12.09 | 1.00 | 0.43 | 0.69 |
| SPBC23G7.10c | 1.00 | 0.45 | 0.62 |
| SPAPB24D3.07c | 1.00 | 0.47 | 1.78 |
| SPAC22F8.03c | 1.00 | 0.48 | 0.71 |
| SPBPB2B2.02 | 1.00 | 0.48 | 0.65 |
| KCl-dependent genes repressed >2-fold in Δatf1 and Δpcr1 cells | |||
| SPBPB10D8.02cb | 1.00 | 0.07 | 0.15 |
| SPBPB2B2.08 | 1.00 | 0.08 | 0.12 |
| SPBPB10D8.01 | 1.00 | 0.08 | 0.18 |
| gpx1 | 1.00 | 0.09 | 0.19 |
| rds1 | 1.00 | 0.31 | 0.42 |
| SPCC965.06 | 1.00 | 0.31 | 0.48 |
| SPAC15E1.02c | 1.00 | 0.37 | 0.48 |
Unstressed values only; see Table S2 in the supplemental material for KCl-induced values. n-fold inductions are indicated using the wild-type strain as a reference (with an assigned value of 1).
The five genes repressed in the Δatf1 and Δpcr1 strains and in boldface type belong to the cadmium response.
DISCUSSION
The participation of the MAPK Sty1 in several cellular functions has been firmly established (for reviews, see references 14 and 40). Cells lacking Sty1 are sensitive to many environmental stresses and are defective in specific cellular programs, such as mating, adaptation to stationary phase, or hot-spot-induced meiotic recombination. Most of the roles of Sty1 are mediated by the transcription factor Atf1, which specifically binds to cis sites on DNA to either activate transcription or promote recombination. There has been controversy about whether Pcr1 is required for these cellular functions; whereas some authors claim that it is dispensable for stress responses (18, 19), most reports related to stress responses suggest, more than demonstrate, that Pcr1 and Atf1 work as a heterodimer to regulate the transcription of all CESR genes. Here we provide evidence demonstrating that Pcr1 is required for the transcriptional activation of most but not all Sty1-dependent genes in response to diverse stress stimuli. From our studies we can conclude that Atf1 and Pcr1 play similar but not identical roles in the stress response of fission yeast. For example, Pcr1 is not required for osmotic or oxidative stress on solid plates but is required for heat shock and oxidative stress in liquid cultures (Fig. 3AB). Furthermore, the phenotype of a strain lacking both Atf1 and Pcr1 is more severe under all stress conditions assayed than that of a single Δatf1 strain (Fig. 3B), which indicates that their roles do not fully overlap.
Our data confirm that the presence of Atf1 is essential for the Sty1-dependent transcriptional response to stress, with the majority of KCl-dependent genes dependent on the presence of Atf1 for their expression (Fig. 7) (see Table S1 in the supplemental material). It was recently suggested that stress induction of CESR genes is critically dependent on the concentration of Atf1 (20). The authors claim that basal Sty1-dependent phosphorylation stabilizes the Atf1 protein, since the steady-state levels of a mutant Atf1 protein lacking all Sty1 phosphorylation sites are 10-fold lower than those of the wild-type protein. However, we have shown here that Atf1 protein steady-state levels, when expressed under the control of the nmt promoter, are not dependent on the presence of Sty1 (Fig. 5B). The same study also proposed that Atf1 stability is regulated by its heterodimeric partner Pcr1, since the expression of Atf1 in a strain lacking Pcr1 is lower than that in wild-type cells (20). Nevertheless, we show here that the levels of the nmt-driven Atf1 protein in Δpcr1 Δatf1 cells are very similar to those in single-mutant Δatf1 cells. Our results suggest that Atf1 protein levels are mainly regulated at the level of transcription and/or translation, rather than by protein stabilization mediated by basal Sty1-dependent phosphorylation or dimerization with Pcr1. We also show here that the role of Pcr1 is not to anchor Atf1 at the nucleus, as previously suggested (11): when GFP-Atf1 is expressed from a heterologous promoter in wild-type and Δpcr1 cells, the fusion protein shows nuclear localization in both cases (Fig. 1C). We suspect that the low expression of Atf1 in cells lacking Pcr1 could explain the lack of nuclear immunofluorescence reported earlier (11). Regarding posttranscriptional regulation of these transcription factors, we demonstrate here that Pcr1 is a phosphoprotein and dephosphorylation occurs upon oxidative, but not osmotic, stress in a Sty1-dependent manner (Fig. 2). Analysis of strains expressing mutant Pcr1 forms with substitutions at its Ser 122 position, the only MAPK phosphorylation site in Pcr1, will help to unravel whether such phosphorylation status is important for Pcr1 function. It is noteworthy, however, that the Sty1-dependent phosphorylation of Atf1 upon stress does not seem to be essential for its role as a transcription factor (20).
Transcriptional profiling analysis revealed that some Sty1-dependent genes are dependent upon Atf1, but dependence upon Pcr1 was not tested (8). Here we show that the overall KCl response is highly dependent on Pcr1, although a subset of 32 genes seems to be induced in Δpcr1 cells as effectively as in wild-type cells. However, we would caution that we have only performed a comparative analysis by microarrays in response to one stress stimulus and at one time point. Even though a wider expression analysis has been performed by Northern blotting regarding different signals and different times after stress imposition (see Fig. S1 in the supplemental material), a more exhaustive global analysis will be required to broaden our knowledge about the participation of Pcr1 in stress-induced transcriptional regulation.
We also show here by chromatin immunoprecipitation that both Atf1 and Pcr1 are bound to most CESR promoters (exemplified by cta3) before and after stress and that binding of Atf1 to these promoter regions requires the participation of Pcr1. The in vivo protein-DNA binding experiments presented here also indicate that in wild-type cells both Atf1 and Pcr1 are bound to the gpd1 and hsp9 promoters, whereas Atf1 but not Pcr1 can be detected at the srx1 promoter. We propose that the Atf1-Pcr1 heterodimer is normally bound at most promoters, with very few exceptions. The small bZIP factor Pcr1 probably stabilizes the interaction of Atf1 with all stress gene promoters, but it is dispensable in others, such as srx1. The lack of Pcr1 would thus weaken Atf1 binding to most but not all of these promoters. We do not know whether in the absence of Pcr1 Atf1 binds to DNA as a homodimer or whether it heterodimerizes with another partner. The gene encoding the bZIP transcription factor Atf21 is strongly expressed under sorbitol stress conditions (8, 28). We postulated that Atf21 might substitute for Pcr1 by heterodimerizing with Atf1 at the promoters of the 32 genes whose expression was independent of Pcr1 upon KCl stress. However, a double-knock-out Δpcr1 Δatf21 strain is still resistant to osmotic stress conditions (data not shown).
Regarding the role of these bZIP factors in the general repression of gene expression, it was reported that under basal conditions Atf1 and Pcr1 are involved in heterochromatin nucleation at the mat locus and that deletion of atf1 or pcr1 in combination with RNA interference mutants fails to promote heterochromatin assembly in this chromosomal region (15). It has also been reported that the Atf1/Pcr1 heterodimer functions as both an inducer and a repressor of chromatin remodeling at the cgs2 promoter (9). According to our data, Atf1 and Pcr1 down-regulate the basal transcription of 4% and 6% of the whole S. pombe genome, respectively. Through their chromatin binding and remodeling activities, these proteins, acting alone or in combination with other factors, may facilitate the assembly of transcription, recombination, or silencing machinery. The involvement of these stress transcription factors in general repression suggests that they might modify chromatin structure as a part of a programmed sequence of events that serves to cushion against the effects of environmental stresses. However, derepression does not seem to be a factor in the roles of Pcr1 and Atf1 on stress activation, since in atf1 or pcr1 deletion mutant strains the basal expression of the subset of genes highly induced by KCl is unaffected or only slightly elevated above wild-type levels (see Table S1). In conclusion, the heterodimer Atf1-Pcr1 seems to bind to most CESR genes and is required for their activation upon stress. However, both proteins can probably work independently of each other, both regarding (i) stress-dependent gene activation (in cells lacking Pcr1, a subset of genes is still triggered by Atf1, which binds to their promoters either alone or forming a heterodimer with a yet-uncharacterized partner) and (ii) basal repression (in cells lacking Pcr1, 6% of the whole genome is derepressed, whereas in the absence of Atf1 only 4% of the total mRNAs show enhanced basal levels).
Supplementary Material
Acknowledgments
We thank Mercè Carmona and Pol Margalef for technical assistance and other members of the laboratory for helpful discussions. We also thank Karin Zueckert-Gaudenz for RNA labeling, hybridization, and quantification of the arrays. We acknowledge Caroline Wilkinson and Nic Jones for communicating results prior to publication.
This work was supported by Dirección General de Investigación of Spain grant BFU2006-02610, by the Spanish program Consolider-Ingenio 2010, grant CSD 2007-0020, and by Distinció de la Generalitat de Catalunya per a la Promoció de la Recerca Universitaria, Joves Investigadors DURSI (Generalitat de Catalunya) to E.H.
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aiba, H., H. Yamada, R. Ohmiya, and T. Mizuno. 1995. The osmo-inducible gpd1+ gene is a target of the signaling pathway involving Wis1 MAP-kinase kinase in fission yeast. FEBS Lett. 376199-201. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 3.Aoyama, K., H. Aiba, and T. Mizuno. 2001. Genetic analysis of the His-to-Asp phosphorelay implicated in mitotic cell cycle control: involvement of histidine-kinase genes of Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 652347-2352. [DOI] [PubMed] [Google Scholar]
- 4.Aoyama, K., Y. Mitsubayashi, H. Aiba, and T. Mizuno. 2000. Spy1, a histidine-containing phosphotransfer signaling protein, regulates the fission yeast cell cycle through the Mcs4 response regulator. J. Bacteriol. 1824868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 2591760-1763. [DOI] [PubMed] [Google Scholar]
- 6.Buck, V., J. Quinn, P. T. Soto, H. Martin, J. Saldanha, K. Makino, B. A. Morgan, and J. B. Millar. 2001. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12407-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo, E. A., J. Ayte, C. Chiva, A. Moldon, M. Carrascal, J. Abian, N. Jones, and E. Hidalgo. 2002. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 45243-254. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson, M. K., H. K. Shandilya, K. Hirota, K. Ohta, and W. P. Wahls. 2004. Atf1-Pcr1-M26 complex links stress-activated MAPK and cAMP-dependent protein kinase pathways via chromatin remodeling of cgs2+. J. Biol. Chem. 27950857-50863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii, Y., T. Shimizu, T. Toda, M. Yanagida, and T. Hakoshima. 2000. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 7889-893. [DOI] [PubMed] [Google Scholar]
- 11.Gaits, F., G. Degols, K. Shiozaki, and P. Russell. 1998. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 121464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaits, F., and P. Russell. 1999. Active nucleocytoplasmic shuttling required for function and regulation of stress-activated kinase Spc1/StyI in fission yeast. Mol. Biol. Cell 101395-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaits, F., K. Shiozaki, and P. Russell. 1997. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J. Biol. Chem. 27217873-17879. [DOI] [PubMed] [Google Scholar]
- 14.Ikner, A., and K. Shiozaki. 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 56913-27. [DOI] [PubMed] [Google Scholar]
- 15.Jia, S., K. Noma, and S. I. Grewal. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 3041971-1976. [DOI] [PubMed] [Google Scholar]
- 16.Kanoh, J., Y. Watanabe, M. Ohsugi, Y. Iino, and M. Yamamoto. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1391-408. [DOI] [PubMed] [Google Scholar]
- 17.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith, and W. P. Wahls. 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 9413765-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kon, N., S. C. Schroeder, M. D. Krawchuk, and W. P. Wahls. 1998. Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol. Cell. Biol. 187575-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, C. L., H. Maekawa, J. L. Worthington, W. Reiter, C. R. Wilkinson, and N. Jones. 2007. Regulation of Schizosaccharomyces pombe Atf1 protein levels by Sty1-mediated phosphorylation and heterodimerization with Pcr1. J. Biol. Chem. 2825160-5170. [DOI] [PubMed] [Google Scholar]
- 21.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123127-130. [DOI] [PubMed] [Google Scholar]
- 22.Millar, J. B., V. Buck, and M. G. Wilkinson. 1995. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 92117-2130. [DOI] [PubMed] [Google Scholar]
- 23.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 24.Nakamichi, N., H. Yamada, K. Aoyama, R. Ohmiya, H. Aiba, and T. Mizuno. 2002. His-to-Asp phosphorelay circuitry for regulation of sexual development in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 662663-2672. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen, A. N., A. Lee, W. Place, and K. Shiozaki. 2000. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 111169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen, A. N., and K. Shiozaki. 1999. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 131653-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, K. H., B. Szamecz, L. Valasek, A. Jivotovskaya, B. S. Shin, and A. G. Hinnebusch. 2004. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 231166-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmiya, R., C. Kato, H. Yamada, H. Aiba, and T. Mizuno. 1999. Isolation of multicopy suppressors of the calcium sensitivity of a mutant lacking the bZIP transcription factor Atf1 in fission yeast. Mol. Gen. Genet. 261297-306. [DOI] [PubMed] [Google Scholar]
- 29.Orlandi, I., P. Cavadini, L. Popolo, and M. Vai. 1996. Cloning, sequencing and regulation of a cDNA encoding a small heat-shock protein from Schizosaccharomyces pombe. Biochim. Biophys. Acta 1307129-131. [DOI] [PubMed] [Google Scholar]
- 30.Samejima, I., S. Mackie, and P. A. Fantes. 1997. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 166162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Piris, M., F. Posas, V. Alemany, I. Winge, E. Hidalgo, O. Bachs, and R. Aligue. 2002. The serine/threonine kinase Cmk2 is required for oxidative stress response in fission yeast. J. Biol. Chem. 27717722-17727. [DOI] [PubMed] [Google Scholar]
- 32.Shieh, J. C., M. G. Wilkinson, V. Buck, B. A. Morgan, K. Makino, and J. B. Millar. 1997. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 111008-1022. [DOI] [PubMed] [Google Scholar]
- 33.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 102276-2288. [DOI] [PubMed] [Google Scholar]
- 34.Shiozaki, K., M. Shiozaki, and P. Russell. 1997. Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol. Biol. Cell 8409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinibaldi, R., C. O'Connell, C. Seidel, and H. Rodriguez. 2001. Gene expression analysis on medium-density oligonucleotide arrays. Methods Mol. Biol. 170211-222. [DOI] [PubMed] [Google Scholar]
- 36.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3Article3. [DOI] [PubMed] [Google Scholar]
- 37.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 146193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivancos, A. P., E. A. Castillo, B. Biteau, C. Nicot, J. Ayte, M. B. Toledano, and E. Hidalgo. 2005. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. USA 1028875-8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vivancos, A. P., E. A. Castillo, N. Jones, J. Ayte, and E. Hidalgo. 2004. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 521427-1435. [DOI] [PubMed] [Google Scholar]
- 40.Vivancos, A. P., M. Jara, A. Zuin, M. Sanso, and E. Hidalgo. 2006. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol. Genet. Genomics 276495-502. [DOI] [PubMed] [Google Scholar]
- 41.Wahls, W. P., and G. R. Smith. 1994. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 81693-1702. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 102289-2301. [DOI] [PubMed] [Google Scholar]
- 44.Wolberger, C. 1998. Combinatorial transcription factors. Curr. Opin. Genet. Dev. 8552-559. [DOI] [PubMed] [Google Scholar]
- 45.Wu, T. C., and M. Lichten. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263515-518. [DOI] [PubMed] [Google Scholar]
- 46.Yamada, T., K. Mizuno, K. Hirota, N. Kon, W. P. Wahls, E. Hartsuiker, H. Murofushi, T. Shibata, and K. Ohta. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 231792-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.