Abstract
Magnaporthe grisea is a model fungus for studying fungus-plant interactions. Two mitogen-activated protein (MAP) kinase genes, PMK1 and MPS1, have been implicated in regulating plant infection processes in M. grisea. However, transcription factors activated by these MAP kinases are not well studied. In this study we functionally characterized the MIG1 gene that encodes a MADS-box transcription factor homologous to Saccharomyces cerevisiae Rlm1. In yeast two-hybrid assays, MIG1 interacts with MPS1, suggesting that MIG1 may function downstream from the MPS1 pathway. The mig1 deletion mutant had a normal growth rate and formed melanized appressoria, but it was nonpathogenic and failed to infect rice leaves through wounds. Appressoria formed by the mig1 mutant developed penetration pegs and primary infectious hyphae, but further differentiation of the secondary infectious hyphae inside live plant cells was blocked. However, the mig1 mutant formed infectious hypha-like structures in heat-killed plant cells or cellophane membranes. In transformants expressing the MIG1-GFP fusion, green fluorescent protein (GFP) signals were not detectable in vegetative hyphae and conidiophores. Mig1-GFP was localized to nuclei in conidia, appressoria, and infectious hyphae. Deletion of the MADS box had no effect on the expression and localization of the MIG1-GFP fusion but eliminated its ability to complement the mig1 mutant. These results suggest that MIG1 may be required for overcoming plant defense responses and the differentiation of secondary infectious hyphae in live plant cells. The MADS-box domain is essential for the function of MIG1 but dispensable for its nuclear localization, which may be associated with the activation of MIG1 by MPS1 during conidiation and plant infection.
Magnaporthe grisea is an economically important fungal pathogen that invades crops such as rice, barley, wheat, and millet. Rice blast, caused by this heterothallic haploid ascomycete, is one of the most severe fungal diseases of rice throughout the world, and it has been developed as a model system to study fungus-plant interactions (11, 39). M. grisea was the first plant pathogenic fungus to be sequenced (8). The infection process of this pathogen is initiated by the germination of conidia attached to the plant surface and the development of a highly specialized infection structure called an appressorium. Direct penetration of the underlying plant surface is achieved by the generation of enormous turgor pressure in the appressorium (9). After penetration, the penetration peg differentiates into an unbranched primary infectious hypha, which in turn differentiates into bulbous, lobed secondary infectious hyphae. Initially, the fungus grows biotrophically in infected plant tissues but eventually causes typical blast lesions (20).
In the past decade, several signal transduction pathways involved in the regulation of different plant infection processes have been characterized in M. grisea (42). The cyclic AMP (cAMP) signaling pathway is required for surface recognition and the initiation of appressorium formation (5). The PMK1 mitogen-activated protein (MAP) kinase cascade regulates appressorium formation and infectious growth (42). In contrast, the MPS1 MAP kinase gene is dispensable for appressorium formation. However, the mps1 deletion mutant is defective in cell wall integrity and fails to successfully penetrate plant cells and develop infectious hyphae (37). A number of upstream components of the MAP kinase and cAMP signaling pathways also have been identified in M. grisea, such as the trimeric G proteins (22, 25) and Ras genes (29). Nevertheless, the downstream transcription factors of these signaling pathways have not been well studied in M. grisea. One putative downstream transcription factor regulated by Pmk1 is Mst12 (a homolog of Saccharomyces cerevisiae Ste12), which is dispensable for appressorium formation but essential for penetration and infectious growth (30). The Mst12 homolog is also required for plant infection but not appressorium formation in Colletotrichum lagenarium (31). In Cryphonectria parasitica, CpSTE12 is down-regulated by hypovirus infection and the cpste12 mutant is reduced in virulence and female sterile (10). In Ustilago maydis, the HMG domain transcription factor Prf1 is involved in both the cAMP signaling and MAP kinase pathways for mating and plant infection (19). However, M. grisea and other sequenced filamentous ascomycetes lack the Prf1 homolog. To our knowledge, no downstream transcription factors of the MPS1 MAP kinase pathway have been characterized and published for plant pathogenic fungi.
To better understand the downstream targets of the Pmk1 and Mps1 MAP kinases in the development and pathogenesis of M. grisea, we aimed in this study to functionally characterize two predicted genes encoding putative MADS-box transcription factors that are homologous to S. cerevisiae Mcm1 and Rlm1. In S. cerevisiae, Mcm1 is an essential gene product that interacts with a number of cofactors to regulate the expression of various genes required for cell growth and proliferation. In haploid cells, Mcm1 interacts with Ste12 to activate genes required for mating and cell fusion (24, 27). Rlm1 is one of the downstream transcription factors of the Slt2 MAP kinase (a Mps1 homolog), which plays a critical role in cell wall integrity (35). While we failed to isolate mutants with the MCM1 homolog deleted in M. grisea, the RLM1 homolog (named MIG1 for MADS-box protein required for infectious growth 1 gene) was found to be required for plant infection. MIG1 directly interacted with MPS1 in yeast two-hybrid assays. The mig1 deletion mutant formed appressoria, penetration pegs, and primary infectious hyphae but failed to develop secondary infectious hyphae in live plant cells. However, it penetrated and developed infectious hypha-like structures in heat-killed plant cells or cellophane membranes, suggesting that MIG1 may function downstream from MPS1 to regulate genes required for overcoming plant defense responses. Nuclear localization of the Mig1-green fluorescent protein (GFP) fusion protein was observed in conidia, appressoria, and infectious hyphae but not in vegetative hyphae and conidiophores. Localization of Mig1 to nuclei is independent of the MADS-box domain but may be related to its activation during conidiation and plant infection.
MATERIALS AND METHODS
Strains and culture conditions.
The wild-type M. grisea strains (Guy11 and 70-15) and transformants generated in this study (Table 1) were grown on oatmeal agar plates at 25°C under fluorescent light for conidiation as previously described (36). Mycelia from 3-day-old 5× YEG (0.5% yeast extract, 2% glucose) and complete medium (CM) cultures shaken at 150 rpm at room temperature were collected by filtration through Miracloth and used for genomic DNA extraction and RNA isolation. Sensitivities to lysing enzymes, calcofluor white (Sigma), and caffeine (Sigma, St. Louis, MO) were assayed as previously described (29, 37, 41). Media were supplemented with 200 μg/ml hygromycin B (Calbiochem, La Jolla, CA) and 200 μg/ml zeocin (Invitrogen, Carlsbad, CA) for selecting hygromycin-resistant and zeocin-resistant transformants, respectively. Genetic crosses, progeny isolations, and assays for conidiation with 10-day-old oatmeal cultures (6 cm in diameter) were performed as described previously (29, 43).
TABLE 1.
Wild-type and mutant strains of Magnaporthe grisea used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| 70-15 | Wild type, MAT1-1 | 4 |
| Guy11 | Wild type, MAT1-2 | 4 |
| M3H51 | mps1 mutant | 37 |
| R116 | mig1 deletion mutant | This study |
| R90 | mig1 deletion mutant | This study |
| R177 | Ectopic transformant of the MIG1 deletion construct | This study |
| C10 | Native promoter fused with MIG1-GFP in mutant R116 | This study |
| M16 | MIG1ΔMADS in mutant R116 | This study |
| O11WT | PRP27-MIG1-GFP in 70-15 | This study |
Appressorium formation and plant infection assays.
Conidia were harvested from 10-day-old oatmeal agar cultures and resuspended to 5 × 104 conidia/ml in sterile distilled water. Drops of conidial suspensions (30 μl) were placed on glass or plastic coverslips (Fisher Scientific Co.) and incubated at room temperature for examining conidium germination and appressorium formation. Appressorial penetration and infectious hypha formation were assayed with onion epidermal cells as described previously (40). For the plant infection assays, conidia were resuspended to 1 × 105 conidia/ml in a 0.25% gelatin solution. Two-week-old seedlings of rice, Oryza sativa cv. Nipponbare, and 8-day-old seedlings of barley, Hordeum vulgare cv. Golden Promise, were used for spray or injection infection assays (28). Plant incubation and inoculation were performed as described previously (33). Lesion formation was examined 5 to 7 days after inoculation.
MIG1 gene replacement construct and mutants.
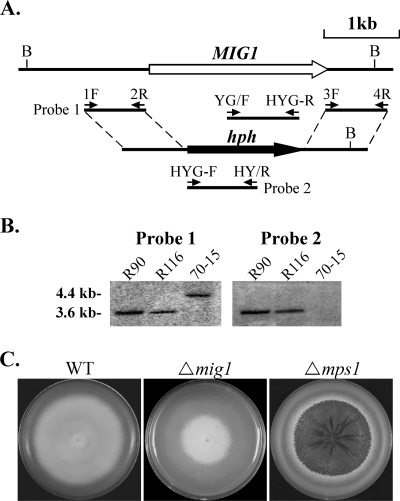
The split-marker approach (3) was used to delete the MIG1 gene. A 759-bp fragment upstream from MIG1 was amplified with primers MIG1-1F and MIG1-2R (Table 2). A 632-bp fragment downstream from MIG1 was amplified with primers MIG1-3F and MIG1-4R. The 5′ and 3′ fragments of the hygromycin phosphotransferase gene (hph) were amplified from pCB1003 with primers HYG-F/HY-R and YG-F/HYG-R (Table 2). Overlapping PCR was used to generate the MIG1-5′-hph and 3′-hph-MIG1 chimeric fragments with primer pairs MIG1-1F/HY-R and YG-F/MIG1-4R (Fig. 1A), respectively. About 10 μl each of the resulting PCR products was mixed and transformed into protoplasts of the wild-type strain 70-15 as described previously (36). Putative mig1 gene replacement mutants were screened by PCR with primers MIG1-6F and MIG1-7R (Table 2) and further confirmed by Southern blot analyses.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| Nat-F1 | CTATAGGGCGAATTGGGTACTCAAATTGGTGGAGATCTATGTCCCAGAGTAC |
| PRY-F | CGGAATTCATGGGTCGCAGGAAGATTGAAATT |
| PRY-R | TCACCGCTCGAGTCAAGAGTCTACTTTGACCCGTT |
| PRY-2R | CGGAATTCTCAAGAGTCTACTTTGACCCGTT |
| MIG1-GFP-R | GAACAGCTCCTCGCCCTTGCTCACAGAGTCTACTTTGACCCGTTTTG |
| MADS-F | CATTCGCGAATAGTCTTCAAGATGTCCGGTGACATGCGGGAGCTTATTAC |
| MIG1-1F | CTCCTGCATGCTACTGGTCA |
| MIG1-2R | TTGACCTCCACTAGCTCCAGCCAAGCCAATGCCTGTCTAGGCTGGTGAG |
| MIG1-3F | GAATAGAGTAGATGCCGACCGCGGGTTATACCACCTTCACGCTTTGGTT |
| MIG1-4R | ATGCAGGGCATGGAGAAAGA |
| MIG1-6F | GATGACGATGATGAGGAGGAAG |
| MIG1-7R | TGAGGTGGCATGGTAGTGTTC |
| Nat-F1 | CTATAGGGCGAATTGGGTACTCAAATTGGTGGAGATCTATGTCCCAGAGTAC |
| GFP-R | CTAAACAGACATTATCATCATCATGC |
| BLEO-F | GTACCTGAAGGAGCATTTTTGGG |
| BLEO-R | GAAAGAAGGATTACCTCTAAACAAGTG |
| GFP-F | GCAAGGGCGAGGAGCTGTT |
| GFP-SEQ-R | CTTGCCGGTGGTGCAGATGAACT |
| HYG-F | GGCTTGGCTGGAGCTAGTGGAGGTCAA |
| HYG-R | GAACCCGCGGTCGGCATCTACTCTATTC |
| YG-F | GATGTAGGAGGGCGTGGATATGTCCT |
| HY-R | GTATTGACCGATTCCTTGCGGTCCGAA |
| ActinQF | AGCACGGTGTCGTTACCAACTG |
| ActinQR | CAACAGAACGGGGTGCTCCT |
| Rlm1QF3 | CCTCGCCTCAAGGTGCAGAT |
| Rlm1QR3 | TCTGTTGAAGCACCTCCTTGCG |
| Slt5E | GGGAATTCATGTCGGATCTCCAGGGC |
| M3N3 | EGGGCGGCCGCTCACCTCCTCTGATCCAAA |
FIG. 1.
Generation of mig1 deletion mutants. (A) The MIG1 gene replacement construct was generated by the split-marker approach. The small arrows mark the positions and directions of primers used in the PCRs. (B) Southern blots of BglII-digested genomic DNAs of mig1 deletion mutants (R90 and R116) and 70-15 were hybridized with probe 1 (left) and probe 2 (right). Probe 1 hybridized to a 4.4-kb fragment in 70-15 and a 3.6-kb fragment in R90 and R116. Probe 2 detected a 3.6-kb band in R90 and R116 but not in 70-15. (C) CM cultures of the wild-type (WT) strain and the mig1 and mps1 deletion mutants. The mig1 mutant was reduced in aerial hyphal growth and conidiation but had no defect in growth rate and cell wall integrity. Colony autolysis was observed only in the mps1 mutant.
Construction of the MIG1-GFP fusion and the MIG1ΔMADS allele.
For generating the MIG1-GFP fusion construct pRM10, the full-length MIG1 gene, including about 1.6 kb of the promoter region, was amplified with primers Nat-F1 and GFP-R (Table 2). The resulting PCR product was cloned into the bleomycin-resistant vector pKB04 by the in vivo yeast homologous recombination approach as described previously (2). To delete the MADS box of MIG1, PCR products amplified with primer pairs Nat-F1/MADS-R and MADS-F/MIG1-GFP-R were cotransformed with XhoI-digested pKB04 into the yeast strain XK-25 (2). Plasmid pRM16 was isolated from Ura+ yeast transformants and confirmed by sequence analysis to carry the deletion of amino acid residues 2 to 58 in the MIG1 gene. The pRM10 (MIG1WT-GFP) and pRM16 (MIG1ΔMADS-GFP) constructs were sequenced with the BigDye Terminator sequencing kit (PE Applied Biosystems) to confirm the in-frame fusion with the EGFP carried by pKB04.
qRT-PCR analysis of gene expression.
Total RNA was isolated with TRIzol reagent (Invitrogen) and purified with the DNA-free kit (Ambion, Austin, TX). The first-strand cDNA synthesized with M-MLV reverse transcriptase (Invitrogen) was used for amplifying the open reading frame (ORF) of MIG1. For quantitative real-time PCR (qRT-PCR), cDNA was synthesized with the StrataScript QPCR cDNA synthesis kit (Stratagene). The Mx3000P QPCR machine (Stratagene) was used for PCRs that consisted of 2 min at 50°C (1 cycle), 10 min at 95°C (1 cycle), and 15 s at 95°C followed by 1 min at 60°C (40 cycles). Each qRT-PCR mixture (final volume of 20 μl) contained 10 μl of QuantiTect Sybr green PCR master mix (Qiagen), forward and reverse primers (500 nM concentrations of each), a cDNA template, and nuclease-free water. PCRs with serial dilutions of cDNA as templates were used to verify that the amplification efficiency was approximately equal for MIG1 (primers Rlm1QF3 and RlmQR3) and MgACT1 (primers ActinQF and ActinQR). The expression level of MIG1 was calculated by the comparative threshold cycle method with ACT1 as the endogenous reference for normalization (23).
Complementation assays with the yeast rlm1 mutant.
The full-length MIG1 cDNA was amplified with primers PRY-F and PRY-2R and cloned into the EcoRI site of pYES2 (Invitrogen). The resulting construct, pRM15, was confirmed by sequence analysis and transformed into the yeast rlm1 mutant DL2447 (18), which was kindly provided by David Levin. Sensitivities of DL2447 and transformants expressing pRM15 to different concentrations of caffeine were assayed on synthetic dextrose (SD) medium as described previously (18).
Yeast two-hybrid assays.
The yeast two-hybrid system III was used to assay protein-protein interactions as described previously (13, 15). The bait construct pNX10 was generated by the amplification of the MPS1 ORF with primer pair Slt5E/M3N3 and cloned between the EcoRI and NotI sites on pEG202. The MIG1 ORF was released from pRM15 (see above) by EcoRI digestion and cloned to the EcoRI site on pJG4-5 to generate the prey construct pRM13. The resulting bait and prey vectors were cotransformed into yeast strain EGY48 with the alkali-cation yeast transformation kit (MP Biomedicals, Solon, OH). The His+ and Trp+ transformants were isolated and assayed for growth on SD-Trp-His-Ura-Leu medium and for galactosidase activity as described previously (15). The positive control for transcriptional activation of the LexA operator-LacZ reporter was generated by transforming EGY48 with pSH17-4, pSH18-34, and pJG4-5. For the negative control, EGY48 was transformed with pRFMH1, pSH18-34, and pJG4-5 as described previously (15).
RESULTS
The M. grisea genome contains two MADS-box transcription factors.
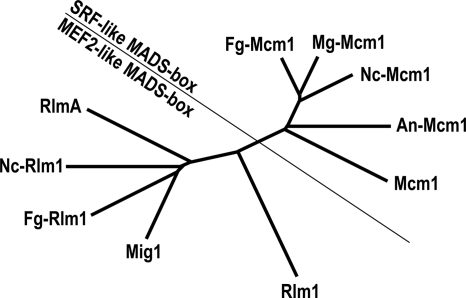
Similarly to other filamentous ascomycetes, such as Neurospora crassa, Fusarium graminearum, and Aspergillus nidulans, M. grisea has two putative MADS-box transcription factor genes (MGG_02773.5 and MGG_01204.5) that are homologous to yeast RLM1 and MCM1 (Fig. 2). By sequence alignment analysis, we found that MGG_02773.5 (temporarily named MgMCM1) appeared to be correctly predicted by automated annotation. However, the annotation of MGG_01204.5 (named MIG1 for MADS-box protein required for infectious growth 1 gene) was problematic. We sequenced the PCR product amplified from the first-strand cDNA of the wild-type strain 70-15 with primers PRY-F and PRY-R. In comparison with the cDNA sequence, the predicted sequences for the third intron and stop codon of MGG_01204.5 by automated annotation were incorrect. The actual ORF of MIG1 (GenBank accession no. EU164776) was 200 bp shorter than the prediction. Phylogenetic analysis revealed that Mig1 groups with yeast Rlm1 in the MEF2-like type II subfamily of MADS-box proteins. MgMcm1 belongs to the SRF-like type I MADS-box proteins (Fig. 2), and its homology with Mig1 was limited to the MADS-box domain.
FIG. 2.
The unrooted neighbor joining tree of the following putative MADS-box transcription factors from selected ascomycetes: Mcm1 (CAA88409) and Rlm1 (AAB68210) from Saccharomyces cerevisiae, MgMcm1 (EAA47530) and Mig1 (EU164776) from Magnaporthe grisea, NcMcm1 (EAA36453) and NcRlm1 (EAA35381) from Neurospora crassa, FgMcm1 (EAA76082) and FgRlm1 (EAA70796) from Fusarium graminearum, and AnMcm1 (EAA63555) and RlmA (EAA60098) from Aspergillus nidulans. SRF, serum response factor; MEF2, myocyte enhancer factor 2.
The mig1 deletion mutant is reduced in aerial hyphal growth and conidiation.
To determine the functions of MgMCM1 and MIG1 in M. grisea, we used the split-marker approach (3) to generate gene replacement constructs for these two genes. The resulting PCR products were transformed directly into protoplasts of strain 70-15. After screening over 150 transformants, we failed to identify any gene replacement mutant for MgMCM1, suggesting that it may be an essential gene in M. grisea. For the MIG1 gene (Fig. 1A), putative gene replacement mutants were identified by screening hygromycin-resistant transformants with primers MIG1-6F and MIG1-7R. Two mig1 deletion mutants, R90 and R116 (Table 1), were further confirmed by Southern blot analysis (Fig. 1B). The two mutants were phenotypically indistinguishable, although data are presented only for transformant R116 in this paper.
The mig1 deletion mutant produced typical gray colonies on oatmeal agar plates (data not shown). The growth rate of the mig1 mutant was similar to that of strain 70-15 on oatmeal agar and CM, but the mutant was reduced in aerial hyphal growth (Fig. 1C) and conidiation compared to the wild type (Table 3). However, the mig1 mutant produced more conidia and aerial hyphae than the mps1 mutant, which barely produces conidia and aerial hyphae on oatmeal agar plates (37). Colonies of the mig1 mutant had no autolysis defects (Fig. 1C). While the mps1 mutant is hypersensitive to cell wall-degrading enzymes (37), vegetative hyphae of the mig1 mutant released only a few protoplasts after digestion with 5 mg/ml lysing enzymes for 20 min (data not shown). These results suggest that MIG1, unlike MPS1, does not play a major role in vegetative growth, cell wall integrity, or conidiation in M. grisea.
TABLE 3.
Defects of the mig1 mutant in conidiation and appressorial penetration
| Strain | Conidiation (105 conidia/plate) | Appressorial penetrationa |
|---|---|---|
| 70-15 (wtb) | 10.6 ± 0.6 | 65.0 ± 13.7 |
| R116 (mig1) | 2.9 ± 0.4 | 3.2 ± 1.2 |
| R177 (ectopic) | NDc | 53.8 ± 19.6 |
| C10 (MIG1/mig1) | 10.5 ± 0.5 | 18.4 ± 14.0 |
Percentages of appressoria that successfully penetrated onion epidermal cells and developed primary and/or secondary infectious hyphae.
wt, wild type.
ND, no data.
MIG1 is essential for plant infection.
In spray inoculation of 2-week-old seedlings, the mig1 mutant failed to cause any lesions on leaves of O. sativa cv. Nipponbare rice at 7 days postinoculation. Under the same conditions, numerous lesions were observed on leaves inoculated with the wild-type strain and the complementation strain C10 (Fig. 3A). Similar results were observed in infection assays with barley seedlings of H. vulgare cv. Golden Promise (Fig. 3B), indicating that the mig1 deletion mutant is nonpathogenic.
FIG. 3.
Infection assays with the wild-type strain (WT), the mig1 mutant R116 (Δmig1), and the complementation transformant of R116 (C10). Inoculation with 0.25% gelatin was used as the control. (A) Spray inoculation with 2-week-old rice seedlings. (B) Spray inoculation with 8-day-old barley seedlings. (C) Rice leaf segments containing the wound sites from injection-inoculation assays were surface sterilized and incubated on water agar for 2 days. Hyphal growth and conidiation were not observed on leaves inoculated with the mig1 mutant.
To determine whether the mig1 mutant is defective in plant infection through wounds, we conducted injection infection assays with rice seedlings. On leaves inoculated with the wild type and transformant R177, typical blast lesions developed beyond the wound sites (data not shown). The mig1 deletion mutant caused limited necrosis at the wound sites but failed to infect unwounded areas of rice leaves. To determine whether M. grisea could be recovered from the necrotic tissue, rice leaf segments containing the wound sites were collected at 7 days postinoculation and surface sterilized. After incubation on water agar for 2 days under high humidity, abundant conidia and hyphal growth were observed over lesions and wound sites on leaves inoculated with the wild type and an ectopic transformant (Fig. 3C). Under the same conditions, we failed to detect fungal growth at the injection sites inoculated with the mig1 mutant (Fig. 3C). Thus, we conclude that MIG1 is required for colonization through wounds and infectious growth.
To confirm that the observed phenotype was directly related to the mig1 gene replacement event, we isolated 21 ascospore progeny from a cross between R116 (MAT1-1) and the wild-type strain Guy11 (MAT1-2). While none of the 12 hygromycin-susceptible progeny was defective in plant infection, all 9 hygromycin-resistant progeny were nonpathogenic (data not shown). These data indicate that MIG1 is dispensable for sexual reproduction and loss of pathogenicity cosegregated with hygromycin resistance. Therefore, deletion of the MIG1 gene is responsible for the defect of the mig1 mutant in plant infection.
MIG1 is dispensable for appressorium formation but required for infectious growth.
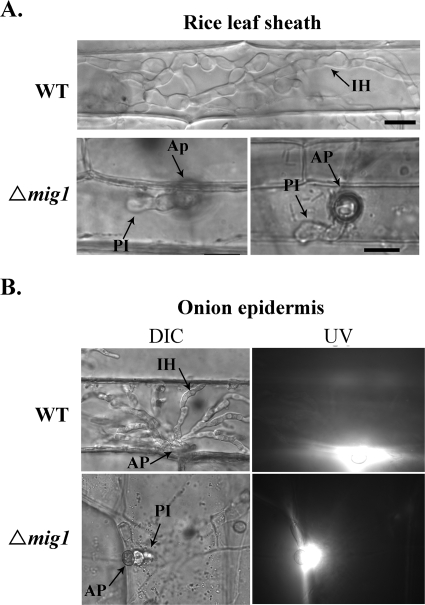
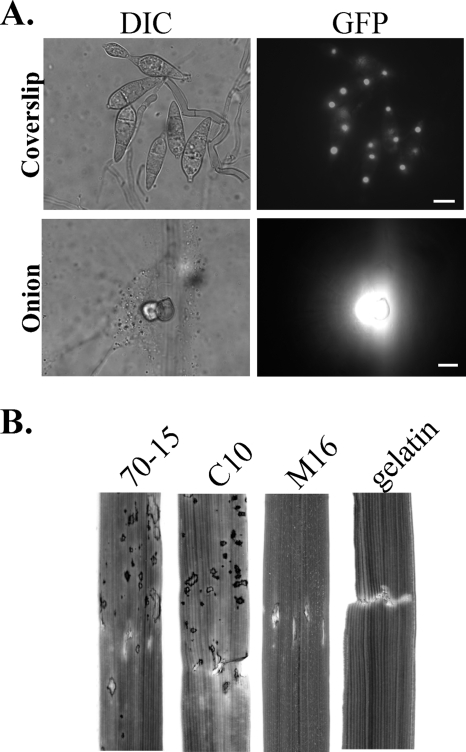
On artificial hydrophobic surfaces, the vast majority (>95%) of the mig1 conidia germinated and developed appressoria by 24 h, which is similar to the wild-type strain. No obvious difference in appressorium size, morphology, or melanization was observed between the mig1 mutant and 70-15 (data not shown). In penetration assays with rice leaf sheaths (Fig. 4A) and onion epidermis (Fig. 4B), the mig1 mutant also efficiently formed melanized appressoria but failed to develop secondary infectious hyphae. Appressoria formed by the mig1 mutant were able to elicit autofluorescence and papilla formation in underlying rice leaf sheath or onion epidermal cells. About 3% of them (Table 3) were able to develop short primary infectious hyphae in underlying rice leaf sheath or onion epidermal cells (Fig. 4). However, branching, bulbous secondary infectious hyphae were not observed in plant cells penetrated by the mig1 deletion mutant by 48 h (Fig. 4) or after prolonged incubation (data not shown). Under the same conditions, the wild type produced bulbous secondary infectious hyphae (Fig. 4). These results indicate that the deletion of the MIG1 gene has no obvious effect on the initial plant penetration processes, such as the development of penetration pegs and primary infectious hyphae, but blocks the differentiation of secondary infectious hyphae.
FIG. 4.
Penetration assays with the wild-type strain (WT) and the mig1 mutant (Δmig1). (A) Bulbous secondary infectious hyphae were observed only in rice leaf sheath epidermal cells penetrated by the wild-type strain. Some appressoria (about 3%) of the mig1 mutant were able to develop short primary infectious hyphae in plant cells. (B) The mig1 mutant elicited autofluorescence and papilla formation in onion epidermal cells but failed to develop secondary infectious hyphae. Images on the left and right were the same field examined under differential interference contrast (DIC) and epifluorescence microscopy (UV). Ap, appressorium; PI, primary infectious hyphae; IH, secondary infectious hyphae.
The mig1 mutant is likely defective in overcoming plant defense responses.
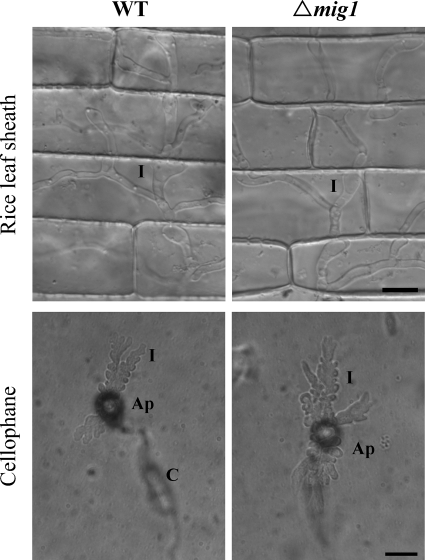
To further characterize the defect of the mig1 mutant in infectious growth, we repeated the penetration assays with rice leaf sheaths and onion epidermis incubated at 75°C for 25 min before inoculation. At 48 h postinfection, about 90% of the appressoria formed by the wild type and the mig1 mutant penetrated heat-killed epidermal cells of rice leaf sheaths and developed hyphae that were thicker than vegetative hyphae but less bulbous and branching than normal infectious hyphae (Fig. 5) and that lacked the regular constrictions or yeast-like growth observed in secondary infectious hyphae (20). No obvious difference was observed between the wild type and the mig1 mutant in penetration efficiency or the extent of hyphal growth in heat-killed rice leaf sheath cells (Fig. 5A). In penetration assays with heat-killed onion epidermal cells, similar infectious hypha-like structures were produced by both the wild type and the mig1 mutant (data not shown). These results indicate that the MIG1 gene is not essential for the differentiation or growth of infectious hyphae. However, it may be required for infectious hyphae to overcome plant defense responses in live plant cells.
FIG. 5.
Penetration assays with heat-killed rice leaf sheaths and cellophane membranes. Both the wild-type strain (WT) and the mig1 mutant R116 (Δmig1) penetrated and developed infectious hypha-like structures in heat-killed rice leaf sheath epidermal cells of rice and cellophane membranes. Ap, appressorium; C, conidium; I, infectious hypha-like structures. Bars = 10 μm.
We repeated the penetration assays with cellophane membranes that are devoid of live cells but can support limited growth of infectious hypha-like structures (1, 6). Similarly to the wild-type strain, the mig1 mutant penetrated the cellophane membranes and formed infectious hypha-like structures (Fig. 5B). These observations further indicate that the defect of the mig1 mutant in infectious hyphal growth and plant infection results from its inability to overcome plant defense responses. Because appressoria formed by the mps1 mutant also elicit plant defense responses in underlying cells (37) and are normal in penetration peg formation (28), we conducted penetration assays with the mps1 mutant in this study for comparison. Like the mig1 mutant, the mps1 mutant penetrated and formed infectious hypha-like structures in heat-killed rice leaf sheath cells and cellophane membranes (data not shown). Therefore, we conclude that the mps1 and mig1 mutants have similar defects in the differentiation and growth of secondary infectious hyphae in healthy, intact rice leaves. Unlike many other known nonpathogenic mutants of M. grisea, they were able to penetrate but appeared to be defective in overcoming plant defense responses.
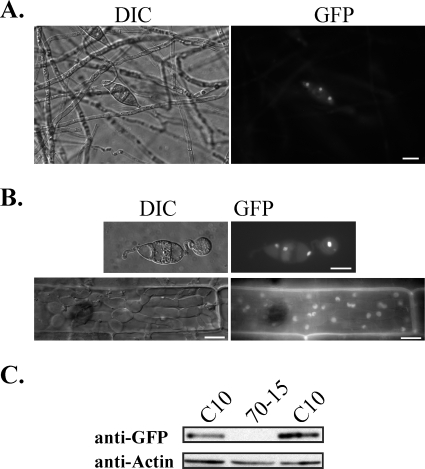
The Mig1-GFP fusion protein localizes to nuclei in conidia, appressoria, and infectious hyphae.
To determine the expression and localization of MIG1, we generated a MIG1-GFP fusion construct, pRM10, and transformed it into the mig1 deletion mutant R116. Transformant C10 was identified by PCR and confirmed by Southern blot analysis to contain a single copy of pRM10 (data not shown). It had no obvious defects in vegetative growth, appressorium formation, or penetration (Table 3). Moreover, the conidiation defect of the mig1 mutant was rescued in transformant C10 (Table 3). In infection assays, C10 was as virulent as the wild-type strain (Fig. 3), indicating that the GFP fusion had no effect on Mig1 function and the MIG1-GFP fusion complemented the mig1 deletion mutant in plant infection.
When cells were grown on oatmeal agar plates, GFP signals were usually not detected in aerial hyphae and conidiophores of transformant C10 (Fig. 6A). Vegetative hyphae grown in liquid CM or 5× YEG medium also lacked detectable GFP signals (data not shown). In freshly harvested conidia, GFP signals were predominantly localized to the nucleus (Fig. 6A). Nuclear localization of Mig1-GFP fusion proteins also was observed in appressoria and infectious hyphae formed by transformant C10 (Fig. 6B). Therefore, MIG1 is expressed in conidia, appressoria, and infectious hyphae. Because nuclear localization of Mig1-GFP may represent its activation status, we conducted Western blot analyses with proteins isolated from vegetative hyphae. The Mig1-GFP fusion protein of expected size was detected in transformant C10 with the anti-GFP antibody (Fig. 6C), indicating that MIG1 also is expressed in vegetative hyphae. However, the expression level of MIG1 appears to be relatively low because of the over 25,000 expressed sequence tags sequenced from various libraries (12) and deposited in GenBank, only one corresponds to MIG1. Therefore, the lack of detectable GFP signals in vegetative hyphae may be related to the low abundance of the MIG1-GFP fusion. It is possible that GFP signals were visible only when Mig1-GFP is activated during conidiation and plant infection and becomes concentrated in nuclei.
FIG. 6.
Expression and localization of Mig1-GFP in transformant C10. (A) GFP signals were observed in the nuclei of conidia but not in aerial hyphae or conidiophores harvested from 9-day-old oatmeal cultures. The same field was examined under differential interference contrast (DIC) and epifluorescence microscopy (GFP). Bar = 10 μm. (B) GFP signals were preferentially localized to the nuclei in appressoria formed on glass coverslips and infectious hyphae produced in rice leaf sheath epidermal cells. Bars = 10 μm. (C) Western blot analysis with proteins isolated from vegetative hyphae of the wild-type strain 70-15 and transformant C10. An 86-kDa band of the expected size of Mig1-GFP fusion was detected with an anti-GFP antibody.
We also used qRT-PCR to determine the expression of MIG1 with the M. grisea actin gene MgACT1 as the endogenous reference for normalization. In the wild-type strain, MIG1 was constitutively expressed in vegetative hyphae and infected rice leaves. No significant change in the expression level of MIG1 was observed between vegetative growth on CM cultures and pathogenic growth during plant infection (Table 4). In comparison with MgACT1, the expression level of MIG1 was relatively low (Table 4). We also assayed the expression level of MIG1 in mycelia harvested from CM cultures of the mps1 mutant. The deletion of MPS1 had no obvious effect on the expression of MIG1 (Table 4).
TABLE 4.
RT-PCR quantification of the expression level of MIG1
| RNA source |
CTa
|
Normalized MIG1 level relative to β-actinb | |
|---|---|---|---|
| MIG1 | β-Actin | ||
| Wild-type vegetative hyphae | 25.82 ± 0.13 | 21.39 ± 0.07 | 1.00 (0.91-1.09) |
| Wild-type infected leaves | 26.21 ± 0.22 | 21.52 ± 0.10 | 1.39 (1.18-1.65) |
| mps1 mutant vegetative hyphae | 25.88 ± 0.11 | 21.68 ± 0.04 | 0.86 (0.79-0.93) |
CT, cycle number at which the fluorescence crossed the threshold. Means and standard deviations were calculated with data from three replicates.
Relative quantification of the MIG1 expression level (23) during plant infection and in the mps1 mutant in comparison to that of vegetative growth. Data are given as the averages (ranges) of three replicates.
The MADS-box domain is essential for MIG1 function.
The MADS-box domain is well conserved in MIG1 and its homologs. We used the overlapping PCR approach to delete amino acid residues 2 to 58 in the MIG1-GFP fusion construct. The resulting MIG1ΔMADS-GFP construct, pRM16, was transformed into the mig1 mutant R116. Transformant M16 (Table 1) was confirmed by Southern analysis to contain an ectopically integrated copy of pRM16 (data not shown). The strength of the GFP signals in the nuclei of conidial cells in transformant M16 was comparable to that of transformant C10 (Fig. 7A), indicating that the deletion of the MADS box had no effect on the expression of MIG1. If nuclear localization is indeed related to its activation, results from this experiment also suggest that the MADS box is dispensable for the activation of MIG1.
FIG. 7.
The MADS box is essential for the function of Mig1. (A) Deletion of the MADS box had no effect on the expression and localization of the MIG1-GFP fusion. Fluorescent signals were observed in the nuclei of conidial cells but not in aerial hyphae of MIG1ΔMADS-GFP transformant M16 (top panels). Appressoria formed by transformant M16 elicited papilla formation and autofluorescence in underlying plant cells but failed to develop infectious hyphae in onion epidermal cells (bottom panels). Bars = 10 μm. (B) Rice leaves wound-inoculated with the wild-type (70-15), complementation transformant C10, MIG1ΔMADS-GFP transformant M16, and 0.25% gelatin.
Similarly to the original mig1 deletion mutant, transformant M16 was reduced in conidiation (Table 3) and defective in plant infection in the spray inoculation and injection assays (Fig. 7B). Appressoria formed by transformant M16 also elicited plant defense responses, such as papilla formation and autofluorescence, but were defective in the differentiation of secondary infectious hyphae (Fig. 7A). Thus, we conclude that the MADS-box domain is essential for the function of MIG1 in M. grisea. Deletion of the MADS box may not affect the activation of Mig1 but likely abolishes its ability to bind with promoter elements of downstream target genes.
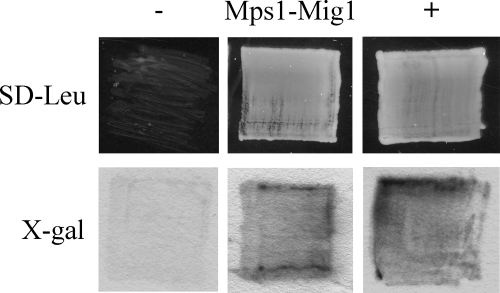
Mig1 interacts with Mps1 in yeast two-hybrid assays.
The direct interaction of Mig1 with the Mps1 MAP kinase was assayed with the interaction trap system (15). The MPS1 bait construct pNX10 and MIG1 prey construct pRM13 were cotransformed into the yeast strain EGY48. The resulting Ura+ Trp+ transformants were confirmed to contain pNX10 and pRM13 by PCR with primer pairs MIG1-6F/MIG1-7R and Slt5E/M3N3, respectively (data not shown). When galactose was used as the carbon source, yeast cells expressing both the MPS1 bait and the MIG1 prey constructs were able to grow on SD-Leu plates and display LacZ activities (Fig. 8). Growth on SD-Leu plates and LacZ activities were not observed in the same transformants when glucose was used as the carbon source (data not shown), indicating that Mig1 interacts directly with Mps1 in yeast cells. MIG1 is likely one of the transcription factors that function downstream from MPS1 in M. grisea.
FIG. 8.
Yeast two-hybrid assays. Yeast transformants containing the bait and prey constructs of MIG1 and MPS1 were able to grow on SD-Leu plates and express β-galactosidase activities when galactose was used as the carbon source. Transformants expressing pRFHM1 and pSH18-34 or pSH17-4 and pSH18-34 were used as the negative (−) and positive (+) controls, respectively. Growth on SD-Leu plates and β-galactosidase activities were examined after incubation for 48 h. X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used as the substrate to assay galactosidase activities.
DISCUSSION
Like many other filamentous ascomycetes, M. grisea contains two well-conserved MADS-box transcription factor genes, MgMCM1 and MIG1. Errors in the automated annotation of MGG_01204.5 may explain why only one MADS-box-containing gene was identified in a previous study (7). Because MADS-box transcription factors are not well characterized in filamentous ascomycetes, in this study we aimed to functionally characterize both MgMCM1 and MIG1 in M. grisea. Unfortunately, we failed to identify the mgmcm1 deletion mutant, suggesting that MgMCM1 may be an essential gene in M. grisea. In S. cerevisiae, the deletion of MCM1 is lethal (24). However, the MCM1 homolog is not an essential gene in Sordaria macrospora (26). Therefore, it remains possible that the mgmcm1 deletion mutant is viable, but the deletion of MgMCM1 may result in a severe growth defect or sensitivity to the transformation process in M. grisea.
The deletion of MIG1 had no effect on growth rate or appressorium formation (Fig. 3) but blocked the differentiation of secondary infectious hyphae (Fig. 4). MIG1 is homologous to yeast RLM1, which is a downstream transcription factor of the SLT2 MAP kinase (35). In M. grisea, the MPS1 MAP kinase gene (an SLT2 homolog) is important for plant infection and conidiation. Interestingly, the mps1 and mig1 mutants differ significantly in colony morphology. Unlike the mps1 mutant (37), the mig1 mutant was only slightly reduced in aerial hyphal growth and conidiation (Table 3). Autolysis, which occurred in the mps1 mutant colonies, was not observed with the mig1 mutant (Fig. 1), indicating that Mig1 is dispensable for cell wall integrity. In S. cerevisiae, the growth defects of the rlm1 and slt2 mutants also are different. While the slt2 mutant has pleiotropic phenotypes, including caffeine sensitivity, temperature-dependent cell lysis, the inability to grow on glycerol, and starvation sensitivity, the rlm1 mutant is only sensitive to caffeine (21).
Nevertheless, the defect of the mig1 mutant in plant infection is similar to that of the mps1 mutant. Both the mps1 and mig1 mutants formed melanized appressoria but failed to develop secondary infectious hyphae in live plant cells (Fig. 4). However, they were able to develop infectious hypha-like structures in heat-killed plant cells and cellophane membranes (Fig. 5), suggesting that the mps1 and mig1 mutants may be defective in overcoming host defense responses. It is possible that Mig1 regulates subsets of genes that are controlled by the Mps1 pathway and required for plant infection. Their defect in infectious growth differs from other known nonpathogenic mutants in M. grisea. Mutants blocked in the PMK1 MAP kinase pathway and the mac1 or mgb1 mutants are defective in appressorium formation (29, 41). MST12 is dispensable for appressorium formation but essential for penetration peg formation (28). The cpkA mutant forms nonfunctional appressoria but still causes occasional lesions and can colonize rice plants through wounds (38). The pls1 mutant is defective in appressorial penetration and fails to develop infectious hypha-like structures in cellophane membranes (6). The atg8 mutant is defective in appressorium turgor generation and infectious growth (34). It has been hypothesized that the transporter gene ABC1 may be involved in pumping out toxic plant compounds in M. grisea, but the abc1 mutant is still pathogenic, is able to form infectious hyphae in rice tissues, and causes few lesions on inoculated leaves (32). The characterization of the defect of the mig1 mutant in this study also indicates that the penetration assay with heat-killed plant cells can be used as one of the approaches to distinguish mutants that are defective in overcoming plant defense responses or the differentiation of infectious hyphae.
When the MIG1-GFP fusion construct was transformed into the mig1 deletion mutant, the resulting transformant C10 was as virulent as the wild type on rice seedlings (Fig. 3). Transformant C10 also had a wild-type level of conidiation (Table 3), indicating that the MIG1-GFP fusion fully complemented the defects of the mig1 mutant in conidiation and rice infection. The efficiency of the appressorial penetration of C10 was over five times higher than that of the mig1 mutant but still less than that of the wild type (Table 3). Therefore, transformant C10 appeared to be only partially complemented by Mig1-GFP in the penetration of onion epidermal cells. Fusion with GFP may have a minor, adverse effect on the activation or function of Mig1 during appressorial penetration. It is also possible that ectopic integration of the single copy of the MIG1-GFP fusion construct has a subtle positional effect. However, the Mig1-GFP fusion must be fully functional during rice infection because no obvious difference in virulence was observed between C10 and the wild-type strain in repeated infection assays. In addition, data from the cosegregation analysis supported our conclusion that the deletion of MIG1 was responsible for the phenotype observed in the mig1 mutant.
To date, Rlm1 homologs in filamentous ascomycetes have been functionally characterized only in Aspergillus niger (7) and Aspergillus nidulans (14). In A. niger, the rlmA deletion mutant has a normal growth rate but is more sensitive to sodium dodecyl sulfate, H2O2, and calcofluor white at 30°C (7). Unlike the yeast rlm1 mutant, the rlmA deletion mutant is not hypersensitive to caffeine. The M. grisea mig1 mutant characterized in this study had no increased sensitivities to cell wall-degrading enzymes, caffeine, sodium dodecyl sulfate, or H2O2 at 25°C or 30°C (data not shown). In A. niger, the induced expression of the gfaA glutamine:fructose-6-phosphate amidotransferase and agsA α-1,3-glucan synthase genes in response to cell wall stress is regulated by RlmA. The promoters of these two cell wall synthesis-related genes have putative Rlm1-binding sites. In A. nidulans, the transcription of agsA and agsB is regulated by rlmA, but RlmA is not essential for their induced expression in response to the β-1,3-glucan synthase inhibitor micafungin (14). The gfaA (MGG_11597.5) and agsA (MGG_09639.5) homologs in M. grisea lack putative Rlm1-binding sites [TA(A/T)4TAG] in 1.5-kb regions upstream from their respective start codon. A preliminary analysis indicated that the expression of the agsA but not the gfaA homolog is regulated by Mig1 in M. grisea (S. Ding and J.-R. Xu, unpublished data). Although vegetative hyphae of the mig1 mutant had no obvious defect in cell wall integrity, some of the genes regulated by MIG1 in M. grisea may be required for cell wall modifications in secondary infectious hyphae, which have distinct morphologies and physiological properties.
In yeast, Slt2 regulates multiple downstream transcription factors, including Swi4 and Swi6 (16, 17). These transcription factors form protein complexes and function in different biological processes. Mig1 interacted with Mps1 in yeast two-hybrid assays (Fig. 8), indicating that Mig1 may be one of the downstream transcription factors regulated by the Mps1 pathway for infectious growth. Additional transcription factors, such as homologs of Swi4 and Swi6, may be responsible for the regulation of cell wall integrity by Mps1 in M. grisea. Because the expression of MPS1 in S. cerevisiae suppresses the growth defect of the slt2 mutant (37), we also cloned the MIG1 ORF into pYES2 (Invitrogen) and transformed it into the rlm1 mutant (18). Our preliminary data indicated that MIG1 failed to complement the rlm1 mutant in its sensitivity to caffeine (data not shown). Unlike highly conserved MAP kinase genes such as Mps1 and Slt2, Mig1 and Rlm1 share only limited homology in the MADS box. Other regions required for protein-protein interactions and activation are not well conserved between these two proteins. Interestingly, the A. nidulans rlmA gene can functionally complement the yeast rlm1 mutant (14). In A. nidulans, the Answi4 and Answi6 deletion mutants have no growth defects. In M. grisea, the deletion of putative homologs of swi4 or swi6 resulted in severe defects in growth and plant infection (Y. Peng, personal communication). Therefore, the functions of these transcription factors must be distinct between M. grisea (a sordariomycete) and A. nidulans (a eurotiomycete).
Besides the MADS box, Rlm1 and Mig1 share only limited homology. Two phosphorylation sites of Rlm1 required for its activation by Slt2 have been identified (18). However, these two phosphorylation sites and surrounding amino acid residues are not well conserved between Rlm1 and Mig1. We also failed to identify a putative MAP kinase docking site in Mig1 based on its sequence homology with Rlm1. It will be important to characterize the phosphorylation sites of Mig1 and the activation mechanism of Mig1 by Mps1 or other products of upstream genes during plant infection. In M. grisea, the Mig1-GFP fusion was expressed and localized to nuclei in the infectious hyphae (Fig. 5), indicating that Mig1 may regulate the transcription of a subset of genes during plant infection. Because the mig1 mutant could colonize heat-killed plant cells, some of these genes regulated by MIG1 may play an important role in overcoming the plant defense responses.
Acknowledgments
We thank Larry Dunkle and Burt Bluhm for critical reading of the manuscript, David Levin for providing the yeast rlm1 mutant, and Youling Peng for insightful discussion.
This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2005-35319-16073.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Bourett, T. M., and R. J. Howard. 1990. In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68329-342. [Google Scholar]
- 2.Bruno, K. S., F. Tenjo, L. Li, J. E. Hamer, and J.-R. Xu. 2004. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 31525-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catlett, N. L., B. Lee, O. C. Yoder, and B. G. Turgeon. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 509-11. [Google Scholar]
- 4.Chao, C. C. T., and A. H. Ellingboe. 1991. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 692130-2134. [Google Scholar]
- 5.Choi, W. B., and R. A. Dean. 1997. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 91973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clergeot, P. H., M. Gourgues, J. Cots, F. Laurans, M. P. Latorse, R. Pepin, D. Tharreau, J. L. Notteghem, and M. H. Lebrun. 2001. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 986963-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damveld, R. A., M. Arentshorst, A. Franken, P. A. vanKuyk, F. M. Klis, C. van den Hondel, and A. F. J. Ram. 2005. The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 58305-319. [DOI] [PubMed] [Google Scholar]
- 8.Dean, R. A., N. J. Talbot, D. J. Ebbole, M. Farman, et al. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434980-986. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, J. C., B. J. McCormack, N. Smirnoff, and N. J. Talbot. 1997. Glycerol generates turgor in rice blast. Nature 389244-245. [Google Scholar]
- 10.Deng, F. Y., T. D. Allen, and D. L. Nuss. 2007. Ste12 transcription factor homologue CpST12 is down-regulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 6235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebbole, D. J. 2007. Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45437-456. [DOI] [PubMed] [Google Scholar]
- 12.Ebbole, D. J., Y. Jin, M. Thon, H. Q. Pan, E. Bhattarai, T. Thomas, and R. Dean. 2004. Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: analysis of expressed sequence tags. Mol. Plant-Microbe Interact. 171337-1347. [DOI] [PubMed] [Google Scholar]
- 13.Finley, R. L., and R. Brent. 1995. Interaction trap cloning with yeast, p. 169-203. In B. D. Hames and D. M. Glover (ed.), DNA cloning, a practical approach, vol. 2. Expression systems. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 14.Fujioka, T., O. Mizutani, K. Furukawa, N. Sato, A. Yoshimi, Y. Yamagata, T. Nakajima, and K. Abe. 2007. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 61497-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golemis, E. A., J. Gyuris, and R. Brent. 2002. Interaction trap/two-hybrid system to identify interacting proteins, p. 19.15-19.26. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 16.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 155001-5013. [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409533-538. [DOI] [PubMed] [Google Scholar]
- 18.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46781-789. [DOI] [PubMed] [Google Scholar]
- 19.Kaffarnik, F., P. Muller, M. Leibundgut, R. Kahmann, and M. Feldbrugge. 2003. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 225817-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kankanala, P., K. Czymmek, and B. Valent. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19706-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 133067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S. H., and R. A. Dean. 1997. G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 101075-1086. [DOI] [PubMed] [Google Scholar]
- 23.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 24.Mead, J., A. R. Bruning, M. K. Gill, A. M. Steiner, T. B. Acton, and A. K. Vershon. 2002. Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol. Cell. Biol. 224607-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura, M., G. Park, and J. R. Xu. 2003. The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol. Microbiol. 50231-243. [DOI] [PubMed] [Google Scholar]
- 26.Nolting, N., and S. Pöggeler. 2006. A MADS box protein interacts with a mating-type protein and is required for fruiting body development in the homothallic ascomycete Sordaria macrospora. Eukaryot. Cell 51043-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oehlen, L. J. W. M., J. D. McKinney, and F. R. Cross. 1996. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol. Cell. Biol. 162830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, G., K. S. Bruno, C. J. Staiger, N. J. Talbot, and J. R. Xu. 2004. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 531695-1707. [DOI] [PubMed] [Google Scholar]
- 29.Park, G., C. Xue, X. Zhao, Y. Kim, M. Orbach, and J. R. Xu. 2006. Multiple upstream signals converge on an adaptor protein Mst50 to activate the PMK1 pathway in Magnaporthe grisea. Plant Cell 182822-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, G., G. Y. Xue, L. Zheng, S. Lam, and J. R. Xu. 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 15183-192. [DOI] [PubMed] [Google Scholar]
- 31.Tsuji, G., S. Fujii, S. Tsuge, T. Shiraishi, and Y. Kubo. 2003. The Colletotrichum lagenarium Ste12-like gene CST1 is essential for appressorium penetration. Mol. Plant-Microbe Interact. 16315-325. [DOI] [PubMed] [Google Scholar]
- 32.Urban, M., T. Bhargava, and J. E. Hamer. 1999. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valent, B., L. Farral, and F. G. Chumley. 1991. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 12787-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veneault-Fourrey, C., M. Barooah, M. Egan, G. Wakley, and N. J. Talbot. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312580-583. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 172615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 102696-2706. [DOI] [PubMed] [Google Scholar]
- 37.Xu, J. R., C. J. Staiger, and J. E. Hamer. 1998. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 9512713-12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, J. R., M. Urban, J. A. Sweigard, and J. E. Hamer. 1997. The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant-Microbe Interact. 10187-194. [Google Scholar]
- 39.Xu, J. R., X. Zhao, and R. A. Dean. 2007. From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv. Genet. 57175-218. [DOI] [PubMed] [Google Scholar]
- 40.Xue, C. Y., G. Park, W. B. Choi, L. Zheng, R. A. Dean, and J. R. Xu. 2002. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 142107-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, X., Y. Kim, G. Park, and J.-R. Xu. 2005. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 171317-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, X., R. Mehrabi, and J.-R. Xu. 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 61701-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, X. H., and J. R. Xu. 2007. A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63881-894. [DOI] [PubMed] [Google Scholar]