Abstract
We characterized the oxidative stress response of Candida glabrata to better understand the virulence of this fungal pathogen. C. glabrata could withstand higher concentrations of H2O2 than Saccharomyces cerevisiae and even Candida albicans. Stationary-phase cells were extremely resistant to oxidative stress, and this resistance was dependent on the concerted roles of stress-related transcription factors Yap1p, Skn7p, and Msn4p. We showed that growing cells of C. glabrata were able to adapt to high levels of H2O2 and that this adaptive response was dependent on Yap1p and Skn7p and partially on the general stress transcription factors Msn2p and Msn4p. C. glabrata has a single catalase gene, CTA1, which was absolutely required for resistance to H2O2 in vitro. However, in a mouse model of systemic infection, a strain lacking CTA1 showed no effect on virulence.
Recent surveys show that Candida species are responsible for about 8% of all hospital-acquired bloodstream infections, and among these species, the two most frequently isolated are Candida albicans and Candida glabrata (60, 73). C. glabrata is an opportunistic fungal pathogen that is a commensal in human gastrointestinal and genitourinary tracts but that also causes severe invasive infections. Phylogenetically, C. glabrata is quite distinct from C. albicans but is closely related to Saccharomyces cerevisiae. The virulence attributes that allow C. glabrata to colonize human tissues and cause disseminated infections have recently started to be identified (reviewed in reference 40).
Phagocytic cells are the first line of defense against fungal infections (49). These cells generate reactive oxygen species (ROS), including superoxide, hydrogen peroxide (H2O2), and hydroxyl radicals, that can damage all biomolecules and destroy phagocytosed pathogens (27, 70). ROS are also by-products of normal aerobic metabolism, and all aerobic organisms possess mechanisms to maintain very low levels of these species. In particular, a variety of small antioxidant molecules, such as glutathione and thioredoxin, are synthesized to scavenge ROS, and even tyrosine has been proposed to have a protective role against oxidative stress (48). In addition, several well-characterized enzymes, such as the superoxide dismutases, catalases, peroxidases, and glutathione peroxidases, are produced to eliminate ROS. Pathogens have coopted these well-conserved antioxidation mechanisms to evade phagocyte defenses (5, 27, 69, 70); thus, the production of these enzymes is directly related to virulence (35, 76).
When cells are under oxidative stress, transcriptional remodeling occurs to ensure the proper response. The enzymes and the regulation of the oxidative stress response (OSR) are well conserved among fungal species. Catalases are well-conserved detoxifying enzymes catalyzing the conversion of H2O2 to H2O and molecular oxygen (reviewed in references 1 and 2). S. cerevisiae has two catalase genes, both of which are required for detoxifying H2O2 (13, 30, 65, 66, 72). Both C. albicans and C. glabrata carry only one catalase gene, and C. albicans catalase has been shown to play an important role in the virulence of C. albicans (56, 74, 76). The OSR in S. cerevisiae is in part under the control of the well-studied transcription factors Yap1p, Skn7p, Msn2p, and Msn4p (21, 43, 44, 46, 53, 64). S. cerevisiae Yap1p (ScYap1p) belongs to the family of basic leucine zipper domain transcription factors and controls the expression of at least 32 proteins of the H2O2 stimulon (46). Strains lacking Yap1p are hypersensitive to H2O2. The Yap1p orthologs in C. albicans (Cap1p) (3, 78), Schizosaccharomyces pombe (Pap1p) (71), and Ustilago maydis (Yap1p) (52) have been characterized previously, and they are involved in the OSR. The C. glabrata Yap1p ortholog is functionally involved not only in the OSR, but also in resistance to different drugs (11). ScSkn7p contains a receiver domain found in the family of two-component signal transduction systems of prokaryotes and a DNA-binding domain similar to that of heat shock factor Hsf1p (6, 54). The target genes of ScSkn7p overlap with those of ScYap1p, and a skn7Δ strain is hypersensitive to H2O2 (46, 53). The C. albicans Skn7p ortholog has been characterized previously, and cells lacking Skn7p are modestly attenuated in virulence (68). ScMsn2p and ScMsn4p are functionally nonredundant Zn2+ finger transcription factors involved in the general stress response, including the response to oxidative stress (reviewed in references 20 and 55). They control the expression of about 27 gene products regulated in response to H2O2 (31). Msn2p and Msn4p play an important role in stationary-phase (SP) survival under oxidative stress. S. cerevisiae cells lacking Msn2p and Msn4p are sensitive to H2O2 (21, 31, 50, 55, 63). Interestingly, C. albicans Msn2p (CaMsn2p) and CaMsn4p play no obvious role in the stress response, including the response to oxidative stress (58).
The OSR of C. glabrata has not been analyzed previously. In this study, we showed that the growth of C. glabrata could withstand higher concentrations of H2O2 than that of S. cerevisiae and even that of C. albicans (see below) (5, 9). This phenotype could be seen in both log-phase (LP) and SP cells, but the phenotype was more profound in the latter case. SP resistance was dependent on the concerted roles of Yap1p, Skn7p, and Msn4p. We also showed that C. glabrata was able to adapt to high levels of H2O2 and that this adaptive response was dependent on the stress-related transcription factors Yap1p and Skn7p and partially on the general stress transcription factors Msn2p and Msn4p. Lastly, we showed that the C. glabrata catalase gene CTA1 was absolutely required for resistance to H2O2 in vitro. However, a strain lacking CTA1 had no obvious phenotype in vivo in a mouse model of systemic infection.
MATERIALS AND METHODS
Strains.
All strains used in this study are summarized in Table 1.
TABLE 1.
Strains used in this study
| Strain | Parent | Genotype and/or description | Reference or source |
|---|---|---|---|
| E. coli strains | |||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara,leu)7697 galU galK λ−rpsL nupG | 7 | |
| JM109 | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | 77 | |
| S. cerevisiae strains | |||
| W303 | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1 ade3::hisG | 51 | |
| YJM128 | Clinical isolate | 12 | |
| YJM336 | Clinical isolate | 12 | |
| C. albicans strains | |||
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 25 | |
| CA5 | Clinical isolate | Lab collection | |
| CA7 | Clinical isolate | Lab collection | |
| C. glabrata strains | |||
| MC7 | Clinical isolate | Lab collection | |
| MC22 | Clinical isolate | Lab collection | |
| BG2 | Clinical isolate (strain B) | 22 | |
| BG14 | BG2 | ura3Δ::Tn903 G418r; wt Ura− strain used in this study | 15 |
| BG462 | BG14 | URA3 | 18 |
| BG1739 (msn2Δ mutant) | BG14 | ura3Δ::Tn903 G418rmsn2Δ Hygs Ura− pRD96/BcgI | R. Domergue and B. Cormack |
| BG1740 (msn4Δ mutant) | BG14 | ura3Δ::Tn903 G418rmsn4Δ Hygs Ura− pRD97/BcgI | R. Domergue and B. Cormack |
| BG1742 (msn2Δ msn4Δ mutant) | BG1739 | ura3Δ::Tn903 G418rmsn2Δ msn4Δ::hph Hygr | R. Domergue and B. Cormack |
| CGM295 (cta1Δ mutant) | BG14 | ura3Δ::Tn903 G418rcta1Δ::hph Hygr pCV15/BcgI | This work |
| CGM297 (yap1Δ mutant) | BG14 | ura3Δ::Tn903 G418ryap1Δ::hph Hygr pCV17/BcgI | This work |
| CGM306 (skn7Δ mutant) | BG14 | ura3Δ::Tn903 G418rskn7Δ::hph Hygr pCV21/BcgI | This work |
| CGM308 (yap1Δ mutant) | CGM297 | ura3Δ::Tn903 G418ryap1Δ Hygs Ura− | This work |
| CGM310 (yap1Δ skn7Δ mutant) | CGM308 | ura3Δ::Tn903 G418ryap1Δ skn7Δ::hph Hygr | This work |
| CGM351 (cta1Δ mutant) | CGM295 | URA3 cta1Δ::hph Hygr | This work |
| CGM385 (yap1Δ skn7Δ mutant) | CGM310 | ura3Δ::Tn903 G418ryap1Δ skn7Δ Hygs Ura− | This work |
| CGM386 (yap1Δ skn7Δ msn2Δ mutant) | CGM385 | ura3Δ::Tn903 G418ryap1Δ skn7Δ msn2Δ::hph Hygr | This work |
| CGM388 (yap1Δ skn7Δ msn4Δ mutant) | CGM385 | ura3Δ::Tn903 G418ryap1Δ skn7Δ msn4Δ::hph Hygr | This work |
| CGM480 (yap1Δ skn7Δ msn4Δ mutant) | CGM388 | ura3Δ::Tn903 G418ryap1Δ skn7Δ msn4Δ Hygs Ura− | This work |
| CGM537 (yap1Δ skn7Δ msn4Δ msn2Δ mutant) | CGM480 | ura3Δ::Tn903 G418ryap1Δ skn7Δ msn4Δ msn2Δ::hph Hygr | This work |
Plasmids.
All plasmids used in this study are summarized in Table 2.
TABLE 2.
Plasmids used in this study
| Plasmid | Description and/or relevant genotype | Reference or source |
|---|---|---|
| Plasmids for cloning, construction, and marker removal | ||
| pGEM-T | Cloning vector; Kmr Apr | Promega |
| pAP599 | Cloning vector with 2 FRT direct repeats flanking the hygromycin marker [FRT-PPGK::hph::(3′ UTR of HIS3)-FRT] for construction of multiple mutants; URA3 Hygr Ampr | 19 |
| pMZ17 | Replicative vector expressing ScFLP1 (recombinase gene) for removing the hygromycin marker; PPGK::FLP1::(3′ UTR of HIS3) CgCEN ARS Ampr | Cormack lab collection |
| pBC34.1 | pUC19::CgURA3; 2.2-kb PstI fragment; Apr | 15 |
| Plasmids for deletion | ||
| pCV15 cta1Δ | A 0.906-kb SpeI/BamHI PCR fragment (corresponding to primers 57 and 58) carrying the promoter region of CTA1 and a 0.682-kb HindIII/KpnI PCR fragment (corresponding to primers 60 and 61) carrying the 3′ UTR of CTA1 were cloned into pAP599 Apr | This work |
| pCV17 yap1Δ | A 0.846-kb XhoI/HindIII PCR fragment (corresponding to primers 7 and 8) carrying the promoter region of YAP1 and a 0.652-kb BamHI/SacI PCR fragment (corresponding to primers 11 and 12) carrying the 3′ UTR of YAP1 were cloned into pAP599 Apr | This work |
| CV21 skn7Δ | A 0.875-kb KpnI/XhoI PCR fragment (corresponding to primers 4 and 5) carrying the promoter region of SKN7 and a 0.929-kb BamHI/SacI PCR fragment (corresponding to primers 1 and 2) carrying the 3′ UTR of SKN7 were cloned into pAP599 Apr | This work |
| pRD96 msn2Δ | A 0.691-kb KpnI/XhoI PCR fragment (corresponding to primers 2984 and 2985) carrying the promoter region of MSN2 and a 0.520-kb BamHI/SacI PCR fragment (corresponding to primers 2986 and 2987) carrying the 3′ UTR of MSN2 were cloned into pAP599 Apr | R. Domergue and B. Cormack |
| pRD97 msn4Δ | A 0.564-kb KpnI/XhoI PCR fragment (corresponding to primers 2990 and 2991) carrying the promoter region of MSN4 and a 0.535-kb BamHI/SacI PCR fragment (corresponding to primers 2992 and 2993) carrying the 3′ UTR of MSN4 were cloned into pAP599 Apr | R. Domergue and B. Cormack |
Primers.
All primers used for cloning are summarized in Table 3.
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequencea | Description or restriction site(s) |
|---|---|---|
| 13 | GTTGTAAAACGACGGCCAGTG | pUC forward |
| 17 | GGAAACAGCTATGACCATG | pUC reverse |
| 1 | CGCGGATCCAAGTATACTGCTATGAGCTAC | BamHI |
| 2 | CAAGGAGCTCTTGTGCAGCGTGTAAGATATGAATCAAGTGAT | SacI/BsgI |
| 4 | TCAGATCTCGAGTTGACCGTGACCGAAAC | XhoI |
| 5 | CCGGGTACCTTGTGCAGTCGAAGTTAGAGTGCCCTATTC | KpnI/BsgI |
| 7 | AATCTCGAGTTGTGCAGTGCGGGTAACAATTCTCGGCG | XhoI/BsgI |
| 8 | CCCAAGCTTTTACTTCCTAGTTCTTGTCTC | HindIII |
| 11 | CGCGGATCCTGTCTATATTATCTCGGTAGATC | BamHI |
| 12 | CAAGGAGCTCTTGTGCAGTCAACTCATAGATCACAACATTAACAC | SacI/BsgI |
| 57 | CGCGGATCCTCAATTGTGGGAAGTTATCTAATAAGCC | BamHI |
| 58 | CACTACTAGTTAAACACTTGTAGGAG | SpeI |
| 60 | CCCAAGCTTTTGAACCACGTAAAGTGCTGT | HindIII |
| 61 | CCGGGTACCTTGTGCAGCCTGTAAGGACTTCTAAACC | KpnI/BsgI |
| 2984 | GGTACCCGAGGGAGTTGCCTTCACTATGTGCGT TGAG | KpnI/BcgI |
| 2985 | CTCGAGCTGTTCTTGTTGATCTGTGTTTGGT | XhoI |
| 2986 | GGATTCTGACAGTGTTCCTTATTTTATCTAG | BamHI |
| 2987 | GAGCTCCGACCGCGGTGCATCTGTTACCAGGTTAGCC | SacI/BcgI |
| 2990 | GGTACCCGACTCCTGTGCCTTGTCGTACCAGAGAAAC | KpnI/BcgI |
| 2991 | CTCGAGGAACAGGAATGGACACTAATATATA | XhoI |
| 2992 | GGATCCCCATTCTTTATTTTATTTTCTGCTA | BamHI |
| 2993 | GAGCTCCGAGGCATCTGCTAATAATTTTCCGTTTCAA | SacI/BcgI |
Restriction sites are indicated in boldface.
Media.
Yeast media were prepared as described previously (67), and 2% agar was added for plates. Yeast extract-peptone-dextrose (YPD) medium contained yeast extract at 10 g/liter and peptone at 20 g/liter and was supplemented with 2% glucose. Synthetic complete medium (SC) was a mixture of yeast nitrogen base, without NH2SO4, at 1.7 g/liter and NH2SO4 at 5 g/liter and was supplemented with 0.6% Casamino Acids, 2% glucose, and when needed, uracil at 25 mg/liter and 5-fluoroorotic acid (5-FOA) at 1.1 g/liter for 5-FOA plates. YPD plates were supplemented with either hygromycin (Invitrogen) at 200 μg/ml or penicillin (100 U/ml)-streptomycin (100 μg/ml) (GIBCO BRL). Bacterial medium was prepared as described previously (4), and 1.5% agar was used for plates. Luria-Bertani medium contained yeast extract at 5 g/liter, Bacto peptone at 10 g/liter, and NaCl at 10 g/liter and was supplemented where needed with either 30 μg of kanamycin (Invitrogen)/ml or 100 μg of carbenicillin (Invitrogen)/ml. Phosphate-buffered saline (PBS) was 8 g of NaCl/ml, 0.2 g of KCl/liter, 1.65 g of Na2HPO4·7H2O/liter, and 0.2 g of KH2PO4/liter.
Transformation.
Yeast transformations with linear or supercoiled plasmid DNA were done as described previously (8).
Sequence analysis.
The amino acid sequence homology analysis was done by ClustalW alignment (33) with the MacVector program (Accelrys).
Construction of deletion strains.
To construct deletion strains in this study, first we PCR amplified the promoter and 3′ untranslated region (3′ UTR) of each gene to be deleted (CTA1, YAP1, and SKN7) and cloned the amplified fragments into pGEM-T (Promega). Both fragments, the 5′ and the 3′ regions of each gene, were subcloned in the same direction, flanking the hygromycin cassette in pAP599 (Table 2). All plasmid constructs were introduced into Escherichia coli DH10 or JM109 by electroporation (4), and plasmids were purified by using Qiagen mini prep kits. The mutant clones were recombined in C. glabrata strains by a one-step replacement procedure. Plasmids were cut with a set of enzymes leaving homologous ends, and the linear fragments were gel purified and used to transform C. glabrata. Transformants were isolated by selecting for the Hygr phenotype on YPD plates with hygromycin at 200 μg/ml. Insertion at the correct locus was verified with locus-specific genomic primers external to the cloned fragments. To make double, triple, and quadruple mutants, C. glabrata mutant strains were transformed with pMZ17, and transformants were isolated by selection for the URA+ phenotype on SC-Ura plates. pMZ17 is a replicative vector expressing the product of ScFLP1, a recombinase that recognizes the two direct repeats, the FLP recognition target (FRT) sites, flanking the hygromycin marker in the plasmid constructs as follows: 5′ region of the gene::FRT-PPGK::hph::(3′ UTR of HIS3)-FRT::3′ region of the gene. After Flp1p recognizes the FRT sites, the hygromycin marker is deleted from the chromosome and lost by dilution through cell division. Transformants were purified and streaked onto SC-Ura plates. Single colonies were then grown on nonselective medium (YPD plates) and screened for Hygs on YPD plates with hygromycin at 200 μg/ml for the loss of the hygromycin cassette and for 5-FOA resistance on SC-5-FOA plates for the loss of the Ura+ plasmid pMZ17. URA+ cells die on SC-5-FOA. This protocol allows the construction of multiple mutants. Plasmids and primers used for this procedure are described in Tables 2 and 3. Strains constructed in this way are described in Table 1.
Mouse infections.
Eight- to 9-week-old BALB/c mice (Taconic) were infected with 2.2 × 107 cells in a volume of 100 μl by tail vein injection. The strains BG462 (wild type [wt] strain expressing URA3) and CGM351 (cta1Δ URA3) were grown overnight in YPD, and the cells were washed with 1× PBS and resuspended in 1× PBS to a concentration of 2.2 × 108 cells/ml. The concentration of cells was determined by reading the optical density at 600 nm (OD600) of the culture (the concentration of BG462 cells at an OD600 of 1 is 4 × 107/ml), counting the cells in a hemocytometer, and plating serial dilutions and confirming the number of cells the following day. Mice were kept in cages in groups of 10 until they were sacrificed at day 7 after infection. Kidneys, livers, and spleens were retrieved from the mice, and the organs were homogenized. Dilutions of the homogenates were plated onto YPD-penicillin-streptomycin plates. CFU were counted the following day; geometric means are reported.
H2O2 sensitivity assays.
All the starting overnight cultures of C. glabrata were grown for 36 h in YPD to an OD600 of 30.0 at 30°C. A 30% (wt/wt) H2O2 solution was obtained from Sigma-Aldrich. All liquid cultures and plates were incubated at 30°C. For H2O2 sensitivity assays of LP cells, overnight cultures were diluted in fresh rich medium (YPD) in such a way that all strains went through seven doublings to reach an OD600 of 0.5. Once the cultures reached an OD600 of 0.5 after seven doublings, the cultures were divided, exposed to different H2O2 concentrations, and incubated with shaking for 3 h. For the adaptation experiments, cells were pretreated for 1 h with a nonlethal H2O2 concentration and then challenged with a lethal concentration of H2O2 for 2 additional hours. After the treatment, H2O2 was removed by centrifugation, the cultures were resuspended in distilled H2O, the OD was adjusted when needed to an OD600 of 0.5, and the cultures were then serially diluted in 96-well plates. Each dilution was spotted onto YPD plates, and the plates were incubated at 30°C. All dilutions had the same amount of cells.
For SP experiments, cell cultures at an OD600 of 30.0 were diluted to an OD600 of 0.5 with distilled water or spent medium from the same strain. Cell cultures were divided into aliquots and treated with H2O2 at different concentrations for 3 h. After the treatment, the cultures remained at the same OD600 of 0.5 and H2O2 was removed by centrifugation. Cells were resuspended in distilled water to an OD600 of 0.5, and the suspensions were diluted in 96-well plates and spotted onto YPD plates.
All manipulations for these assays were performed in a 30°C temperature-controlled room to prevent abrupt changes in temperature. It has been reported previously that cold shock has an impact on H2O2 resistance in S. cerevisiae. Since there were small variations among the results of these experiments, experiments were repeated at least four times.
RESULTS
C. glabrata is resistant to high levels of hydrogen peroxide.
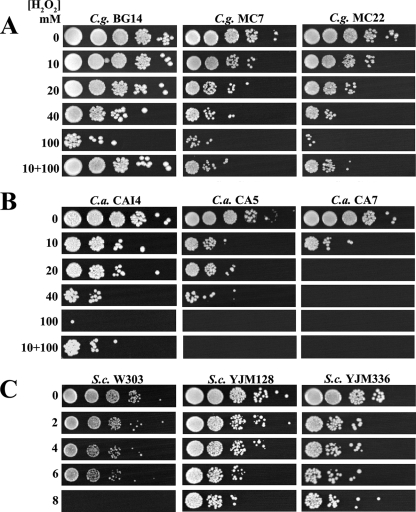
In order to characterize the OSR in C. glabrata, we investigated the resistance of C. glabrata LP cells to H2O2. C. glabrata strains (BG14 [lab reference strain] and clinical isolates [CI] MC7 and MC22) were treated with H2O2 as described in the legend to Fig. 1 and Materials and Methods. As shown in Fig. 1A, C. glabrata BG14 was able to survive exposure to H2O2 at concentrations up to 40 mM. There was a severe loss of viability at 100 mM and a complete loss at 200 mM (data not shown), while CI MC7 and MC22 showed reduced resistance at 40 mM. We also performed an adaptation experiment by pretreating the LP cells with 10 mM H2O2 for 1 h and then treating them with 100 mM H2O2 for 2 additional hours. As shown in Fig. 1A (lane 10+100), the survival rate of BG14 cells pretreated with a low dose of H2O2 was much higher than that of cells without this treatment, and CI MC7 and MC22 showed reduced adaptation compared to that of BG14. Experiments similar to those described above were also performed with C. albicans strains (CAI4 [lab reference strain] and CI CA5 and CA7) and S. cerevisiae strains (W303 [lab reference strain] and CI YJM128 and YJM336). C. albicans CAI4 was resistant to 40 mM H2O2 and showed complete sensitivity to 100 mM H2O2, while C. albicans CI showed reduced resistance compared to that of CAI4 (Fig. 1B). As shown in Fig. 1C, S. cerevisiae was resistant only to 6 mM H2O2 and completely sensitive to 8 mM H2O2. Interestingly, CI YJM128 and YJM336 were more resistant than the W303 lab reference strain. These experiments indicate that LP C. glabrata cells are modestly more resistant to H2O2 than LP C. albicans cells and significantly more resistant than LP S. cerevisiae cells. In addition, C. glabrata and C. albicans cells were able to adapt, though to different levels, suggesting that adaptation is a common trait for both pathogens (Fig. 1A and B). Similarly, S. cerevisiae has been shown previously to be able to adapt to higher concentrations of H2O2 when pretreated with a lower dose (14, 17, 37, 38).
FIG. 1.
C. glabrata, C. albicans, and S. cerevisiae LP resistance to H2O2. Saturated cultures of C. glabrata (C.g.) strain BG14 and CI MC7 and MC22 (A), C. albicans (C.a.) strain CAI4 and CI CA5 and CA7 (B), and S. cerevisiae (S.c.) strain W303 and CI YJM128 and YJM336 (C) were diluted with fresh medium (YPD) so that all strains reached an OD600 of 0.5 after seven doublings at 30°C. C. glabrata and C. albicans strains were divided and exposed to 0, 10, 20, 30, 40, 50, and 100 mM H2O2 and S. cerevisiae strains were exposed to 2, 4, 6, and 8 mM H2O2 for 3 h. For adaptation experiments, C. glabrata and C. albicans cells were pretreated for 1 h with 10 mM H2O2 and then with 100 mM H2O2 for 2 additional hours. After the treatment, H2O2 was removed by centrifugation. The cultures were resuspended in distilled water, and the OD600s were adjusted when needed to 0.5. Cultures were serially diluted, and each dilution was spotted onto YPD plates, ensuring that the same amounts of cells were plated. Plates were incubated at 30°C. See Materials and Methods.
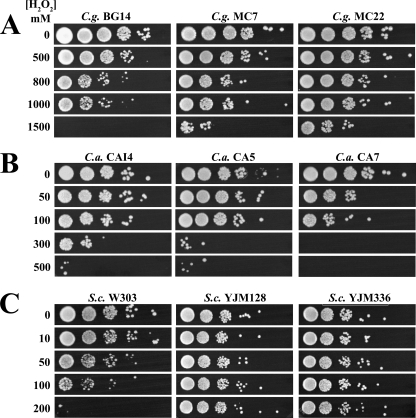
It has been reported previously that SP cells of various bacterial and fungal pathogens are more resistant to oxidants than LP cells (16, 29). To determine if this is true also for C. glabrata, we treated C. glabrata strain BG14 and CI MC7 and MC22 as described in the legend to Fig. 2 and Materials and Methods. As shown in Fig. 2A, C. glabrata SP cells exhibited resistance to H2O2 at concentrations up to 1,000 mM, significantly higher than those to which LP cells showed resistance. The same experiments were performed with C. albicans strain CAI4 and CI CA5 and CA7 and S. cerevisiae strain W303 and CI YJM128 and YJM336. As shown in Fig. 2B, C. albicans CAI4 showed resistance to up to 300 mM H2O2 while CI showed resistance to up to 100 mM. S. cerevisiae W303 SP cells showed an increase in resistance compared to that of LP cells, exhibiting resistance to up to 100 mM H2O2 and complete sensitivity to 200 mM. Interestingly, S. cerevisiae CI were more resistant than the lab reference strain W303 (Fig. 2C). Overall, the SP levels of resistance were much higher than those in LP. These data are in agreement with previous findings that SP cells are more resistant to oxidative stress than LP cells. Moreover, our data confirm that C. glabrata in SP is intrinsically more resistant to H2O2 than S. cerevisiae and C. albicans in SP.
FIG. 2.
C. glabrata, C. albicans, and S. cerevisiae SP resistance to H2O2. Saturated cultures of C. glabrata (C.g.) strain BG14 and CI MC7 and MC22 (A), C. albicans (C.a.) strain CAI4 and CI CA5 and CA7 (B), and S. cerevisiae (S.c.) strain W303 and CI YJM128 and YJM336 (C) were diluted to an OD600 of 0.5 with spent medium from the same cultures. The cells were divided into aliquots and treated for 3 h with H2O2 at different concentrations: for C. glabrata, 500, 800, 1,000, and 1,500 mM; for C. albicans, 0, 50, 100, 300, and 500 mM; and for S. cerevisiae, 0, 10, 50, 100, and 200 mM. After the treatment, the cultures remained at an OD600 of 0.5, oxidant was removed by centrifugation, cells were resuspended in distilled water, and suspensions were diluted and spotted onto YPD plates. Plates were incubated at 30°C. See Materials and Methods.
Role of the transcription factors Skn7p, Yap1p, Msn2p, and Msn4p in the OSR.
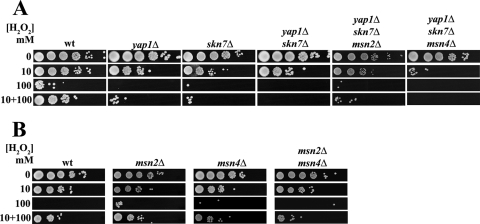
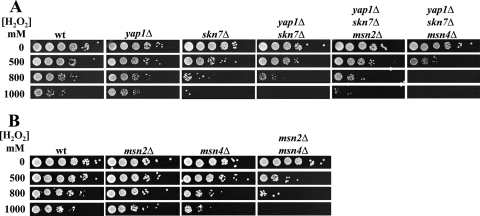
The key regulators of the OSR in S. cerevisiae, such as Skn7p, Yap1p, Msn2p, and Msn4p, are well characterized. C. glabrata orthologs of S. cerevisiae genes encoding these transcription factors were identified in the C. glabrata genome database (http://cbi.labri.fr/Genolevures/elt/CAGL), and an analysis of the amino acid sequence homology of C. glabrata and S. cerevisiae transcription factors was done by ClusalW alignment, with the following results: C. glabrata Skn7p (CgSkn7p; CAGL0F09097g), 48% identical and 15% similar to ScSkn7p; CgMsn2p (CAGL0F05995g), 25% identical and 16% similar to ScMsn2p; CgMsn4p (CAGL0M13189g), 22% identical and 13% similar to ScMsn4p; and CgYap1p (CAGL0H04631g), 36% identical and 12% similar to ScYap1p (11). We simply asked whether these transcription factors participate in the OSR in C. glabrata. We constructed C. glabrata mutant strains containing single (yap1Δ or skn7Δ), double (yap1Δ skn7Δ), triple (yap1Δ skn7Δ msn2Δ or yap1Δ skn7Δ msn4Δ), and quadruple (yap1Δ skn7Δ msn2Δ msn4Δ) deletions. Additional null strains including msn2Δ, msn4Δ, and msn2Δ msn4Δ strains were provided by R. Domergue and B. Cormack. We first characterized the sensitivities to H2O2 treatment of the LP cells of the various C. glabrata deletion strains described above. As shown in Fig. 3A, the yap1Δ and skn7Δ strains showed reduced resistance to 10 mM H2O2 compared to that of the parental wt strain. When treated with 100 mM H2O2, both the yap1Δ and skn7Δ strains lost viability, in contrast with the parental strain. The same phenotype was seen in the adaptation experiment. As shown in the bottom panel of Fig. 3A, when pretreated with a low dose of H2O2 of 10 mM, the yap1Δ and skn7Δ strains could not adapt to the stress, in contrast with the parental strain. A yap1Δ skn7Δ strain showed the same phenotype as the strains carrying a single deletion of either gene, indicating that the corresponding transcription factors are needed for adaptation and resistance in LP and that they do not compensate for each other. Furthermore (Fig. 3A), since (i) the yap1Δ skn7Δ msn2Δ triple mutant had the same reduced resistance phenotype at 10 mM as the single and double mutants (yap1Δ, skn7Δ, and yap1Δ skn7Δ strains), (ii) the yap1Δ skn7Δ msn4Δ triple mutant was more sensitive than the single and double mutants to 10 mM, and (iii) the quadruple mutant (yap1Δ skn7Δ msn2Δ msn4Δ) behaved the same as the yap1Δ skn7Δ msn4Δ triple mutant (see Fig. 6A), Skn7p, Yap1p, and Msn4p together, but not Msn2p, coordinate the overall resistance in LP cells. Msn2p and Msn4p may play a role in adaptation since the msn2Δ msn4Δ double mutant but not the corresponding single mutants showed a subtle but reproducible defect in adaptation (Fig. 3B). These results indicate, first, not only that C. glabrata has conserved the adaptive response to oxidative stress in LP cells but also that this adaptation is dependent mainly on Skn7p and Yap1p and partially on both Msn2p and Msn4p and, second, that Skn7p, Yap1p, and Msn4p coordinate the control of the OSR since the triple and quadruple mutants were almost as sensitive as the cta1Δ mutant in LP (see below and Fig. 6A).
FIG. 3.
Regulation of the OSR to H2O2 in LP. The wt (BG14) and single, double, and triple mutants with yap1Δ, skn7Δ, msn2Δ, and msn4Δ mutations were grown and treated with H2O2 as described in the legend to Fig. 1. See Materials and Methods.
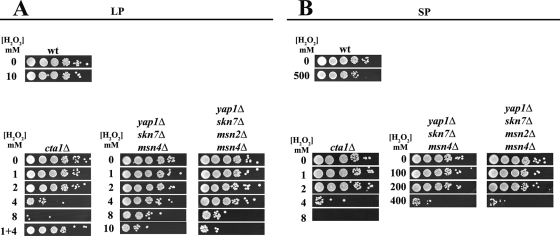
FIG. 6.
Analysis of Cta1p in the OSR. Experiments with both LP and SP wt (BG14), cta1Δ, yap1Δ skn7Δ msn4Δ, and yap1Δ skn7Δ msn2Δ msn4Δ cells were performed as described in the legends to Fig. 1 and 2. See Materials and Methods.
We then tested the role of these transcription factors for resistance in SP. Saturated cultures (OD600, 30.0) of the parental strain BG14 and single (yap1Δ, skn7Δ, msn2Δ, and msn4Δ), double (yap1Δ skn7Δ and msn2Δ msn4Δ), triple (yap1Δ skn7Δ msn2Δ and yap1Δ skn7Δ msn4Δ), and quadruple (yap1Δ skn7Δ msn2Δ msn4Δ) mutants were treated as described in the legend to Fig. 2. Figure 4A shows that the yap1Δ mutant behaved as the parental strain, that the skn7Δ mutant was more sensitive to H2O2 than the wt, and that the skn7Δ mutation is epistatic to yap1Δ (compare the data for the yap1Δ skn7Δ double mutant and the single yap1Δ and skn7Δ mutants). This finding indicates that, surprisingly, Skn7p but not Yap1p is required in SP. As in LP cells, Msn4p was required for resistance in SP cells: an msn4Δ single mutant and a triple mutant with msn4Δ in combination with skn7Δ showed reduced resistance to H2O2 (Fig. 4). Interestingly, Msn2p has a role in SP since the msn2Δ msn4Δ double mutant was more sensitive than the msn2Δ and msn4Δ single mutants (Fig. 4B). These data suggest that Msn2p and Msn4p are both important and act independently of each other. The analysis of the triple and quadruple mutants (skn7Δ yap1Δ msn2Δ, skn7Δ yap1Δ msn4Δ, and skn7Δ yap1Δ msn2Δ msn4Δ strains) confirms the roles in SP of both Skn7p and Msn4p (Fig. 4A; also see Fig. 6B).
FIG. 4.
Regulation of the OSR to H2O2 in SP. The wt (BG14) and single, double, and triple mutants with yap1Δ, skn7Δ, msn2Δ, and msn4Δ mutations were grown and treated with H2O2 as described in the legend to Fig. 2. See Materials and Methods.
CTA1 is absolutely required for resistance to H2O2 in vitro.
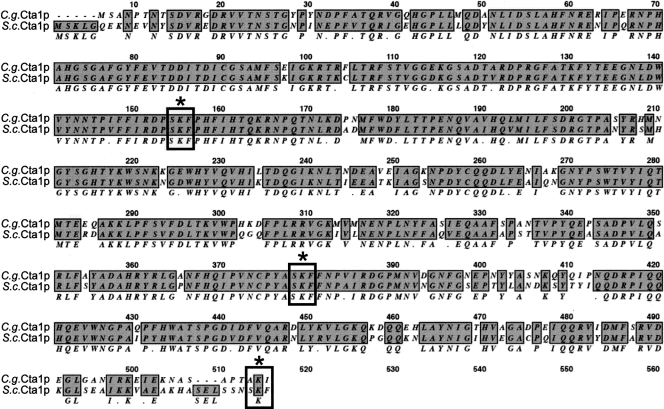
Catalase plays a central role in the cell response against oxidative stress. We characterized the function of C. glabrata catalase Cta1p in the resistance to H2O2 treatment. S. cerevisiae has two catalases, cytoplasmic (Ctt1p) and peroxisomal (Cta1p) (13, 30, 65, 66, 72), and C. albicans has only one catalase (Ctt1p) (76). Interestingly, the single C. glabrata catalase gene, CTA1 (CAGL0K10868g), is the ortholog of S. cerevisiae CTA1, the gene for the peroxisomal catalase. CgCta1p and ScCta1p are 85% similar (78% identical and 7% similar) over the entire lengths of the proteins, and CgCta1p has two putative internal peroxisomal targeting signals (SKF) (59) (Fig. 5). It is not known whether CgCta1p is targeted to the peroxisome, the mitochondria, or the cytoplasm.
FIG. 5.
ScCta1p and CgCta1p alignment. ScCta1p and CgCta1p are 85% similar across the entire lengths of the proteins. Identical residues are boxed and shaded. SKF (indicated by boxes and asterisks) is the peroxisomal targeting signal.
We tested whether CTA1 is required in C. glabrata for the high-level resistance to H2O2 displayed by both the LP and SP cells. A CTA1 null strain (cta1Δ) was constructed. cta1Δ LP cells (Fig. 6A) and SP cells (Fig. 6B) behaved in the same way: cta1Δ cells completely lost their ability to survive at high concentrations of H2O2 (>4 mM) (Fig. 6). Interestingly, cta1Δ LP cells were still able to adapt but only when exposed to low levels of H2O2 (Fig. 6A, lane 1 + 4). This result may indicate the presence of a catalase-independent pathway to respond to H2O2. In SP, the cta1Δ mutant also became sensitive to H2O2 (Fig. 6B). These results clearly indicate that this single Cta1p plays a central role in the resistance of C. glabrata to H2O2 either in LP or in SP cells. In addition, CTA1 regulation in LP is likely controlled primarily by Skn7p, Yap1p, and Msn4p since the removal of these transcription factors rendered the cells almost as sensitive to H2O2 as those of the cta1Δ strain (Fig. 6A). In SP, by contrast, the regulation was more complex since the triple and quadruple mutants lacking three or four of the transcription factors (skn7Δ yap1Δ msn4Δ and skn7Δ yap1Δ msn2Δ msn4Δ strains), while being less resistant than the wt, were still able to grow at 200 mM H2O2, displaying a level of resistance well above that of the cta1Δ strain (Fig. 6B). This result suggests the possibility of additional regulators of CTA1 or an independent pathway to respond to H2O2.
Cta1p is not necessary for virulence in C. glabrata.
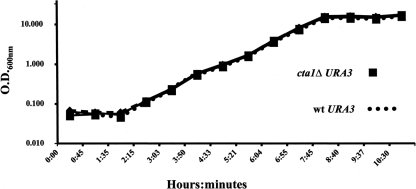
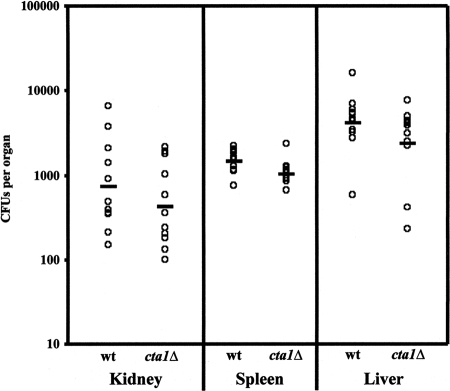
Since Cta1p is entirely responsible for the extremely high level of resistance to H2O2 in vitro (Fig. 6), CTA1 is induced in macrophages (41), and CaCTA1 is required for virulence (56, 76), we investigated whether C. glabrata catalase plays a role during disseminated infection. Prior to the in vivo analysis, the parental and the mutant strains were made to express the Ura+ phenotype by restoring URA3 at the URA3 locus to generate BG462 and CGM351 (see Materials and Methods). The two strains grew with identical doubling times in rich medium at 30 or 37°C (Fig. 7). To test for the virulence of the cta1Δ strain, we infected groups of 10 mice by tail vein injection using 2 × 107 cells of strain BG462 (wt expressing URA3) or CGM351 (cta1Δ URA3). Mice were sacrificed on day 7 after infection, and CFUs in kidneys, spleens, and livers were enumerated (Materials and Methods). The average numbers of CFU of the mutant strain and the wt strain recovered from the three organ types showed no difference (Fig. 8). The results of these experiments indicate that Cta1p is required in vitro but is dispensable in the murine disseminated-infection model.
FIG. 7.
Growth curves of the cta1Δ mutant versus the wt (BG14) at 37°C.
FIG. 8.
Cta1p is dispensable in vivo. Numbers of CFU in kidney, spleen, and liver tissues 7 days postinfection with the wt (BG462) and the cta1Δ mutant (CGM351) are shown. Individual datum points represent results for individual mice in groups of 10 mice. The bars indicate the geometric mean for each group. See Materials and Methods.
DISCUSSION
Higher eukaryotes use ROS through the oxidative burst to eliminate invading pathogens (42, 57). During the coevolution of pathogens and their hosts, pathogens have coopted the antioxidant enzymes and molecules for normal ROS elimination to evade oxidative killing so that survival and persistence are ensured. C. glabrata is no exception; it possesses a defined genetic program to respond to oxidative killing by the host. C. glabrata adapts by reprogramming its gene expression, providing a clear advantage to this fungal pathogen upon infection. In this paper, we present evidence that the OSR in C. glabrata is controlled in part by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. C. glabrata is resistant to extreme concentrations of H2O2, and this resistance is mediated in vitro by the single catalase Cta1p. This single catalase is, however, dispensable in vivo.
Resistance and adaptation to H2O2.
Both the enzymes and the regulation of the OSR are well conserved among S. cerevisiae, C. albicans, and C. glabrata. Searching for genes involved in oxidative stress in the genome databases (http://www.yeastgenome.org, http://www.candidagenome.org, and http://cbi.labri.fr/Genolevures/elt/CAGL), we found that of 96 oxidative stress-related genes in S. cerevisiae, 67 are present in C. glabrata and 49 are present in C. albicans. Additionally, six oxidative stress-related genes are present only in C. albicans. In S. cerevisiae, the OSR is partly under the control of the well-studied transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. These transcription factors have orthologs in C. glabrata, as follows: Skn7p (63% similar), Msn2p (41% similar), Msn4p (35% similar), and Yap1p (48% similar). From the results of our study, the roles of these transcription factors seem to be conserved as well.
C. glabrata was naturally resistant to higher levels of H2O2 than C. albicans and S. cerevisiae (Fig. 1 and 2). LP cells of C. glabrata were able to detect low levels of oxidant and induce a set of enzymes that would allow resistance to high levels and adaptation to the new environment. This response was mediated mainly by the transcription factors Skn7p and Yap1p but also by Msn2p and Msn4p (Fig. 3). This adaptation was present among cells of both C. albicans (Fig. 1) and S. cerevisiae, in which both catalases and the transcription factors Yap1p and Skn7p are required for this response (14, 17, 23, 27, 37, 39). Furthermore, Yap1p, Skn7p, and Msn4p coordinated the response to oxidative stress in LP cells, which required at least the activation and induction of the catalase gene (Fig. 3). The roles of Msn2p and Msn4p are interesting in two ways: first, CaMsn2p and CaMsn4p have no obvious role in oxidative stress (58), and second, ScMsn2p and ScMsn4p perform nonredundant functions depending on the stress (reviewed in references 20 and 55). In C. glabrata upon oxidative stress, these two transcription factors appeared to work independently of each other. Both were needed for SP resistance (see below), and Msn4p was required for LP resistance, along with Skn7p and Yap1p (Fig. 3 and 4).
SP is important for resistance.
It has been shown previously that not only yeast but other microorganisms in SP are more resistant to oxidants than the same organisms in LP (38, 61, 62), and C. glabrata is no exception. C. glabrata in SP was naturally resistant to high levels of H2O2, up to1,000 mM, compared to about 100 mM H2O2 for S. cerevisiae and 300 mM H2O2 for C. albicans (Fig. 2) (5, 9). This resistance was controlled by Msn2p, Msn4p, and Skn7p (Fig. 4), whereas Yap1p did not appear to have a central role. This high-level resistance suggests that C. glabrata has an extremely efficient Cta1p and that it is probably able to repair efficiently the damage generated by the oxidant. It is possible that C. glabrata catalase activity may increase post-exponential phase. For example, S. cerevisiae catalase activity increases in SP (37), C. albicans shows growth phase-dependent resistance to H2O2 (38, 75), and the C. albicans Mn-SOD3 superoxide dismutase is induced in SP and is needed for the OSR (45). This high-level natural resistance to H2O2 may, in part, explain why C. glabrata is able to evade elimination by macrophages (41). Another clear advantage would be that C. glabrata can compete with H2O2-generating pathogens for specific niches inside the host. It has been shown previously that H2O2-producing bacteria inhibit the proliferation of C. albicans (75). Interestingly, CI of the nonpathogenic S. cerevisiae showed increased resistance to H2O2 relative to that of the reference strain (Fig. 1C and 2C). This result is consistent with the idea that pathogens, in order to survive, require a proper response to oxidative stress.
Catalase and virulence.
The C. glabrata single catalase (Cta1p) was absolutely required to confer resistance on LP and SP cells in vitro. cta1Δ cells were extremely sensitive (Fig. 6) to H2O2. CgCta1p is the ortholog of the S. cerevisiae peroxisomal catalase (ScCta1p; 85% similar) (Fig. 5), which converts H2O2 formed by acyl coenzyme A oxidase (Pox1p) during fatty acid beta-oxidation (34) to H2O and O2 in the peroxisomal matrix. CgCta1p is a monofunctional 57-kDa protein classified as a group III catalase (42). It is interesting that S. cerevisiae has two catalases, cytoplasmic (Ctt1p) and peroxisomal (Cta1p), and that S. cerevisiae was about 10 times less resistant than C. glabrata (Fig. 2). Surprisingly, it has been shown previously that both catalases in S. cerevisiae (Ctt1p and Cta1p) are dispensable in growing cells and that glutathione compensates for the lack of the catalases (37). The fungal pathogens C. albicans and C. glabrata, though distantly related phylogenetically, show increased resistance to oxidative stress relative to that of S. cerevisiae.
The results presented in Fig. 6 suggest that in LP cells CTA1 may be controlled by the concerted actions of Yap1p, Skn7p, and Msn4p. The triple (yap1Δ skn7Δ msn4Δ) and quadruple (yap1Δ skn7Δ msn2Δ msn4Δ) mutants behaved exactly the same, rendering LP cells almost as sensitive as the cta1Δ strain (Fig. 6A). In fact, bioinformatic analyses of the CgCTA1 promoter have previously revealed putative conserved cis-acting elements for each of these transcriptional regulators (reviewed in references 24, 32, and 36). In SP, however, both triple (yap1Δ skn7Δ msn4Δ) and quadruple (yap1Δ skn7Δ msn2Δ msn4Δ) mutants showed increased resistance to H2O2 relative to that of the reference strain W303. (Fig. 6B). These mutants could still grow at elevated levels of H2O2 (up to 200 mM). This finding would indicate that in SP but not in LP, other transcriptional regulators of the CTA1 gene besides Yap1p, Skn7p, Msn2p, and Msn4p are in play.
Is the catalase a virulence factor? (i) cta1Δ strains are extremely sensitive to H2O2 in vitro, (ii) CaCta1p is important for virulence (56, 76; reviewed in reference 10), and (iii) CTA1 is induced after phagocytosis (26, 41, 47). We therefore assayed cta1Δ cells in a mouse model of systemic infection. The experiment showed no difference in the colonization of the kidney, spleen, and liver by the cta1Δ strain (Fig. 8). This finding is in strong contrast with the results of the in vitro experiments, in which cta1Δ cells were extremely sensitive to H2O2. Our results suggest either that the catalase is not important in the OSR in vivo or that there are additional factors that may compensate for the lack of Cta1p in vivo. One possibility is that glutathione may mediate H2O2 resistance in vivo, since it has been shown previously that both catalases and glutathione provide an overlapping antioxidant defense system in S. cerevisiae (28). The results also suggest that these additional factors are silent in vitro. In fact, the cta1Δ strain was still able to adapt to oxidative stress in vitro, though at low levels of H2O2 (Fig. 6A). This finding indicates that there may be an additional catalase-independent pathway to respond to H2O2. Currently, we are working to identify these additional regulators/effectors of the OSR.
Acknowledgments
We thank B. Cormack and B. Ma for careful reading of the manuscript. We thank B. Cormack, A. Johnson, M. Christman, and C. Gross for kindly providing strains. We are grateful to W. Hansberg, J. Aguirre, M. Soberón, A. Bravo, G. Soberón, P. León, M. Zurita, X. Soberón, A. González, J. Folch, E. Merino, B. Valderrama, and F. Sánchez for providing reagents and supplies. We thank B. Cormack, S. Pan, and M. Zupancic for helping with the in vivo experiments. We thank R. López-Revilla and L. Salazar for providing space and equipment.
This work was funded by CONACyT fellowships to M.C.-C. (163140), M.B.-M.-D.-C. (209276), I.C.-V.(224300), and J.M.-A.(209255) and by CONACyT grant no. CB-2005-48279 to A.D.L.P.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Aguirre, J., W. Hansberg, and R. Navarro. 2006. Fungal responses to reactive oxygen species. Med. Mycol. 44(Suppl.)101-107. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, J., M. Rios-Momberg, D. Hewitt, and W. Hansberg. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13111-118. [DOI] [PubMed] [Google Scholar]
- 3.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 5.Avery, A. M., and S. V. Avery. 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 27633730-33735. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. L., H. Bussey, and R. C. Stewart. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 135186-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvin, N. M., and P. C. Hanawalt. 1988. High-efficiency transformation of bacterial cells by electroporation. J. Bacteriol. 1702796-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castano, I., R. Kaur, S. Pan, R. Cregg, A. De Las Penas, N. Guo, M. C. Biery, N. L. Craig, and B. P. Cormack. 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauhan, N., D. Inglis, E. Roman, J. Pla, D. Li, J. A. Calera, and R. Calderone. 2003. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell 21018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan, N., J. P. Latge, and R. Calderone. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4435-444. [DOI] [PubMed] [Google Scholar]
- 11.Chen, K. H., T. Miyazaki, H. F. Tsai, and J. E. Bennett. 2007. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 38663-72. [DOI] [PubMed] [Google Scholar]
- 12.Clemons, K. V., J. H. McCusker, R. W. Davis, and D. A. Stevens. 1994. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J. Infect. Dis. 169859-867. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, G., W. Rapatz, and H. Ruis. 1988. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur. J. Biochem. 176159-163. [DOI] [PubMed] [Google Scholar]
- 14.Collinson, L. P., and I. W. Dawes. 1992. Inducibility of the response of yeast cells to peroxide stress. J. Gen. Microbiol. 138329-335. [DOI] [PubMed] [Google Scholar]
- 15.Cormack, B. P., and S. Falkow. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cyrne, L., L. Martins, L. Fernandes, and H. S. Marinho. 2003. Regulation of antioxidant enzymes gene expression in the yeast Saccharomyces cerevisiae during stationary phase. Free Radic. Biol. Med. 34385-393. [DOI] [PubMed] [Google Scholar]
- 17.Davies, J. M., C. V. Lowry, and K. J. Davies. 1995. Transient adaptation to oxidative stress in yeast. Arch. Biochem. Biophys. 3171-6. [DOI] [PubMed] [Google Scholar]
- 18.De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg, and B. P. Cormack. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 172245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell, J. R. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308866-870. [DOI] [PubMed] [Google Scholar]
- 20.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24469-486. [DOI] [PubMed] [Google Scholar]
- 21.Estruch, F., and M. Carlson. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 133872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidel, P. L., Jr., J. L. Cutright, L. Tait, and J. D. Sobel. 1996. A murine model of Candida glabrata vaginitis. J. Infect. Dis. 173425-431. [DOI] [PubMed] [Google Scholar]
- 23.Flattery-O'Brien, J., L. P. Collinson, and I. W. Dawes. 1993. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J. Gen. Microbiol. 139501-507. [DOI] [PubMed] [Google Scholar]
- 24.Folch-Mallol, J. L., A. Garay-Arroyo, F. Lledias, and A. A. Covarrubias Robles. 2004. The stress response in the yeast Saccharomyces cerevisiae. Rev. Latinoam. Microbiol. 4624-46. (In Spanish.) [PubMed] [Google Scholar]
- 25.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fradin, C., P. De Groot, D. MacCallum, M. Schaller, F. Klis, F. C. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56397-415. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Parraga, P., J. A. Hernandez, and J. C. Arguelles. 2003. Role of antioxidant enzymatic defences against oxidative stress (H2O2) and the acquisition of oxidative tolerance in Candida albicans. Yeast 201161-1169. [DOI] [PubMed] [Google Scholar]
- 28.Grant, C. M., G. Perrone, and I. W. Dawes. 1998. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253893-898. [DOI] [PubMed] [Google Scholar]
- 29.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartig, A., and H. Ruis. 1986. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur. J. Biochem. 160487-490. [DOI] [PubMed] [Google Scholar]
- 31.Hasan, R., C. Leroy, A. D. Isnard, J. Labarre, E. Boy-Marcotte, and M. B. Toledano. 2002. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol. 45233-241. [DOI] [PubMed] [Google Scholar]
- 32.He, X. J., and J. S. Fassler. 2005. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 581454-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266383-402. [DOI] [PubMed] [Google Scholar]
- 34.Hiltunen, J. K., A. M. Mursula, H. Rottensteiner, R. K. Wierenga, A. J. Kastaniotis, and A. Gurvitz. 2003. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2735-64. [DOI] [PubMed] [Google Scholar]
- 35.Hwang, C. S., G. E. Rhie, J. H. Oh, W. K. Huh, H. S. Yim, and S. O. Kang. 2002. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 1483705-3713. [DOI] [PubMed] [Google Scholar]
- 36.Ikner, A., and K. Shiozaki. 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 56913-27. [DOI] [PubMed] [Google Scholar]
- 37.Izawa, S., Y. Inoue, and A. Kimura. 1996. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 32061-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamieson, D. J. 1992. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J. Bacteriol. 1746678-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamieson, D. J., S. L. Rivers, and D. W. Stephen. 1994. Analysis of Saccharomyces cerevisiae proteins induced by peroxide and superoxide stress. Microbiology 1403277-3283. [DOI] [PubMed] [Google Scholar]
- 40.Kaur, R., R. Domergue, M. L. Zupancic, and B. P. Cormack. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8378-384. [DOI] [PubMed] [Google Scholar]
- 41.Kaur, R., B. Ma, and B. P. Cormack. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 1047628-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klotz, M. G., and P. C. Loewen. 2003. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 201098-1112. [DOI] [PubMed] [Google Scholar]
- 43.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29327-334. [DOI] [PubMed] [Google Scholar]
- 44.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamarre, C., J. D. LeMay, N. Deslauriers, and Y. Bourbonnais. 2001. Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J. Biol. Chem. 27643784-43791. [DOI] [PubMed] [Google Scholar]
- 46.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 27416040-16046. [DOI] [PubMed] [Google Scholar]
- 47.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 31076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupo, S., C. Aranda, L. Miranda-Ham, H. Olivera, L. Riego, L. Servin, and A. Gonzalez. 1997. Tyrosine is involved in protection from oxidative stress in Saccharomyces cerevisiae. Can. J. Microbiol. 43963-970. [DOI] [PubMed] [Google Scholar]
- 49.Mansour, M. K., and S. M. Levitz. 2002. Interactions of fungi with phagocytes. Curr. Opin. Microbiol. 5359-365. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 152227-2235. [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 1471557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molina, L., and R. Kahmann. 2007. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 192293-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 161035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan, B. A., N. Bouquin, G. F. Merrill, and L. H. Johnston. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 145679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moye-Rowley, W. S. 2002. Transcription factors regulating the response to oxidative stress in yeast. Antioxid. Redox Signal. 4123-140. [DOI] [PubMed] [Google Scholar]
- 56.Nakagawa, Y., T. Kanbe, and I. Mizuguchi. 2003. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol. Immunol. 47395-403. [DOI] [PubMed] [Google Scholar]
- 57.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 978841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholls, S., M. Straffon, B. Enjalbert, A. Nantel, S. Macaskill, M. Whiteway, and A. J. Brown. 2004. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot. Cell 31111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrova, V. Y., D. Drescher, A. V. Kujumdzieva, and M. J. Schmitt. 2004. Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem. J. 380393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schenberg-Frascino, A. 1972. Lethal and mutagenic effects of elevated temperature on haploid yeast. II. Recovery from thermolesions. Mol. Gen. Genet. 117239-253. [DOI] [PubMed] [Google Scholar]
- 62.Schenberg-Frascino, A., and E. Moustacchi. 1972. Lethal and mutagenic effects of elevated temperature on haploid yeast. I. Variations in sensitivity during the cell cycle. Mol. Gen. Genet. 115243-257. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 935777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnell, N., B. Krems, and K. D. Entian. 1992. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr. Genet. 21269-273. [DOI] [PubMed] [Google Scholar]
- 65.Seah, T. C., A. R. Bhatti, and J. G. Kaplan. 1973. Novel catalatic proteins of bakers' yeast. I. An atypical catalase. Can. J. Biochem. 511551-1555. [DOI] [PubMed] [Google Scholar]
- 66.Seah, T. C., and J. G. Kaplan. 1973. Purification and properties of the catalase of bakers' yeast. J. Biol. Chem. 2482889-2893. [PubMed] [Google Scholar]
- 67.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 68.Singh, P., N. Chauhan, A. Ghosh, F. Dixon, and R. Calderone. 2004. SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect. Immun. 722390-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Temple, M. D., G. G. Perrone, and I. W. Dawes. 2005. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15319-326. [DOI] [PubMed] [Google Scholar]
- 70.Thorpe, G. W., C. S. Fong, N. Alic, V. J. Higgins, and I. W. Dawes. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 1016564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toda, T., M. Shimanuki, and M. Yanagida. 1991. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 560-73. [DOI] [PubMed] [Google Scholar]
- 72.Traczyk, A., T. Bilinski, J. Litwinska, M. Skoneczny, and J. Rytka. 1985. Catalase T deficient mutants of Saccharomyces cerevisiae. Acta Microbiol. Pol. 34231-241. [PubMed] [Google Scholar]
- 73.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35627-630. [DOI] [PubMed] [Google Scholar]
- 74.Vermitsky, J. P., K. D. Earhart, W. L. Smith, R. Homayouni, T. D. Edlind, and P. D. Rogers. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61704-722. [DOI] [PubMed] [Google Scholar]
- 75.Westwater, C., E. Balish, and D. A. Schofield. 2005. Candida albicans-con-ditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot. Cell 41654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wysong, D. R., L. Christin, A. M. Sugar, P. W. Robbins, and R. D. Diamond. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 661953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36618-629. [DOI] [PubMed] [Google Scholar]