Abstract
Efg1p is a key transcriptional regulator in Candida albicans which controls various aspects of morphogenesis and metabolism in this organism. Efg1p contains a central basic helix-loop-helix (bHLH) domain, flanked by sequences highly conserved in fungal APSES proteins, as well as polyglutamine stretches at the N- and C-terminal ends. A systematic deletion approach to specify functional domains of Efg1p revealed that the APSES domain is essential for morphogenesis of the normal yeast and true hyphal cell forms and that bHLH flanking sequences are needed for Efg1p stability. Additional C-terminal sequences were required for hypha formation on some inducing media, and most Efg1p sequences were needed for chlamydospore morphogenesis. Overexpression of EFG1 led to pseudohypha formation only if a functional APSES domain was present, while a switch from the opaque to the white cell type in addition depended on the presence of certain N- and C-terminal segments. Yeast two-hybrid experiments revealed that binding of Efg1p to its antagonist Czf1p required two regions outside of the APSES domain, which did not coincide with Efg1p sequences needed for its transcriptional repressor activity. Binding of the Flo8 transcription factor to Efg1p did not require the APSES domain but appeared to occur at two or more redundant domains. In contrast, DNA binding of Efg1p to an MluI cell cycle box (MCB) element solely required the APSES domain. Overall, these results suggest that functional domains of Efg1p are spread throughout most of its sequences, including the central APSES domain involved in DNA binding, as well as flanking regions required for various protein interactions and regulatory activities.
In the last few years, the so-called APSES class of transcriptional regulators has been identified in ascomycetes and has mainly been described as comprising important regulators of morphological processes. All APSES proteins share a highly conserved DNA-binding domain (APSES domain) of about 100 amino acids, whose central domain is predicted to form a typical basic helix-loop-helix (bHLH) structure (6, 23). In Candida albicans, two APSES proteins have been identified, Efg1p and Efh1p (5). While the function of Efh1p is still unclear, Efg1p has been found to be a central regulator of numerous cellular processes in the human fungal pathogen C. albicans. Efg1p is required for the development of a true hyphal growth form, considered to be essential for interactions with human host cells, as a platform for the presentation of virulence-related components (16, 23), and for chlamydospore formation (20). On the other hand, overexpression of EFG1 results in a pseudohyphal growth form rather than true hyphae (23, 24) and forces opaque-form cells to switch to the white cell form, consistent with low levels of EFG1 expression in opaque-form cells (21). In addition, recent results indicate that Efg1p has a significant effect on metabolism by inducing glycolysis and blocking respiratory activities during normoxia, while in hypoxia it regulates other specific subsets of genes (5, 18).
bHLH proteins are known to bind E-box sequences (CANNTG), and binding of Efg1p to a CATTTG-containing fragment has been verified by in vitro experiments (13). Furthermore, within its basic portion of the bHLH domain, APSES proteins also show homology to the yeast Mbp1 protein, which in combination with Swi6p binds to an “MCB (MluI cell cycle box)” sequence (ACGCGT) (8). Interestingly, the APSES protein StuA of Aspergillus nidulans has also been found to bind to an MCB sequence by in vitro complex formation and by one-hybrid experiments (6). The importance of the APSES domain for Efg1p function was further stressed by the identification of a potential phosphorylation site for an A-type kinase within the bHLH domain (residue T206) that upon mutation to an alanine residue negatively affected hyphal morphogenesis and chlamydospore formation, while an exchange to a glutamic residue increased hypha formation (2, 20). However, the degree and sites of Efg1p phosphorylation have still not been demonstrated directly. Outside of the APSES domain, Efg1p contains extensive polyglutamine (polyQ) stretches at the N- and C-terminal ends and an alanine- and proline-rich low-complexity region C-terminal to the APSES domain. While Efh1p but not Efg1p was found to form homodimers (5), the putative transcription factor Czf1p was found to physically interact with Efg1p, possibly to counteract the repressor action of Efg1p under microaerobic conditions (9). In addition, the function of Efg1p may be aided by the transcription factor Flo8p, since both proteins were shown to interact directly and to similarly regulate filamentation as inducers or repressors, depending on environmental conditions (4).
Since little is known about structure-function relationships of Efg1p or any other APSES protein, we started a systematic deletion approach to identify regions necessary for the various functions of Efg1p. Our results further stress the functional importance of the APSES domain but also demonstrate that the N- and C-terminal ends contribute essential functions for specific phenotypes.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains used are listed in Table 1. Strains were grown in YPD medium or on supplemented SD minimal medium at 30°C (19). Transformation of C. albicans strains was carried out using the spheroplast method (19). For hyphal induction, the strains were grown for 3 to 4 days at 37°C either on Lee′s medium (12) or on 2% agar containing 5% horse serum. To induce chlamydospore formation, cells were streaked out lightly on chlamydospore induction medium (cornmeal agar [Merck]-0.5% Tween 80), covered with a coverslip, and incubated for 5 days at 20°C (20). Colonies of white and opaque-phase cells were visualized on SD medium containing 5 μg/ml phloxine B (21). To induce pseudohyphal growth, the PCK1 promoter in transformants expressing EFG1 variants was induced in SCAA medium (0.67% yeast nitrogen base, 2% Casamino Acids) (14).
TABLE 1.
C. albicans strains
| Strain | Genotype | Reference |
|---|---|---|
| CAF2-1 | URA3/ura3::imm434 | 7 |
| CAI4 | ura3::imm434/ura3::imm434 | 7 |
| BWP17 | ura3Δ::λimm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 26 |
| TS3.3 | opaque TS3.3 ura− | 22 |
| HLC52 | Like CAI4 but efg1::hisG/efg1::hisG-URA3-hisG | 16 |
| HLC67 | Like CAI4 but efg1::hisG/efg1::hisG | 16 |
| HLCE | Like HLC67 but efg1::hisG/efg1::[EFG1p-URA3] | 18 |
| CRC106 | Like CAI8 but with lexA-ADH1-lacZ::ADE2 | 5 |
| AS1 | Like CAI4 but tpk2::hisG/tpk2::hisG | 2 |
| IIHH6-4a | Like CAI4 but tpk1::hisG/tpk1::hisG | 2 |
| C4/d63 | Like CAI4 but efh1::hisG/efh1::hisG-URA3-hisG | 5 |
| HLCEEFG1 | Like HLC67 but efg1::hisG/efg1::[EFG1p-HA-EFG1-URA3] (pTD38-HA/PacI integrated in EFG1p) | This work |
| HLCEEFG1-D1 | Like HLCEEFG1 but encoding Δ(Y9-G74) Efg1p deletion D1 | This work |
| HLCEEFG1-D2 | Like HLCEEFG1 but encoding Δ(Q75-Y137) Efg1p deletion D2 | This work |
| HLCEEFG1-D3 | Like HLCEEFG1 but encoding Δ(N131-R202) Efg1p deletion D3 | This work |
| HLCEEFG1-D4 | Like HLCEEFG1 but encoding Δ(YP203-V306) Efg1p deletion D4 | This work |
| HLCEEFG1-D5 | Like HLCEEFG1 but encoding Δ(I291-V383) Efg1p deletion D5 | This work |
| HLCEEFG1-D6 | Like HLCEEFG1 but encoding Δ(P382-S478) Efg1p deletion D6 | This work |
| HLCEEFG1-D7 | Like HLCEEFG1 but encoding Δ(Y465-N541) Efg1p deletion D7 | This work |
| HLCEEFG1-D8 | Like HLCEEFG1 but encoding Δ(R202-I236) Efg1p deletion D8 | This work |
| HLCEEFG1-D9 | Like HLCEEFG1 but encoding Δ(P297-T312) Efg1p deletion D9 | This work |
| HLCEEFG1-D10 | Like HLCEEFG1 but encoding Δ(A316-K352) Efg1p deletion D10 | This work |
| HLCEEFG1-D11 | Like HLCEEFG1 but encoding Δ(I350-V383) Efg1p deletion D11 | This work |
| AVL1 | Like AVL12 but encoding Δ(Y9-G74) Efg1p deletion D1 | This work |
| AVL2 | Like AVL12 but encoding Δ(Q75-Y137) Efg1p deletion D2 | This work |
| AVL3 | Like AVL12 but encoding Δ(N131-R202) Efg1p deletion D3 | This work |
| AVP4 | Like AVL12 but encoding Δ(YP203-V306) Efg1p deletion D4 | This work |
| AVL5 | Like AVL12 but encoding Δ(I291-V383) Efg1p deletion D5 | This work |
| AVL6 | Like AVL12 but encoding Δ(P382-S478) Efg1p deletion D6 | This work |
| AVL7 | Like AVL12 but encoding Δ(Y465-N541) Efg1p deletion D7 | This work |
| AVL8 | Like AVL12 but encoding Δ(R202-I236) Efg1p deletion D8 | This work |
| AVL9 | Like AVL12 but encoding Δ(P297-T312) Efg1p deletion D9 | This work |
| AVL10 | Like AVL12 but encoding Δ(A316-K352) Efg1p deletion D10 | This work |
| AVL11 | Like AVL12 but encoding Δ(I350-V383) Efg1p deletion D11 | This work |
| AVL12 | Like BWP17 but efg1::hisG/efg1::[EFG1p-HA-EFG1-URA3] (pTD38-HA/PacI integrated in EFG1p) | This work |
| HLCPEFG1 | Like HLC67 but efg1::hisG/efg1::[PCK1p-HA-EFG1-URA3] (pBI-HAHYD/KpnI integrated in LEU2) | This work |
| HLCP | Like HLC67, but efg1::hisG/efg1::[PCK1p -URA3] (pBI/KpnI integrated in LEU2) | This work |
| HLCPEFG1-D1 | Like HLCPEFG1 but encoding Δ(Y9-G74) Efg1p deletion D1 | This work |
| HLCPEFG1-D2 | Like HLCPEFG1 but encoding Δ(Q75-Y137) Efg1p deletion D2 | This work |
| HLCPEFG1-D3 | Like HLCPEFG1 but encoding Δ(N131-R202) Efg1p deletion D3 | This work |
| HLCPEFG1-D4 | Like HLCPEFG1 but encoding Δ(YP203-V306) Efg1p deletion D4 | This work |
| HLCPEFG1-D5 | Like HLCPEFG1 but encoding Δ(I291-V383) Efg1p deletion D5 | This work |
| HLCPEFG1-D6 | Like HLCPEFG1 but encoding Δ(P382-S478) Efg1p deletion D6 | This work |
| HLCPEFG1-D7 | Like HLCPEFG1 but encoding Δ(Y465-N541) Efg1p deletion D7 | This work |
| HLCPEFG1-D8 | Like HLCPEFG1 but encoding Δ(R202-I236) Efg1p deletion D8 | This work |
| HLCPEFG1-D9 | Like HLCPEFG1 but encoding Δ(P297-T312) Efg1p deletion D9 | This work |
| HLCPEFG1-D10 | Like HLCPEFG1 but encoding Δ(A316-K352) Efg1p deletion D10 | This work |
| HLCPEFG1-D11 | Like HLCPEFG1 but encoding Δ(I350-V383) Efg1p deletion D11 | This work |
| LexA-EFG1-D1 | Like CRC106 but containing Clp-LexA-EFG1-D1 | This work |
| LexA-EFG1-D2 | Like CRC106 but containing Clp-LexA-EFG1-D2 | This work |
| LexA-EFG1-D3 | Like CRC106 but containing Clp-LexA-EFG1-D3 | This work |
| LexA-EFG1-D4 | Like CRC106 but containing Clp-LexA-EFG1-D4 | This work |
| LexA-EFG1-D5 | Like CRC106 but containing Clp-LexA-EFG1-D5 | This work |
| LexA-EFG1-D6 | Like CRC106 but containing Clp-LexA-EFG1-D6 | This work |
| LexA-EFG1-D7 | Like CRC106 but containing Clp-LexA-EFG1-D7 | This work |
| LexA-EFG1-D8 | Like CRC106 but containing Clp-LexA-EFG1-D8 | This work |
| LexA-EFG1-D9 | Like CRC106 but containing Clp-LexA-EFG1-D9 | This work |
| LexA-EFG1-D10 | Like CRC106 but containing Clp-LexA-EFG1-D10 | This work |
| LexA-EFG1-D11 | Like CRC106 but containing Clp-LexA-EFG1-D11 | This work |
Construction of strains expressing EFG1 deletions.
The plasmid pTD38-HA was constructed by ligating a ∼2.4-kb partial BglII fragment from the plasmid pBI-HAHYD (23), containing a hemagglutinin (HA)-tagged version of Efg1, into the BglII site of plasmid pTD38 containing ∼2 kb of the promoter region and the 5′ untranslated region of the EFG1 gene (T. Doedt and J. F. Ernst, unpublished results). Eleven specific deletions were introduced into the EFG1 coding region by site-specific mutagenesis using the QuikChange kit (Stratagene). Primers used for construction of deletions D1 to D11 are listed in Table S1 in the supplemental material. Following linearization with PacI and transformation, plasmid pTD38-HA and its deletion derivatives were integrated by transformation into the EFG1 locus of strain HLC67 (efg1 mutant lacking the EFG1 open reading frame [ORF]) via the PacI site, which is located 1.9 kb upstream of the ATG translational initiation codon. A scheme demonstrating chromosomal integration of plasmids shown in Fig. S1A in the supplemental material. Correct plasmid integration in selected transformants was verified by Southern blotting of genomic DNA of transformants after digestion with XbaI (data not shown).
Construction of strains overexpressing EFG1 deletions.
Plasmid pBI-HAHYD, encoding a HA-tagged version of EFG1 derived from C. albicans ATCC 10231 under control of the PCK1 promoter, has been described earlier (Swissprot P43064) (23). As described above, 11 deletion variants of this plasmid were generated in the EFG1 ORF by in vitro mutagenesis using oligonucleotide primers (see Table S1 in the supplemental material). Plasmids were integrated by transformation into the LEU2 locus of the Δefg1 mutant strain HLC67 after digestion with KpnI (see Fig. S1B in the supplemental material), and correct integration was verified by Southern blottings of genomic DNA of selected transformants after digestion with KpnI and XmnI (data not shown). Similarly, plasmids were integrated into the genome of strain CAI4 (EFG1) to examine if variant phenotypes were dominant.
To analyze the forced switch from the opaque to the white growth form induced by overexpression of EFG1 or its variants, the pBI-HAHYD series of plasmids was also transformed into the opaque form of strain TS3.3 (22). Six transformants for each plasmid were grown in SD medium at 25°C. One hundred cells were plated on SCAA medium containing phloxine B (5 μg/ml) to activate the PCK1 promoter. Plates were grown for 4 days at 25°C and scored for differences in coloration (21, 22).
Assay for repressor activity of Efg1p.
A one-hybrid system for analyzing repressor activity of Efg1p in C. albicans has been described (5, 17). Plasmid Clp-LexA-EFG1, encoding a LexA-Efg1 fusion protein, was transformed into strain CRC106 carrying a lacZ reporter gene fused to a lexA operator fused to the basal ADH1 promoter (17). In order to identify domains of Efg1p that are important for repressor activity, the EFG1 ORF of plasmid Clp-LexA-EFG1 was mutagenized by in vitro mutagenesis using oligonucleotide primers to introduce 11 specific deletions into the Efg1 protein, as described above (see Table S1 in the supplemental material). The resulting plasmids were transformed into strain CRC106, and β-galactosidase activity was determined through 5-bromo-4-chloro-3-indolyl-β-galactopyranoside overlay plate assays and by liquid β-galactosidase assays as previously described (5).
To examine Efg1p repressor activity in various genetic backgrounds, we generated a plasmid (pMC2) containing both the LexA-Efg1 fusion and the lacZ reporter gene construct on a single URA3-containing plasmid. For this purpose, the 3.2-kb NotI fragment from pOPlacZ containing the lexA-operator-basal ADH1 promoter fusion to lacZ was inserted into the NotI site of plasmid Clp-LexA-EFG1. A control plasmid, pMC1, lacking the lexA operator, was constructed by a similar approach, inserting the NotI fragment of pCRLacbasal into Clp-LexA-EFG1 (17).
Yeast one-hybrid assays.
Plasmid pDB16 encodes a fusion of the transcriptional activation domain of the Saccharomyces cerevisiae Gal4 protein fused to Efg1p (5). The EFG1 ORF of plasmid pDB16 was mutagenized by in vitro mutagenesis using oligonucleotide primers (see Table S1 in the supplemental material). The resulting plasmids were transformed into S. cerevisiae strain BY600 (MATa swi6::TRP1-197 ade2 ho::lacZ ura3 his3 leu2-3,112 trp1-1 can1-100 met) containing a reporter plasmid carrying three MluI sites (ACGCGT) upstream of the yeast minimal CYC1 promoter (pMCB) (6).
To construct the plasmid pAP-swap, a ∼340-bp fragment of EFH1 containing the APSES domain was amplified by PCR from genomic C. albicans DNA using primers EFH-AP1 (5′-AACCCGGGATTCGACCAAAAGTTGC) and EFH-AP2 (5′-GACCACCGGTTAAAAAGTATTGC). The fragment was digested with the restriction endonucleases XmaI and AgeI (boldface type in primer sequences) and ligated into the single XmaI restriction site of plasmid pDB16-ΔAP that was introduced during the deletion of the APSES domain by site-directed mutagenesis. Presence and proper orientation of the fragment were checked through a novel NdeI restriction site present at the 3′ end of the EFH1 fragment introduced into pDB16-ΔAP. β-Galactosidase activity was determined as described previously (5).
Yeast two-hybrid assays.
To determine Efg1-Czf1 interactions, S. cerevisiae strain Ol1 (MATα leu2 his3 ura3; kindly supplied by W. Tanner) was transformed with the lacZ reporter plasmid pSH18-34 (10), pBOM1a, encoding a LexA-Czf1 fusion protein (9), and the above-described series of pDB16 derivatives encoding Efg1p variants. Additional fusions of the GAL4 activation domain sequence to specific parts of the EFG1 coding region were constructed by homologous recombination. pVL2 encoding residues 131 to 202 of Efg1 (deletion 3 sequence) was generated by PCR amplification of EFG1 sequences using primers pGAD-EFG-D3for and pGAD-EFG-D3rev (see Table S2 in the supplemental material); the PCR fragment was cotransformed with plasmid pGAD-C1 (11) linearized by digestion with BamHI into strain Ol1. Homologous recombination between ends of the PCR fragment and vector sequences generated pVL2, which was recovered from yeast transformants and verified. Likewise, plasmid pVL3, encoding residues 382 to 478 of Efg1p (deletion 6 sequence), was constructed using a PCR fragment obtained with primers pGAD-EFG-D6for and pGAD-EFG-D6rev (see Table S2 in the supplemental material).
To determine Efg1-Flo8 interactions, we used the two hybrid system of James et al. (11). The full-length FLO8/orf19.8695 coding region (4) was amplified by PCR using primers pGBD-FLO8-for and pGBD-FLO8-rev (see Table S2 in the supplemental material) and genomic DNA of strain CAI4 as the template. The PCR fragment was cotransformed with plasmid pGBD-C1 (linearized with BamHI) into strain PJ69-4A, generating FLO8 inserts by homologous recombination (because ends of PCR fragments were homologous to pGBD-C1). Plasmids were recovered from yeast transformants and verified, resulting in plasmid pVL1. Likewise, a part of the FLO8 coding region encoding residues 1 to 601 was amplified by PCR using primers pGBD-FLO8-for and pGBD-FLO8bisNLS-rev and inserted into pGBD-C1 to generate pVL4. Again, β-galactosidase activity was determined as described previously (5).
Protein methods.
Strains producing HA-tagged Efg1p variants under control of the authentic chromosomal EFG1 promoter were grown at 30°C in YPD to an optical density at 600 nm of 1 before harvesting and preparation of cell extracts (2). Strains producing HA-tagged Efg1p variants under transcriptional control of the PCK1 promoter were grown in SCAA medium, and crude cell extracts were isolated. A mix of protease inhibitors was added to the extraction buffer before breakage with glass beads (one tablet Complete Mini protease inhibitor mix per 10 ml; Roche). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblottings were performed using peroxidase-coupled anti-HA antibodies (1:2,500 dilution; Sigma) or rat anti-HA high-affinity antibody (1:1,000; Roche) as described previously (2). Protein extracts (50 μg) were dephosphorylated using 2 U of alkaline phosphatase (Roche) for 30 min at 37°C. Proteins were quantified by the Bradford method using the Bio-Rad protein assay dye reagent, with bovine serum albumin as a standard.
RESULTS
Structure of Efg1p.
Following the initial description of the EFG1 gene from C. albicans ATCC 10231 (23), two additional alleles of EFG1 derived from strain SC5314, orf19.610 and orf19.8243, have been described in the Candida genome sequencing project (1). The ATCC 10231-derived allele encodes a protein of 552 amino acids and is very similar to the orf19.8243-encoded protein, which has a length of 549 residues (differences in the latter allele are due to missing Gln residues in polyQ stretches at positions 83 and 438 to 441, two additional Ala residues following position 54, and A391T and I394T exchanges [ATCC 10231 protein numbering]). In contrast, the orf10.610-derived protein is shortened to 525 residues and differs significantly from ATCC 10231- and orf19.8243-predicted proteins, from residue 450 to the C-terminal end, due to a frameshift mutation. For structure-function analyses, we concentrated on the ATCC 10231-derived allele encoding the most extensive natural Efg1p variant, which was shown to fully complement all phenotypes of efg1 mutant strains (see below).
A diagram of the predicted functional regions in the Efg1 protein is shown in Fig. 1 (top). The APSES domain, which is highly conserved among fungal APSES proteins, spans the region from residues 203 to 306 including the core bHLH domain located between residues 236 and 297. Both the N and C termini contain polyQ stretches, and a low-complexity domain containing a stretch of alanines (residues 315 to 326) and a stretch of prolines (residues 332 to 338) is located C-terminal to the APSES domain. The GlobPlot structural prediction program (15) suggests two globular domains between residues 24 to 120 and 204 to 326 of Efg1p, which coincide with the N-terminal polyQ stretch and the central APSES domain region, respectively.
FIG. 1.
Proposed structural domains of Efg1p and corresponding phenotypes. Top, proposed structural domains of Efg1p. The conserved APSES domain consists of the bHLH region (hatched) and adjacent regions (gray). Polyglutamine stretches (Q) are indicated, as well as a low-complexity region containing a stretch of alanines (A) and prolines (P) (checkered). Deletions D1 to D11 are marked by amino acid positions within and framing the deletions. Two Efg1 regions predicted by the GlobPlot program (http://globplot.embl.de) to form globular domains are shown (A). Sequences essential for the wild-type phenotypes listed (B, C, and D) are indicated by black bars; sequences partially required are shown as gray bars.
Efg1p variant levels in C. albicans transformants.
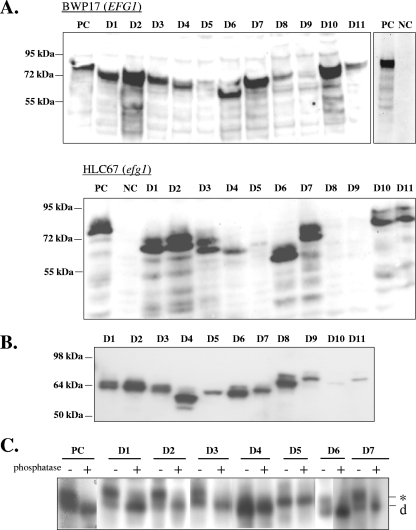
Since little is known about structure-function relationships of Efg1p or any other APSES protein, we started a systematic deletion approach to identify regions necessary for the various functions of Efg1p. Efg1p deletions are designated D1 to D11, and their position is shown in Fig. 1. First, to analyze the effect of specific deletions in the EFG1 ORF on Efg1's regulatory functions, plasmids encoding Efg1p variants were integrated into the chromosomal EFG1 promoter region of efg1 mutant strain HLC67 (whose EFG1 ORF has been deleted), expressing the different EFG1 alleles from the endogenous EFG1 promoter. In addition, these constructs were inserted similarly into the EFG1 locus of the EFG1+ strain BWP17. Correct integration as a monomer at the EFG1 locus was verified by Southern blotting (data not shown). The presence of the HA tag at the N-terminal end of the encoded Efg1 proteins allowed their detection by immunoblotting in cell extracts of selected transformants. The wild-type Efg1 protein migrated with an apparent molecular mass greater than the theoretical size of 60 kDa (80 to 90 kDa depending on the gel system), and with few exceptions, the deletion variants were also detectable both in an EFG1+ and an efg1 mutant genetic background (strains BWP17 and HLC67) (Fig. 2A). Interestingly, very low protein levels were observed for the D8 and D9 variants, in which conserved regions flanking the bHLH domain are missing (they were detected in overexpressing strains, however; see below). This result suggests that Efg1 regions surrounding the bHLH domain, in particular the C-terminal flanking region, are important for Efg1 protein levels, possibly to maintain proper folding and prevent proteolytic breakdown. Since the more extensive D4 deletion variant, however, is produced at normal levels, it appears that the stabilizing role of the D8 or D9 sequence requires interaction with additional sequences contained in D4 sequences. Relatively low levels of the D5 variant (but not of the nested D10 and D11 variants) confirm the importance of the C-terminal flanking region for Efg1p levels. As described earlier, Efg1p migrates as a doublet of proteins after prolonged electrophoresis (2), which was also observed for most variants (shown for strain HLC67 in Fig. 2A). The lack of a protein doublet for the D4 and D5 variants is consistent with the absence of phosphorylation (see below).
FIG. 2.
Presence of Efg1p variants in C. albicans transformants. (A) EFG1 and its deletion derivatives carrying deletions D1 to D11 were inserted downstream of the chromosomal EFG1 promoter in an EFG1+ strain (BWP17; resultant strain series AVL1 to -12) and an efg1 mutant (HLC67; resultant strain series HLCEEFG1-D1 to -D11). Strains carrying the respective wild-type EFG1 constructs (PC) or no EFG1 (NC) were used as controls. Efg1 proteins were detected in cell extracts (45 μg protein) of all strains by immunoblottings using anti-HA antibody. Note that prolonged electrophoresis of HLC67 extracts led to a doublet of two comigrating Efg1 bands. The migration of molecular mass standards is indicated (kDa). (B) Strains expressing EFG1 and its derivatives under transcriptional control of the PCK1 promoter were constructed by integrating PCK1-EFG1 fusions into the chromosomal LEU2 locus. The resulting HLCPEFG1-D1 to -D11 series of strains was grown in PCK1p-inducing SCAA medium, and extracts were analyzed as with panel A. (C) Phosphorylation of Efg1p variants. Strain HLCPEFG1 (PCK1p-EFG1) (PC) and corresponding strains carrying EFG1 deletions D1 to D7 were grown in SCAA medium. Cell extracts (50 μg protein) were not treated (−) or treated (+) with alkaline phosphatase (2 U), separated by SDS-PAGE, and immunoblotted using anti-HA antibody. *, phosphorylated form; d, dephosphorylated Efg1 form.
Morphogenetic phenotypes of Efg1p deletion variants.
Transformants containing EFG1 derivatives in the efg1 mutant background were analyzed for their ability to execute various morphological changes typical of C. albicans, including true hypha formation or chlamydospore formation. Strain HLCEEFG1, containing a wild-type EFG1 gene, and strain HLCE, lacking the EFG1 ORF in corresponding plasmid integration constructs (18), were used as controls.
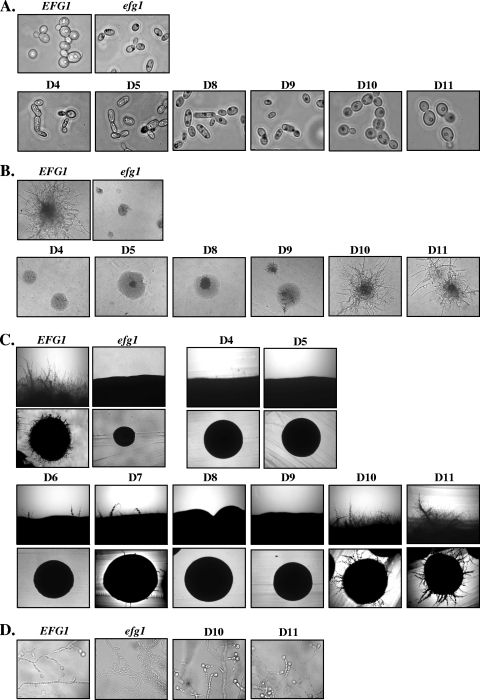
C. albicans grown in standard SD medium at 30°C usually shows a regular yeast-like cell shape. While strain HLCEEFG1 and strains expressing the D1 to D3 Efg1p variants, as well as the D6, D7, D10, and D11 variants, grew normally as spherical yeast cells (Fig. 3A), the efg1 deletion strain HLCE and strains producing Efg1p D4, D5, D8, and D9 variants formed elongated rod-like yeast cells, which are characteristic of efg1 mutants (23). Considering the normal level of the D4 variant protein but the absence or low protein level of the D8, D9, and D5 variants (Fig. 2), we conclude that the APSES domain spanning residue 203 to 306 is essential to allow normal yeast growth (Fig. 1).
FIG. 3.
Effects of Efg1p deletions on morphogenesis. (A) Yeast growth form after growth in SD medium at 30°C for 12 h. Deletions D1 to D3, D6, and D7 showed the wild-type phenotype (not shown). (B) Hyphal induction on agar containing 5% horse serum (1 day at 37°C). The D1 to D3, D6, and D7 deletion variants showed the wild-type phenotype (not shown). (C) Hyphal induction on Lee′s medium (2 days at 37°C); the upper photograph shows the rim of individual colonies. The D1 to D3 deletion variants showed the wild-type phenotype (not shown). (D) Chlamydospore formation on cornmeal agar containing 0.5% Tween 80. Streaks of cells were incubated under a coverslip at room temperature for 5 days. All deletions except D10 and D11 showed the mutant phenotype (not shown). EFG1, strain HLCEEFG1; efg1, strain HLCE.
Efg1p-dependent true hyphal growth of C. albicans is efficiently induced by serum or by certain inducing media, including Lee′s medium. On solid medium containing 5% serum, control strain HLCEEFG1 and strains producing the Efg1p D1 to D3, D6, D7, D10, and D11 variants produced colonies with lateral hyphae while strain HLCE and strains producing the Efg1p D4, D5, D8, and D9 variants produced smooth colonies without visible hyphae (Fig. 3B). In addition, in liquid medium containing 5% serum, the latter strains were also incapable of germ tube/hypha formation. After 60 min of hyphal induction at 37°C, the following percentages of hyphal cells were observed: 70% (HLCEEFG1), 0% (HLCE), 30% (HLCEEFG1-D1), 60% (HLCEEFG1-D2), 65% (HLCEEFG1-D3), 2% (HLCEEFG1-D4), 0% (HLCEEFG1-D5), 70% (HLCEEFG1-D6), 55% (HLCEEFG1-D7), 0% (HLCEEFG1-D8), 0% (HLCEEFG1-D9), 60% (HLCEEFG1-D10), and 60% (HLCEEFG1-D11). Hyphal formation on Lee′s medium revealed a requirement for the presence of additional Efg1p sequences, because the D6 and D7 variants were significantly defective in hypha formation (Fig. 3C). We conclude from these data that the APSES domain, as for normal morphogenesis of yeast cells, is also required for true hyphal formation. In addition, on certain inducing media, such as Lee′s medium, that are less effective in induction than serum, loss of the C-terminal polyQ stretches is already sufficient to abrogate hyphal induction.
Chlamydospore formation depends on Efg1p (20), and therefore, we examined strains producing Efg1p variants for this phenotype. With the exception of strains producing the D10 and D11 variants, all tested variants were defective in chlamydospore formation and merely showed filaments on chlamydospore-inducing medium (Fig. 3D). Thus, of all morphogenetic phenotypes tested, chlamydospore formation requires the most structural information on Efg1p.
Overexpression of deleted EFG1 alleles.
Overexpression of EFG1 has been shown to induce pseudohyphal filamentation with highly elongated cells (23). To evaluate whether Efg1p structures that were found to be involved in true hypha formation (see above) are also required for pseudohypha formation, we constructed deleted derivatives of EFG1 in plasmid pBI-HAHYD (23), in which EFG1 transcription is driven by the strong regulable PCK1 promoter (14). The series of plasmids containing the D1 to D11 deletions was integrated into the chromosomal LEU2 locus of the Δefg1 strain HLC67. As controls, the parental plasmid pBI-HAHYD and plasmid pBI-1 lacking the EFG1 ORF were used accordingly. Correct plasmid integration was verified by Southern blotting (data not shown). Transformants were grown in PCK1p-inducing SCAA medium, and the presence of Efg1 variant proteins was examined by immunoblotting (Fig. 2B). First, the results indicate that all Efg1 variants were produced under overexpressing conditions, including the D5 and D8 variants, which were hardly detectable using the authentic EFG1 promoter (Fig. 2A), although levels of the D10 and D11 variants were relatively low in this case.
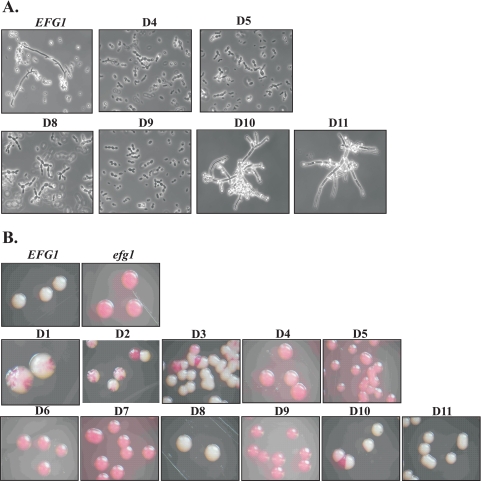
Growth of the selected transformants in SCAA medium, which induces the PCK1 promoter, led to the frequent appearance of elongated pseudohyphae in the pBI-HAHYD transformant and its D1-D3, D6, D7, D10 and D11 deletion derivatives. On the other hand, no pseudohyphae were observed in transformants carrying the pBI-1 plasmid and the D4, D5, D8, and D9 derivatives (Fig. 4A). Thus, these results indicate that pseudohyphal formation triggered by EFG1 overexpression and true hyphal induction share common requirements with respect to Efg1p structure.
FIG. 4.
Phenotypes of strains overproducing Efg1p variants. (A) Pseudohyphal formation induced by overexpression of EFG1 variants by the PCK1 promoter. Plasmid pBI-HAHYD and its D1 to D11 deletion derivatives were integrated into the LEU2 locus of strain HLC67, and strains were grown in PCK1p-inducing SCAA medium. Transformants carrying pBI-HAHYD (EFG1) and deletions D1 to D3, D6, and D7 showed occasional pseudohyphae (not shown), while deletion variants D4, D5, D8, and D9 had lost this ability. (B) Forced opaque-to-white switching. The opaque form of strain TS3.3 was transformed with the pBI-HAHYD series of plasmids, and transformants were plated on SCAA medium containing 5 μg/ml phloxine B. Transformants carrying empty control vector pBI-1 (efg1) remained in the opaque form and were stained red by phloxine B. Note that partial switching activity is apparent in some Efg1p deletion variants by colony sectorings.
Overexpression of EFG1 has been described to enforce a phenotypic switch of C. albicans opaque cells to their corresponding white growth form, which is consistent with low expression levels of the major EFG1 transcript in opaque cells and a morphology and transcriptome of opaque cells similar to those of efg1 mutant cells (21). In order to identify Efg1p regions that are important for Efg1p-driven white-opaque switching, plasmid pBI-HAHYD and its derivatives described above were therefore transformed into the opaque form of strain TS3.3 and selected transformants were subsequently analyzed for their switching ability (see Methods for details). To prevent heat-induced switching of opaque cells to the white cell form, strains were maintained and grown at 25°C. After incubation of plates at 25°C for 4 days, it was apparent that transformants carrying the negative control plasmid pBI-1, lacking the EFG1 ORF, remained in the opaque form (rod-like cells by microscopy), while those transformed with plasmid pBI-HAHYD grew as a white colony (normal yeast cells by microscopy) (Fig. 4B). Transformants carrying deletions D4 to D7 and D9 completely lost their ability to switch to the white form, while strains carrying deletions D1 to D3 showed a reduced switching frequency. Switching was generally normal in strains carrying D8, D10, and D11 deletion derivatives. These results indicate that a large portion of the APSES domain and N- and C-terminal regions of the Efg1 protein are required to promote phenotypic switching in C. albicans.
To check if the tested EFG1 alleles encoded loss-of-function or dominant gain-of-function mutated proteins, we subsequently transformed the pBI-HAHYD-series of plasmids in strain CAI4, which contains both EFG1 alleles. As described before, plasmids were chromosomally integrated into the LEU2 locus, and the resulting transformants were tested for their morphologies in different growth media. Transformants carrying any of the 11 deletion derivatives were normal with respect to growth, vegetative yeast cell shape, and their ability to generate hyphae. Similarly, no morphological phenotypes were observed in the EFG1+ strain BWP17 if EFG1 variants were put under control of the authentic EFG1 promoter by integration into one of the EFG1 alleles. Thus, these results suggest that all deleted Efg1p forms represent loss-of-function proteins unable to interfere with functions of the intact Efg1 protein.
Repressor function of deleted Efg1p derivatives.
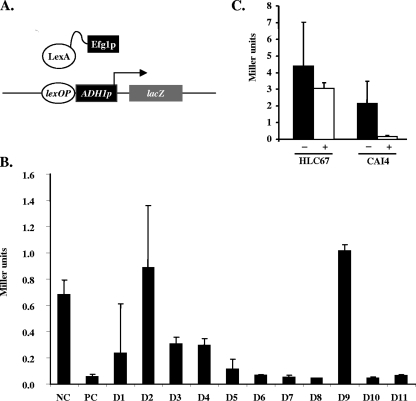
We recently described a one-hybrid system that can be used to monitor the function of Efg1p as a transcriptional repressor in C. albicans (5). In principle, one plasmid encodes a LexA-Efg1 fusion (plasmid Clp-LexA-EFG1), while a second plasmid present in the same cell (strain CRC106) encodes a lacZ reporter gene that is driven by the basal ADH1 promoter fused to the lexA operator (Fig. 5A). The same series of 11 deletions described so far was introduced into Clp-LexA-EFG1 to generate plasmids Clp-LexA-EFG1-D1 to -D11, and the resulting plasmids were transformed into strain CRC106 (17). Unmodified plasmid Clp-LexA-EFG1 was used as a positive control, while Clp-LexA lacking the EFG1 ORF served as a negative control (lack of repression). Our results confirm previous findings that the presence of the LexA-Efg1p fusion represses expression of the reporter gene about 10-fold in comparison to results for the negative control (Fig. 5B). In contrast, deletion plasmids Clp-LexA-EFG1-D2 and Clp-LexA-EFG1D9 were completely unable to repress the reporter gene, while all other deletions showed at least some repressing activity (incomplete in the cases of the D1, D3, and D4 variants). Overall, our experiments indicate that a short segment of the N-terminal region of Efg1p and the highly conserved amino acids at the C-terminal end of the APSES domain are reqired for Efg1p repressor activity in this system. On the other hand, the core bHLH DNA-binding domain seems to be completely dispensable for repressor function. However, since DNA binding of the Efg1 fusion protein depends on LexA-lexA operator interactions in the one-hybrid system used for this analysis, it remains unclear whether the DNA-binding properties of Efg1 are necessary for Efg1 repressor function in vivo.
FIG. 5.
Repressor activity of Efg1p variants in C. albicans. (A) Principle of one-hybrid assay. Basal activity of the lexA operator-ADH1p-lacZ fusion is repressed by the presence of the LexA-Efg1p fusion. (B) Strain CRC106 containing the lexA operator-ADH1p-lacZ fusion was transformed with plasmid Clp-LexA-EFG1 (PC) and its deletion derivatives (D1 to D11); a transformant carrying Clp-LexA was used as a negative control (NC). β-Galactosidase activities of three separate transformants were determined in duplicate assays, and standard deviations were calculated. (C) Plasmid pMC2 carrying expression units for both a LexA-Efg1p fusion protein and a lexA operator-ADH1p-lacZ reporter gene was integrated in the efg1 mutant HLC67 or wild-type strain CAI4. β-Galactosidase activity was determined and compared to the activity of transformants with control plasmid pMC1 lacking the lexA operator sequence. −, pMC1 transformants; +, pMC2 transformants.
Efg1 function in vivo is mainly regulated through the protein kinase A signaling pathway (2, 3). Therefore, we were interested in whether components of any signaling cascade regulating Efg1 would be able to modulate the repressor activity of Efg1p. To test this hypothesis, we constructed a single plasmid, pMC2, carrying both the expression unit for LexA-Efg1p production and the lacZ reporter gene. A corresponding plasmid lacking the lexA operator (pMC1) was used as a negative control. These plasmids were introduced into C. albicans mutant strains lacking either of the two isoforms of protein kinase A, tpk1 (strain IIHH6-4a) or tpk2 (strain AS1) (2), as well as an efg1 (strain HLC67) or efh1 mutant strain (C4/d63) (5). In contrast to the case with transformants carrying the negative control plasmid pMC1, downregulation of lacZ activity was observed for pMC2 transformants in the tpk1, tpk2, and efh1 genetic backgrounds at levels comparable to those for the wild-type strain, CAI4 (data not shown). However, in the efg1 mutant strain HLC67, repressor activity of the LexA-Efg1p fusion was surprisingly lost (Fig. 5C). We first conclude from these data that phosphorylation by a specific protein kinase A isoform is not required for repressor activity of Efg1p. Furthermore, authentic (unfused) Efg1p in combination with the LexA-Efg1 fusion protein appears to be necessary to trigger repressor activity, suggesting the formation of a heterodimer of these proteins, while homodimerization of the LexA-Efg1p fusion protein may not readily occur.
Efg1p binds to an MCB in vitro.
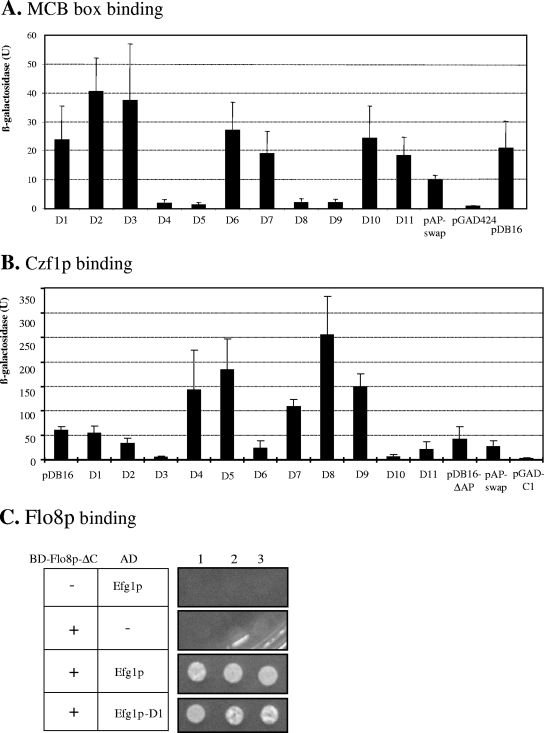
The APSES protein StuA of A. nidulans has been found to bind to an MCB sequence (ACGCGT) (6), and by yeast one-hybrid and gel retardation experiments, we recently established that Efg1p also is able to bind MCB sequences (M. Bussmann and J. F. Ernst, unpublished results). In the one-hybrid assay, a fusion of Efg1p to the Gal4 activation domain (encoded by plasmid pDB16) was shown to activate a MCB-dependent lacZ reporter construct, which is also demonstrated in Fig. 6A. Subsequently we introduced the above-described deletions D1 to D11 into the EFG1-GAL4 fusion construct and analyzed the ability of the mutant Efg1 proteins to activate the MCB-dependent promoter. Our results show that both the bHLH core region (D4 and D5 sequences) and the conserved flanking sequences that are typical of the APSES domain (D8 and D9 sequences) are required for Efg1 binding to the MCB, suggesting that the core bHLH DNA-binding motif by itself is not sufficient for this activity.
FIG. 6.
Properties of Efg1 variants in DNA binding and protein interactions. (A) Activation of an MCB-containing promoter by Efg1p deletion variants. S. cerevisiae strain BY600 containing reporter plasmid pMCB (3×MCB-CYC1p-lacZ) was transformed with plasmid pDB16 [ADH1p-GAL4(AD)-EFG1] and corresponding deletion variants, and β-galactosidase activities of selected transformants (Miller units) were determined. Values were derived from four independent transformants in duplicate measurements (± standard deviation). pGAD, negative control vector [ADH1p-GAL4(AD)]; pAP-swap, exchange of Efg1p APSES domain with Efh1 APSES domain. (B) Binding of Efg1p to Czf1p. Transformants of S. cerevisiae Ol1 encoding a LexA-Czf1 fusion protein (pBOM1a) and Efg1 fusions to the Gal4 activation domain (pDB16 or its deletion derivatives) were tested for activation of the lacZ reporter construct (present on pSH18-34). β-Galactosidase activity was determined as with panel A. (C) Binding of Efg1p to Flo8p. Transformants of strain PJ69-4A encoding a fusion of the Gal4 DNA binding domain to a shortened Flo8 protein (BD-Flo8p-ΔC, encoded by pVL4) and Efg1 fusions to the Gal4 activation domain (pDB16 or its deletion derivatives) were tested for activation of the ADE2 and HIS3 reporter genes by growth on supplemented SD medium lacking adenine and histidine.
In C. albicans, two APSES proteins, Efg1 and Efh1, have been identified that share a highly similar APSES domain (5). The Efh1p APSES domain contains about 40 mostly conservative exchanges compared to the Efg1p APSES domain. We were interested to know whether the APSES domain of the Efg1p homologue Efh1p would be functional in binding to MCB sequences when placed in the Efg1 protein background. We therefore replaced the entire APSES domain of the Efg1-Gal4 fusion construct (pDB16) with the corresponding Efh1p APSES domain. Interestingly, the resulting Efg1-Efh1 hybrid protein was still able to activate the MCB-dependent promoter but with an about twofold lower efficiency than wild type Efg1 (Fig. 6A). This result suggests that MCB binding may be a general property of APSES domains.
Efg1p domains required for binding to the Czf1 transcription factor.
Czf1p is a putative transcription factor able to bind to Efg1p directly and to modify its phenotypes (9, 25). To explore the structural requirements of Efg1p-Czf1p interactions, we performed yeast two hybrid-experiments, using the above-described series of pDB16-derived plasmids encoding fusions of Efg1 variants to the Gal4p activation domain. These plasmids were transformed into S. cerevisiae strain Ol1, along with plasmid pBOM1a, encoding a LexA-Czf1 fusion protein (9), and plasmid pSH18-34, encoding the lacZ reporter under transcriptional control of the lexA operator (10).
Transformants producing the Czf1 bait and full-length Efg1 prey fusions showed β-galactosidase activity, while a control strain containing the empty vector pGAD-C1 lacked activity, as expected (9) (Fig. 6B). Deletions D3 and D6 completely prevented Czf1-Efg1 interactions, while deletions of the APSES domain (deletions D4, D8, and D9) and of the adjacent C-terminal region (deletion D5) even enhanced this interaction. We propose that the sequences corresponding to deletions D4 and D5 hinder access of Czf1p in the native Efg1 protein, because they are flanked by the D3 and D6 sequences that are absolutely required for interaction. We observed one discrepancy in this set of data in that deletion D10 (lacking essentially the stretch of prolines) (Fig. 1) was inactive while the more extensive deletion D5 was active in interaction (Fig. 6B). This result indicates that D5 sequences are not required for binding of Czf1 and suggests that deletion of the nested proline stretch may have altered protein conformation more profoundly than the D5 deletion.
Efg1p binding to the Flo8 transcription factor.
The Flo8 transcription factor participates in many of the morphogenetic functions described for Efg1p, including the regulation of hypha formation under normoxic and hypoxic conditions. Furthermore, complex formation of Flo8p and Efg1p was shown by immunoprecipitation. (4). We used the yeast two-hybrid system to explore which Efg1p sequences are required for Flo8p interactions. Because Flo8p contains a transcriptional activation domain at its C-terminal end (4), we used a fusion of a C-terminally truncated version of Flo8p to the Gal4 binding domain in combination with Efg1-GAD variants encoded by the above-described pDB16 series of plasmids. The expression of CaFLO8 fusions led to strong flocculation of the S. cerevisiae host, preventing accurate measuring of optical densities; therefore, we used growth in the absence of adenine and histidine as the readout of reporter gene activation.
Using this experimental setup, we could clearly verify Flo8p-Efg1p interactions (Fig. 6C). Interestingly, all 11 Efg1p deletion constructs tested were still capable of interacting with Flo8p-ΔC (in Fig. 6C, shown only for the D1 variant), suggesting that no single Efg1p region, including the APSES domain, is responsible for this interaction. Instead, at least two redundant Efg1p sequences appear to independently mediate the interaction with Flo8p.
Modification of Efg1p variants.
In SDS-PAGE experiments, Efg1p usually migrates as two bands after prolonged electrophoresis, with an apparent molecular mass of approximately 80 to 90 kDa instead of its theoretical mass of 60 kDa, suggesting that Efg1 is phosphorylated and/or modified extensively by other yet-undefined posttranslational modifications. We sought to identify regions of Efg1p that contribute significantly to the deviation in molecular mass of Efg1p by either phosphorylation or other modifications by analyzing the Efg1p-producing strain HLCPEFG1 (PCK1p-EFG1) and corresponding strains carrying the Efg1 D1 to D7 deletion variants (see above) using immunoblotting techniques.
First, we treated cellular extracts of cells expressing the wild-type Efg1 protein with phosphatase to determine its degree of phosphorylation. The 80-kDa species present in untreated extracts shifted quantitatively to a faster-migrating protein of approximately 75 kDa, indicating that Efg1p in fact is a phosphoprotein. Performing similar experiments for the Efg1p deletion derivatives, we found that the D4 and D5 variants migrated as single bands and did not migrate differently after phosphatase treatment, suggesting an absence of phosphorylation, while all other deletion variants still appeared to be phosphorylated (Fig. 2C). These results indicate that Efg1 phosphorylation depends on the integrity of the APSES domain and adjacent C-terminal sequences, although the actual phosphorylated residues cannot be predicted with this knowledge. In addition, the results suggest that phosphorylation by itself is not the reason for the difference found between the theoretical and experimentally determined molecular masses of Efg1p. Furthermore, large differences between predicted and apparent molecular masses persist in all deletion variants, suggesting that no specific region of Efg1p is responsible for its unusual migration in SDS-PAGE.
DISCUSSION
APSES proteins in fungi contain a highly conserved domain of about 100 amino acids, the so-called APSES domain, containing a central bHLH motif, believed to mediate binding to MCB and/or E-box sequences, while outside sequences differ significantly and have no known function. Here we present the first evidence for a modular structure of any APSES protein, demonstrating that both the APSES domain and outside regions of the C. albicans Efg1 protein contribute to specific Efg1 functions that are required for various phenotypes. While the APSES domain itself was involved in maintaining the normal yeast morphology and hyphal induction by serum, hypha formation on Lee′s medium had an additional requirement for C-terminal sequences that may be involved in protein-protein interactions. In addition, chlamydospore formation also required segments in the N-terminal sequences apart from the APSES domain. PolyQ stretches may be one key element in these N- and C-terminal regions. EFG1 overexpression phenotypes also had various structural requirements. Thus, pseudohypha formation by Efg1p overproduction (23) required the entire APSES domain, while opaque-to-white switching (21) required additional N- and C-terminal sequences outside of the APSES domain. The results obtained may serve as a framework to focus future research on Efg1p subdomains required for specific morphogenetic requirements and their molecular functions.
Transcription factors in general and especially bHLH-type proteins in particular are known to form homo- and heterodimers and are able to interact with numerous regulatory cofactors. Dimerization of bHLH-type transcription factors is usually promoted through the helix-loop-helix region. Interestingly, evidence for homodimerization of the Efh1p but not the Efg1 protein was obtained by two-hybrid analyses (5), but surprisingly, dimerization of Efh1p was found to occur via sequences outside the APSES domain. While homodimerization of Efg1p was not found in two-hybrid experiments (5), heterodimerization to another bHLH-type protein is still speculative. With regard to interactions of Efg1 with other regulatory factors, interactions with the Czf1 and Flo8 proteins have been reported. Czf1p is a putative transcription factor regulating hyphal morphogenesis under certain environmental conditions (9, 25). Czf1p and Efg1p appear closely related functionally, because Czf1p relieves Efg1p-mediated repression while Efg1p regulates and binds to the CZF1 promoter (9, 25). Importantly, direct interaction of both proteins has been shown by the yeast two-hybrid assay (9). We show here that Czf1p binding requires two regions of Efg1p outside of the APSES domain (D3 and D6 sequences). Because each of these sequences individually is unable to bind to Czf1p, we propose that both sequences and/or additional Efg1 sequences are required for binding. We also speculated that the known ability of Czf1p to block the repressor activity of Efg1p (9) was caused by the direct binding of Czf1p to those Efg1 regions required for transcriptional repression (5). To determine Efg1 sequences involved in repression, we used the previously established C. albicans one-hybrid assay (5) and identified two Efg1 regions outside of the APSES domain (D2 and D9) whose presence was essential for repressing activity. Because deletion of the D9 region was associated with low Efg1 protein levels in C. albicans, we cannot determine its functional role, but we conclude that region D2 (coinciding with a globular domain) is important for the regulatory potential of Efg1p. Importantly, sequences required for repression and for binding to Czf1 do not coincide, indicating that Czf1 antagonism to Efg1 is not simply explained by the coverup of repressing sequences or by interactions with the APSES domain. Flo8p is another transcription factor known to interact with Efg1p and to perform similar functions during morphogenesis (4). Thus, as with Efg1p, Flo8p is required for hypha formation under normoxic conditions and it represses filamentation under hypoxia or during agar embedding. Furthermore, Flo8p regulates a subset of genes also known to be regulated by Efg1p (4). An Efg1p-Flo8p interaction was previously shown by coimmunoprecipitation, and we could confirm this result here with yeast two-hybrid results, using a Flo8 protein deleted for its inherent transcriptional activation domain. None of the Efg1p variants tested here prevented Flo8p interactions, suggesting that at least two redundant Efg1p segments ensure binding. Interestingly, the bHLH domain, which frequently mediates protein dimerization activity, was not required for Efg1p-Flo8p interactions.
bHLH-type transcription factors typically bind to E-box sequences (CANNTG) and the transcriptional regulator Efg1p of C. albicans, a member of the APSES-protein bHLH subfamily, indeed binds to a fragment containing a CATTTG sequence in vitro (13). We have obtained evidence that Efg1p can also attach to an MCB element (ACGCGT) (M. Bussmann and J. F. Ernst, unpublished results), and we show here that its conserved APSES domain is solely responsible for MCB binding, as expected, since deletions of all other Efg1 sequences had no effect. The binding specificity of Efg1p is very likely due to its similarity with the Mbp1 and Swi4 proteins, as well as other bHLH proteins (8). In agreement with this finding, in vitro binding of the StuA APSES protein of A. nidulans to an MCB element was also reported (6). Recently we obtained evidence that Efg1p is binding yet another target sequence (M. Bussmann and J. F. Ernst, unpublished results). Unraveling the physiological significance of Efg1 binding specificities, which may be paralleled by binding of numerous regulatory factors, will be an exciting challenge for future research.
Supplementary Material
Acknowledgments
We thank C. Kumamoto and H. Liu for plasmids.
This project was funded by the Deutsche Forschungsgemeinschaft (SFB590 and DFG priority program 1160) and by EU project “Galar Fungail II” (MRTN-CT-2003-504148).
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bockmühl, D. P., and J. F. Ernst. 2001. A potential phosphorylation site for an A-kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 1571523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockmühl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 421243-1257. [DOI] [PubMed] [Google Scholar]
- 4.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Liu. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doedt, T., S. Krishnamurthy, D. P. Bockmühl, B. Tebarth, C. Stempel, C. L. Russell, A. J. P. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 153167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutton, J. R., S. Johns, and B. L. Miller. 1997. StuA is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 165710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Biol. Cell 142100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 1601749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golemis, E. A. and R. Brent. 1997. Searching for Interacting proteins with the two-hybrid system. In P. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, New York, NY.
- 11.James, P., J. Haladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13148-153. [DOI] [PubMed] [Google Scholar]
- 13.Leng, P., P. R. Lee, H. Wu, and A. J. Brown. 2001. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 1834090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leuker, C. E., A. Sonneborn, S. Delbrück, and J. F. Ernst. 1997. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192235-240. [DOI] [PubMed] [Google Scholar]
- 15.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences of globularity and disorder. Nucleic Acids Res. 133701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo, H. J., J. R. Köhler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 17.Russell, C. L., and A. J. P. Brown. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol. 8676-683. [DOI] [PubMed] [Google Scholar]
- 18.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen-sensing and Efg1p-regulatory networks. J. Mol. Biol. 361399-411. [DOI] [PubMed] [Google Scholar]
- 19.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Sonneborn, A., D. P. Bockmühl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 675514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonneborn, A., B. Tebarth, and J. F. Ernst. 1999. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 674655-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srikantha, T., L. K. Tasai, K. Daniels, and D. R. Soll. 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 1821580-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human fungal pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 161982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tebarth, B., T. Doedt, S. Krishnamurthy, M. Weide, F. Monterola, A. Dominguez, and J. F. Ernst. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329949-962. [DOI] [PubMed] [Google Scholar]
- 25.Vinces, M. D., C. Haas, and C. A. Kumamoto. 2006. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot. Cell 5825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.