Abstract
The heat shock transcription factor Hsf1 and the general stress transcription factors Msn2 and Msn4 (Msn2/4) are major regulators of the heat shock response in Saccharomyces cerevisiae. Here, we show that transcriptional activation of their target genes, including HSP104, an antistress chaperone gene, is obligatory for thermotolerance. Although Hsf1 activity might be necessary before the exposure of cells to high temperature, severe heat shock induced the binding of hyperphosphorylated Hsf1 to its target promoters. However, promoter-bound, phosphorylated Hsf1 was inactive for transcription because RNA polymerase II was inactive at high temperatures. Rather, our results suggest that Hsf1 activates the transcription of most of its target genes during the recovery period following severe heat shock. This delayed upregulation by Hsf1, which would be induced by misfolded proteins that accumulate in severely heat-shocked cells, is required for the resumption of normal cell growth. In contrast, the factors Msn2/4 were not involved in the delayed upregulation of genes and were dispensable for cell growth during the recovery period, suggesting that they play a role before the exposure to high temperature. These results show that Hsf1 and Msn2/4 act differentially before and after exposure to extreme temperatures to ensure cell survival and growth.
Expression of a set of proteins known as the heat shock proteins (Hsps) is rapidly induced when cells are challenged with elevated temperatures (48). Many of these proteins function as molecular chaperones in the synthesis, folding, trafficking, maturation, and degradation of proteins (4, 12, 52). Several Hsps are essential for cell growth and are expressed constitutively under physiological conditions; however, they are only slightly induced by heat shock. Others are not essential for growth at normal temperatures but play vital roles in helping cells survive exposure to elevated temperatures; in contrast to the previous mentioned Hsps, however, these Hsps are strongly induced by heat shock (4, 48).
Heat-induced transcription of HSP genes in Saccharomyces cerevisiae is governed by the transcription factors Hsf1, Msn2, and Msn4 (46). The heat shock transcription factor Hsf1, a protein evolutionarily conserved from yeast to humans, is indispensable for cell viability at physiological temperatures (approximately 25 to 30°C), and hsf1 mutations cause a growth defect at moderately elevated temperatures (approximately 33 to 38°C) (46). Hsf1 binds to heat shock elements (HSEs) within the promoter regions of target genes, including HSP genes. Other target genes encode proteins involved in a broad range of biological functions, including detoxification, protein degradation, energy generation, carbohydrate metabolism, and maintenance of cell integrity (7, 17, 51). The transcriptional activity of Hsf1 is induced by oxidative stress, ethanol treatment, and glucose starvation, as well as by heat (15, 19, 26, 44, 50). In yeast cells, Hsf1 binds with low affinity to many target promoters under physiological conditions and with high affinity to these targets upon stress (17, 19, 50). The active form of Hsf1, which is hyperphosphorylated, is involved in HSE architecture-dependent gene transcription (18-20).
A pair of partially redundant transcription factors, Msn2 and Msn4 (Msn2/4), regulate the general stress response induced by heat shock, osmotic shock, oxidative stress, low pH, and glucose starvation (8, 27, 37). The target genes of Msn2/4 partially overlap with those of Hsf1 and encode several Hsps, enzymes involved in carbohydrate metabolism, and proteins involved in protection against oxidative stress (3, 5, 9, 45). The nuclear translocation of Msn2/4 is controlled by the cyclic AMP-dependent protein kinase and the TOR signaling pathway (2, 13), and the binding of Msn2/4 to stress-responsive DNA elements is regulated by the protein kinase GSK3 (21).
Although Hsf1 and Msn2/4 are transcriptional regulators of cytoplasmic and mitochondrial HSP genes, the expression of molecular chaperones in the endoplasmic reticulum (ER) is also activated by the Hac1 transcription factor (29). In the unfolded protein response (UPR), perturbation of the ER folding apparatus with consequent accumulation of unfolded proteins results in the production of active Hac1 protein. In addition to ER-resident chaperones, the UPR regulates the expression of genes involved in the biosynthesis or secretion of secretory organelles (32).
Prior induction of the heat shock response enables cells to survive subsequent exposure to lethal high temperatures. When S. cerevisiae cells are grown at physiological temperatures and then subjected to mild heat shock (37°C), the fraction of the cells that is able to survive exposure to 50°C increases markedly (31, 33). Thermotolerance requires expression of an antistress chaperone Hsp104, which intervenes in cellular protein homeostasis by mediating the renaturation of aggregated proteins (25, 35). Both Hsf1 and Msn2/4 are positive regulators of HSP104 expression (1, 14). The expression of several Hsps, including Hsp104, is robustly induced in cells recovering from severe heat shock. This type of regulation, called delayed upregulation, is suggested to be necessary for the refolding of heat-denatured proteins and thus for survival (39, 40).
In the present study, we analyzed the relationship between thermotolerance and the transcriptional activities of Hsf1 and Msn2/4. Cells with an hsf1 mutation that inhibits heat-induced transcription of Hsf1 target genes were unable to survive short exposures to extreme temperatures, while cells with msn2/4 null mutations had reduced survival rates when exposed for longer time periods. The transcriptional activity of Hsf1, but not that of Msn2/4, was induced in cells recovering from severe heat shock. Hence, it is conceivable that Hsf1 and Msn2/4 differentially function to cope with heat-induced damage.
MATERIALS AND METHODS
Yeast strains and media.
Strain HS170T (MATα ade2 his3 leu2 trp1 ura3 can1 hsf1::HIS3 YCp-TRP1-HSF1), which contains a null mutation of the chromosomal HSF1 gene and bears wild-type HSF1 on a TRP1-containing centromeric plasmid, was used as the wild-type cell (50). Strain YAY21 is a derivative of HS170T harboring YCp-TRP1-hsf1-R206S-H220R instead of YCp-TRP1-HSF1 (49). The hsf1-R206S-H220R mutation contains amino acid substitutions in the DNA-binding domain: arginine to serine at residue 206 and histidine to arginine at residue 220. Cells with null mutations in both MSN2 and MSN4 (strain HS176) were derived from HS170T (50). Cells with a null mutation in ERG6 (strain W303erg6Δ) were kindly provided by T. Inada (22). Cells were grown in rich glucose (YPD) medium as described previously (49, 50). The reporter gene YEp-URA3-HSE4Ptt-CYC1-lacZ, which contains a 4Ptt-type HSE oligonucleotide upstream of the CYC1-lacZ construct, was introduced into strain HS170T, and transformed cells were grown in enriched synthetic glucose (ESD) medium lacking uracil (20). The coding region of HSP104 was cloned downstream of the ADH1 promoter in plasmid pK538 (YCp-URA3-PADH1-TADH1), a derivative of pRS316 containing the ADH1 promoter-terminator fragment (34). The plasmid was introduced into strains HS170T, HS176, and YAY21, and transformed cells were grown in ESD medium lacking uracil.
RNA analysis.
mRNA levels were analyzed by reverse transcription-PCR (RT-PCR) as described previously (18, 20, 50). The amounts of PCR products were compared after normalizing RNA samples to the levels of control ADH1 mRNA (encoding alcohol dehydrogenase). The experiments were performed at least three times with similar results.
Immunoblot analysis.
Cell extracts were prepared and subjected to immunoblot analysis with an anti-Hsf1 serum as described previously (18). The experiments were performed at least twice with similar results.
Chromatin immunoprecipitation analysis.
Cells were treated with 1% formaldehyde for 6 to 10 min at the culture temperature. Glycine was added to reach a final concentration of 175 mM, and incubation was continued for 5 min at 28°C. Cells were collected, and chromatin samples were subjected to immunoprecipitation analysis with an anti-Hsf1 antiserum, as described previously (19, 50). For immunoprecipitation of RNA polymerase II, an anti-Rpb3 antibody (NeoClone) and protein G Sepharose (Sigma) were used. The primers for PCR analysis were described previously (19, 50) except for those for the PHO5 promoter (GGTCCCTGTTTTCGAAGA and GCTTGCTCTATTTGTTGTTG) and those for the telomere on the right arm of chromosome VI (GCACTAGTTGCACTAGGC and GCCGCTTGTTAACTCTCC). The experiments were performed at least twice with similar results.
RESULTS
Transcription of Hsf1 target genes under various heat shock conditions.
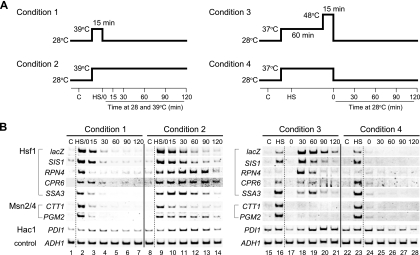
To study the changes in the transcriptional activity of Hsf1, we cultured cells under various heat shock conditions (Fig. 1A) and analyzed the mRNA levels of Hsf1 target genes by RT-PCR. Hsf1-mediated transcription was also evaluated by measuring lacZ mRNA levels of the HSE4Ptt-CYC1-lacZ reporter gene, whose expression is directed by the HSE (20). A temperature shift of the cell culture from 28°C to 39°C induced a rapid accumulation of transcripts of HSE4Ptt-CYC1-lacZ, SIS1, RPN4, CPR6, and SSA3, which are all Hsf1 target genes (Fig. 1B, lanes 2 and 9). A shift back to 28°C resulted in a dramatic decrease in the mRNA levels (condition 1; lanes 3 to 7), indicating the cessation of mRNA synthesis and the degradation of the transcripts. In contrast, mRNA levels remained elevated for at least 30 min when the culture was kept at 39°C (condition 2; lanes 10 to 14). Therefore, transcriptional activation by Hsf1 reflected shifts in the growth temperature of the cultures.
FIG. 1.
Changes in mRNA levels under various heat shock conditions. (A) Schematic representation of the thermal treatments. (B) RT-PCR analysis of various mRNAs. Wild-type cells (strain HS170T) bearing the HSE4Ptt-CYC1-lacZ reporter gene were grown in ESD medium lacking uracil and were heat-shocked as shown in panel A. Control cells (C) were grown at 28°C, and heat-shocked cells (HS) were incubated at 37 or 39°C for 15 min. Total RNA prepared from the cells was subjected to RT-PCR analysis. Transcription factors that activated the mRNA synthesis from each gene are shown to the left of each panel. The ADH1 gene encoding alcohol dehydrogenase was used as a control.
Cells cultured under condition 3 (Fig. 1A) acquired thermotolerance upon mild heat shock at 37°C for 60 min (preconditioning) and were subsequently able to survive at an extreme temperature (48°C) (see below). In severely heat-shocked cells (0 min recovery), the mRNA levels of HSE4Ptt-CYC1-lacZ were reduced to those of the control cells grown at 28°C (Fig. 1B; compare lanes 15 and 17). After a 30- to 60-min recovery period at 28°C, however, the mRNA levels robustly increased to levels that were higher than those of the preconditioned cells, indicating de novo synthesis of mRNA (lanes 18 and 19). Delayed upregulation was also observed for SIS1, RPN4, CPR6, and SSA3, the Hsf1 target genes. Exposure to extreme temperatures is required for delayed upregulation, as shown by the absence of delayed mRNA accumulation under condition 4 (Fig. 1A and B, lanes 24 to 28). In delayed upregulation, therefore, transcriptional activation by Hsf1 does not reflect that of the temperature shift and occurs at normal growth temperatures.
Requirement of Hsf1 for thermotolerance.
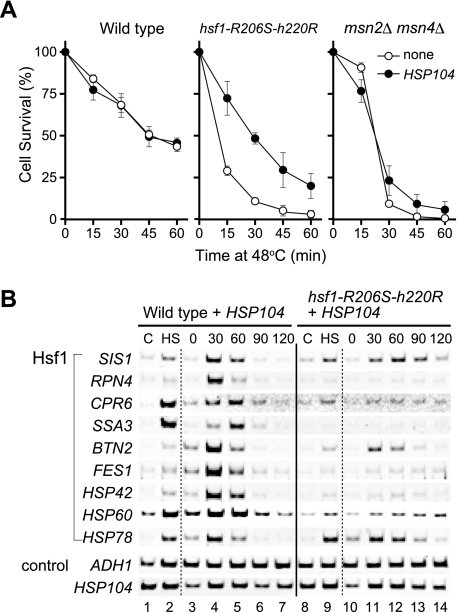
The role of Hsf1 in thermotolerance was analyzed by using hsf1-R206S-H220R cells. R206S and H220R substitutions in the DNA-binding domain of Hsf1 reduce its binding affinity for HSEs in vitro, and cells expressing Hsf1 with these mutations are defective in both heat-induced transcription of various target genes and growth at temperatures above 35°C (49). When hsf1-R206S-H220R cells were preconditioned at 37°C for 1 h and then exposed to 48°C for various periods of time, the cells died at early exposure times (Fig. 2A). The survival rate of the mutant cells was significantly lower than that of wild-type cells at every time point. It has been shown that Hsp104 plays an important role in the induced thermotolerance of S. cerevisiae cells (35). The introduction of a plasmid harboring a constitutively transcribed HSP104 gene did not affect the survival rate of the wild-type cells, suggesting that Hsp104 was adequately expressed in these cells (Fig. 2A). The expression of HSP104 improved the survival of hsf1-R206S-H220R cells but was not enough to restore wild-type thermotolerance. In wild-type cells expressing HSP104, the mRNA levels of various Hsf1 target genes were increased in the preconditioning period and in the recovery period after a 15-min exposure to 48°C (Fig. 2B, lanes 1 to 7). However, transcriptional activation was significantly inhibited by the hsf1-R206S-H220R mutation (lanes 8 to 14). These results show that cell survival requires expression of Hsf1 target genes, including HSP104, prior and/or subsequent to exposure to severe heat shock.
FIG. 2.
Effects of transcription factor mutations on induced thermotolerance. (A) Induced thermotolerance of wild-type and mutant cells. Wild-type, hsf1-R206S-H220R, and msn2Δ msn4Δ cells (open circles) were grown in ESD medium at 28°C. For analysis of cells harboring the ADH1-HSP104 expression plasmid (filled circles), ESD medium lacking uracil was used. Cell cultures were shifted to 37°C for 60 min and then to 48°C. At the indicated times, aliquots of cells were diluted into ESD medium and plated onto YPD medium to determine cell survival rates. Values represent the means and standard deviations of the results of at least three experiments. (B) RT-PCR analysis of various mRNAs. Wild-type HSF1 cells (lanes 1 to 7) and hsf1-R206S-H220R cells (lanes 8 to 14) expressing HSP104 were grown in ESD medium lacking uracil under condition 3. Total RNA prepared from the cells was subjected to RT-PCR analysis. C, control cells; HS, heat-shocked cells.
Role of delayed upregulation in the recovery of severely heat-shocked cells.
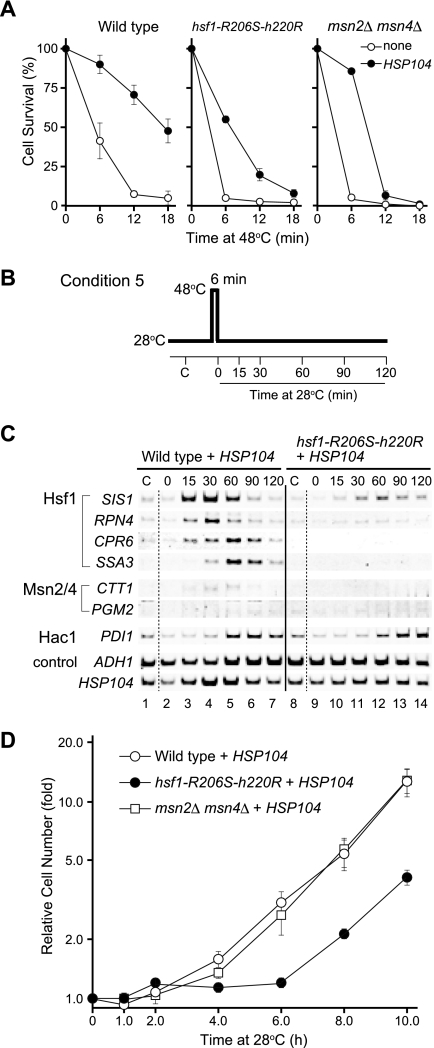
When wild-type cells were directly exposed to 48°C without preconditioning, the survival rate sharply declined (Fig. 3A). As shown previously (25), expression of HSP104 improved basal thermotolerance (Fig. 3A). In order to analyze the transcriptional changes, cells expressing HSP104 were subjected to a 6-min heat shock pulse at 48°C and were then allowed to recover at 28°C for various periods of time (condition 5; Fig. 3B). The mRNA levels of Hsf1 target genes gradually increased during these periods, and their levels peaked after 30 to 60 min of recovery (Fig. 3C, lanes 1 to 7). Essentially the same results were obtained when cells without the HSP104 expression plasmid were subjected to the heat shock pulse (data not shown). The kinetics of transcriptional activation under these conditions were slower than those of the heat shock response, in which mRNA levels peak within 15 min (46). The hsf1-R206S-H220R mutation almost completely inhibited basal thermotolerance, and only half of the mutant cells expressing HSP104 survived a 6-min exposure to 48°C (Fig. 3A). The mRNA levels of Hsf1 target genes in these cells were not significantly increased during the recovery period compared to the mRNA levels of these genes in wild-type cells (Fig. 3C, lanes 8 to 14). Therefore, severe heat shock without preconditioning induces delayed upregulation by Hsf1.
FIG. 3.
Effects of transcription factor mutations on basal thermotolerance. (A) Basal thermotolerance of wild-type and mutant cells. Cell survival was analyzed as described for Fig. 2A except that cells were directly exposed to 48°C without preconditioning. Values represent the means and standard deviations of the results of at least three experiments. (B) Schematic representation of the thermal treatments. (C) RT-PCR analysis of various mRNAs. Wild-type HSF1 cells (lanes 1 to 7) and hsf1-R206S-H220R cells (lanes 8 to 14) expressing HSP104 were grown in ESD medium lacking uracil and were heat-shocked as shown in panel B. Total RNA prepared from the cells was subjected to RT-PCR analysis. C, control cells. (D) Growth of wild-type and mutant cells after severe heat shock. Wild-type (open circles), hsf1-R206S-H220R (filled circles), and msn2Δ msn4Δ (open squares) cells expressing HSP104 were grown in ESD medium lacking uracil at 28°C. Cell cultures were shifted to 48°C for 6 min and then returned to 28°C. At the indicated times, aliquots of cells were diluted into ESD medium and plated onto YPD medium to determine cell growth. Cell numbers were plotted relative to those of the 28°C incubation at 0 min. Values represent the means and standard deviations of the results of at least three experiments.
As delayed upregulation is expected to be an important component of recovery from severe heat shock, cells expressing HSP104 were exposed to 48°C for 6 min and then incubated at 28°C for various periods of time. Changes in the number of viable cells during the recovery period were then determined by plating the cells on solid medium (Fig. 3D). In the case of the wild-type cells, exponential growth resumed after a slight increase in cell number during the first 3 to 4 h of recovery, indicating that cell division was inhibited for approximately 3 h after return of the cells to 28°C. In contrast, hsf1-R206S-H220R cells resumed logarithmic growth only after 6 h. These results show that delayed upregulation by Hsf1 is necessary for the resumption of normal growth after the exposure of the cells to high temperatures.
Requirements of Msn2 and Msn4 for thermotolerance.
We analyzed the role of factors Msn2/4 in cell tolerance to thermal stress. As shown in Fig. 1B, the mRNA levels of CTT1 and PGM2, Msn2/4 target genes, were increased by transient or continuous incubation at 39°C (conditions 1 and 2; lanes 1 to 14). However, Msn2/4-mediated transcription was not significantly upregulated during the recovery period at 28°C after exposure to 48°C under conditions 3 (Fig. 1B, lanes 15 to 21) and 5 (Fig. 3C, lanes 1 to 7). These results show that factors Msn2/4 are not involved in delayed upregulation, although they are potent heat-induced transcription factors.
When cells containing null mutations in both MSN2 and MSN4 were preconditioned at 37°C for 1 h, approximately 90% of the mutant cells survived at 48°C for 15 min (Fig. 2A). However, longer exposures caused a sharp decrease in the survival rate, indicating that factors Msn2/4 are necessary for the induction of thermotolerance. The lower survival rate was slightly improved by the constitutive expression of HSP104. In basal thermotolerance, most msn2Δ msn4Δ cells were unable to survive direct exposure to a 6-min heat shock pulse at 48°C (Fig. 3A). HSP104 expression improved the survival of mutant cells that were exposed for short, but not for long, time periods. Therefore, Hsp104 is essential for survival at early exposure times. Because factors Msn2/4 are not involved in delayed upregulation, constitutive transcription of other Msn2/4 target genes before exposure would also appear to be needed for cell survival at longer exposure times. However, the surviving cells resumed growth as rapidly as the wild-type cells (Fig. 3D), suggesting that Msn2/4 functions are dispensable for the recovery of normal growth.
Activator function of Hsf1 in severely heat-shocked cells.
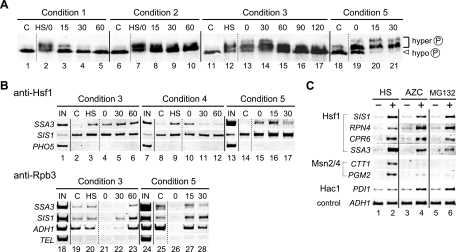
We next analyzed molecular changes in Hsf1 upon heat shock. Treatment of cells with various stresses, including heat shock, induces the hyperphosphorylation of Hsf1, which causes the protein to migrate more slowly than its hypophosphorylated counterpart on denaturing gels (15, 18, 26, 43). As shown by immunoblot analysis with an anti-Hsf1 serum, a slowly migrating form of Hsf1 was observed in extracts of cells that had been transiently or continuously incubated at 39°C (Fig. 4A, lanes 2 and 7 to 10). However, in cells that had been returned to 28°C, Hsf1 migrated at the same position as that of Hsf1 in unstressed cells, indicating temperature-dependent phosphorylation of Hsf1 (lanes 3 to 5) (18, 43). Under condition 3, Hsf1 was continuously hyperphosphorylated during preconditioning at 37°C (lane 12), severe heat treatment at 48°C (lane 13), and recovery at 28°C (lanes 14 to 17). In cells expressing HSP104, a 6-min heat shock pulse at 48°C led to extensive phosphorylation of Hsf1 (lane 19), and the hyperphosphorylation remained stable during early recovery periods (lanes 20 and 21). Therefore, transcriptionally active Hsf1 is hyperphosphorylated, although it is unable to induce transcription in severely heat-shocked cells.
FIG. 4.
Regulation of activator function of Hsf1. (A) Immunoblot analysis of Hsf1. Wild-type cells were grown in YPD medium under conditions 1 (lanes 1 to 5), 2 (lanes 6 to 10), and 3 (lanes 11 to 17). Wild-type cells expressing HSP104 were grown in ESD medium lacking uracil under condition 5 (lanes 18 to 21). Extracts were prepared from the cells and subjected to immunoblot analysis with an anti-Hsf1 serum. The positions of hyper- and hypophosphorylated Hsf1 are shown to the right. C, control cells; HS, heat-shocked cells. (B) Chromatin immunoprecipitation analysis of Hsf1 target promoters. Wild-type cells were grown in YPD medium under conditions 3 (lanes 1 to 6 and 18 to 23) and 4 (lanes 7 to 12). Wild-type cells expressing HSP104 were grown in ESD medium lacking uracil under condition 5 (lanes 13 to 17 and 24 to 28). Chromatin immunoprecipitation analysis was carried out with an anti-Hsf1 serum (lanes 2 to 6, 8 to 12, and 14 to 17) or an anti-Rpb3 antibody (lanes 19 to 23 and 25 to 28). The input (IN) lanes (lanes 1, 7, 13, 18, and 24) show the PCR products amplified from the extracts before immunoprecipitation (1.0% of each sample used for immunoprecipitation). For analysis of the anti-Rpb3 immunoprecipitates, the telomere on the right arm of chromosome VI (TEL) was used as a control. (C) RT-PCR analysis of various genes in cells treated with AZC or MG132. Wild-type cells grown in YPD medium (lanes 1 and 3) were heat shocked at 39°C for 20 min (lane 2) or were treated with 20 mM AZC for 1 h (lane 4). erg6 null mutant cells (strain W303erg6Δ) were grown in ESD medium in the presence of either 50 μM MG132 (lane 6) or 0.1% dimethyl sulfoxide (lane 5) for 30 min. Total RNA prepared from the cells was subjected to RT-PCR analysis.
The interaction of Hsf1 with target promoters was investigated by chromatin immunoprecipitation analysis (Fig. 4B). In extracts of cells grown at 28°C, an anti-Hsf1 serum did not appreciably precipitate the promoter fragment of SSA3 or that of a nontarget gene, PHO5, suggesting that Hsf1 has little binding affinity for the SSA3 promoter in unstressed cells (lanes 2 and 8). However, binding was notably stimulated by mild heat shock (lanes 3 and 9). Elevated binding of Hsf1 to SSA3 was also observed in cells exposed to 48°C for 15 min (lane 4) and in cells that were allowed to recover at 28°C for 30 and 60 min (lanes 5 and 6). Hsf1 binding was reduced when severe heat shock was omitted (lanes 10 to 12). Even without preconditioning (condition 5), a 6-min heat shock pulse at 48°C induced Hsf1 binding (lanes 14 to 17). Similar results were obtained with the SIS1 promoter, but the changes in Hsf1 binding were less noticeable, probably because Hsf1 binds with higher affinity to the SIS1 promoter than to the SSA3 promoter even in unstressed cells (Fig. 4B). Therefore, hyperphosphorylated Hsf1 is active for HSE binding during preconditioning, severe heat treatment, and recovery periods.
The results presented above suggested that the transcriptional defects in Hsf1 target genes in severely heat-shocked cells are due to defects in factors other than Hsf1. Therefore, we analyzed the recruitment of RNA polymerase II to various promoters by using an antibody directed against its Rpb3 subunit. In extracts of cells grown under condition 3, the binding of Rpb3 to the SSA3 and SIS1 promoters was slightly increased by preconditioning (Fig. 4B, lane 20), decreased by exposure to 48°C (lane 21), and induced during the recovery period at 60 and 30 min (lanes 22 and 23). Under condition 5, binding was inhibited in severely heat-shocked cells (lane 26) but was elevated during the recovery period (lanes 27 and 28). The recruitment of RNA polymerase II to promoters correlated with the mRNA levels of the genes. The absence of RNA polymerase II binding in severely heat-shocked cells was also found to be the case with the constitutively transcribed ADH1 promoter (lanes 21 and 26). Therefore, severe heat shock generally inhibits promoter-RNA polymerase II interactions.
Activation of Hsf1 by the accumulation of misfolded proteins.
Transcriptionally active Hsf1 is present in cells recovering from severe heat shock, but not in cells recovering from mild heat shock, suggesting that temperature shifts are not a direct cause of delayed upregulation. It is possible that misfolded proteins accumulating at extreme temperatures induce the transcriptional activity of Hsf1. To test this, cells were treated with the amino acid analog azetidine-2-carboxylic acid (AZC), whose incorporation into proteins in place of proline results in protein misfolding (30, 47). The mRNA levels of SIS1, RPN4, CRP6, and SSA3 were robustly increased by AZC, indicating Hsf1 activation (Fig. 4C, lane 4). In contrast, transcription of CTT1 and PGM2 by factors Msn2/4, which are not subject to delayed upregulation, was not activated by AZC treatment. Inhibition of proteasome function can also lead to the accumulation of misfolded proteins in cells. Cells containing a mutation in ERG6 (a gene involved in ergosterol synthesis) can be permeated by the proteasome inhibitor MG132 (24). Treatment of erg6 mutant cells with MG132 caused the activation of Hsf1 target genes but not of Msn2/4 target genes (Fig. 4C, lane 6). Therefore, only Hsf1 acquires transcriptional activity when misfolded proteins accumulate in cells.
Misfolded proteins in the ER trigger the UPR, in which Hac1 activates transcription of a set of genes, including PDI1 (29). PDI1 mRNA levels were increased by treatment with AZC and MG132 (Fig. 4C, lanes 4 and 6). However, a transient, mild heat shock had little effect on PDI1 mRNA levels (see Fig. 1B, lanes 1 to 7). Importantly, exposure of cells to 48°C, irrespective of preconditioning, caused PDI1 activation during subsequent recovery at 28°C (Fig. 1B, lanes 15 to 21; Fig. 3C, lanes 1 to 7). These results suggest that delayed upregulation by Hsf1 and Hac1 is induced by the accumulation of misfolded proteins.
DISCUSSION
Even though HSP104 is constitutively expressed, mutations in HSF1 and MSN2/4 reduce cell survival at extreme temperatures. Hsf1 activates the transcription of most of its target genes during the recovery period. Delayed upregulation appears to be triggered by misfolded proteins accumulating in severely heat-shocked cells, and this response is essential for the resumption of normal cell growth. In contrast, factors Msn2/4, which are not involved in delayed upregulation, are dispensable for cell survival following short (but not long) exposures to extreme temperatures, as well as for cell growth during the recovery period. These observations demonstrate that the transcriptional activities of Hsf1 and Msn2/4 are differentially regulated under various stress conditions and suggest that their target genes participate in different pathways for cell survival and growth.
The roles of Hsf1 target genes other than HSP104 in thermotolerance remained obscure. In investigations using an hsf1 mutation named hsf1-m3, it has been reported that high-level induction of Hsps is not required for the acquisition of thermotolerance (42). Later analysis showed that Hsp104 expression is sufficient for thermotolerance (25). Our results show that cells containing the hsf1-R206S-H220R mutation are more sensitive to severe heat shock than wild-type cells, even though HSP104 is constitutively expressed, and that target genes other than HSP104 appear to be involved in the thermotolerance mediated by Hsf1. Thermotolerance can be considered to be the result of both increased protection of cellular components at extreme temperatures and increased reactivation and/or degradation of heat-inactivated components (Fig. 5). Genetic analysis has shown that, in the absence of Hsp104, the Hsp70 family is very important for thermotolerance (36). In mitochondria, a set of chaperones is crucial for the maintenance of respiratory competence and mitochondrial DNA synthesis (10, 38). These results suggest that Hsf1-mediated expression of various Hsps is needed to protect proteins against thermal inactivation and aggregation. In addition to HSP genes, Hsf1 regulates the transcription of genes involved in ubiquitination and proteolysis (17, 51). Furthermore, RPN4 encodes a transcriptional activator of genes coding for proteasome subunits, and heat- and AZC-induced transcription of RPN4 is dependent on Hsf1 (16). The genes under the control of Hsf1 and Rpn4 would be involved in the removal of inactivated and aggregated proteins in recovering cells.
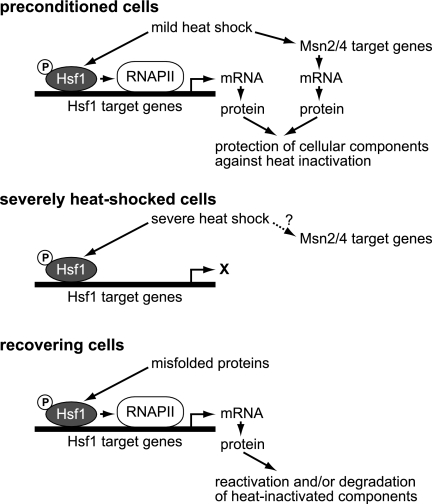
FIG. 5.
Schematic representation of regulation of thermotolerance by Hsf1 and Msn2/4. Cells acquire thermotolerance upon mild heat shock, during which Hsf1 and Msn2/4 induce expression of proteins involved in protection of cellular components against heat inactivation. In severely heat-shocked cells, promoter-bound, hyperphosphorylated Hsf1 is unable to activate transcription because RNA polymerase II (RNAPII) is inactive. It is unknown whether the transcriptional activity of Msn2/4 is activated by severe heat shock. In recovering cells, misfolded proteins appear to activate Hsf1, which induces expression of proteins involved in reactivation and/or degradation of heat-inactivated components. The transcriptional activity of Msn2/4 is not subject to delayed upregulation. More details are described in the text.
Although cells lacking Msn2/4 do not exhibit temperature sensitivity at 38°C, they do exhibit a thermotolerance defect (3, 27). The Msn2/4 function was dispensable during the early phase of exposure to extreme temperatures but was indispensable later on. Because the transcriptional activity of Msn2/4 is not subject to delayed upregulation, it would be required for the expression of proteins which have a protective role before exposure to high temperatures (Fig. 5). The constitutive expression of HSP104 improved basal thermotolerance in msn2Δ msn4Δ cells (Fig. 3A), and the factors Msn2/4 are activators of HSP104 transcription (1, 14), suggesting that one of the roles of Msn2/4 is the transcriptional regulation of HSP104. In addition, Msn2/4 regulates the transcription of genes encoding enzymes for metabolism of trehalose, a disaccharide that plays a pivotal role in thermotolerance (3, 41). This implies that the trehalose content controlled by Msn2/4 affects cell survival at later times during exposure. In this regard, it is interesting that Hsf1 also regulates the transcription of genes encoding trehalose-metabolizing enzymes (51) and that the transcriptional activity of Hsf1 is positively regulated by trehalose (6).
The transcriptional activity of Hsf1 changes in response to stress. Binding of Hsf1 to HSEs was rapidly induced by a shift to elevated temperatures and was reduced by a shift back to normal growth temperatures (Fig. 4B) (11). Hyperphosphorylation of Hsf1 paralleled these temperature shifts (Fig. 4A) (18, 43). The transcriptional activity would also be regulated by conformational changes of Hsf1 and by the cellular level of trehalose (6, 20, 49). The exposure to extreme temperature resulted in the activation of Hsf1 with respect to both its HSE-binding activity and its phosphorylation status. However, activated Hsf1 failed to induce transcription because of heat-inactivation of RNA polymerase II. In cells recovering from severe heat shock, Hsf1 was hyperphosphorylated, bound to the HSE, and robustly induced the transcription of its target genes. The hyperphosphorylation of Hsf1 was positively regulated by its C-terminal basic domain, which is required for the heat-, ethanol-, and oxidative stress-induced phosphorylation of Hsf1 (18-20, 50) (data not shown). Exposure to extreme temperatures results in protein misfolding. The transcriptional activities of Hsf1 and Hac1, but not of Msn2/4, were induced under conditions that resulted in the accumulation of misfolded proteins, suggesting that misfolded proteins in recovering cells trigger delayed upregulation by Hsf1 and Hac1.
It has been shown that translocation of proteins to the ER is blocked by severe heat shock but that translocation resumes upon the synthesis of the ER Hsp70 protein Kar2 in recovering cells (39). Expression of KAR2 and ERO1, encoding ER oxidoreductin, is regulated by both Hac1 and Hsf1 (28, 44), suggesting that the delayed upregulation of several ER proteins by these transcription factors is necessary for recovery. The activity of mitogen-activated protein kinase Mpk1, which is involved in regulating cell wall integrity and progression through the cell cycle, is also implicated in thermotolerance (23). We analyzed the mRNA levels of PST1 and SED1, two Mpk1-controlled genes, and found that their transcription was slightly induced during the recovery period (data not shown). Our study findings suggest that thermotolerance results from a precise balance in the amounts and activities of various cellular components not only just prior to but also after severe heat shock. More studies will be required to further elucidate this issue.
Acknowledgments
We thank Toshifumi Inada for providing the yeast strain.
The work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.S.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Amorós, M., and F. Estruch. 2001. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 391523-1532. [DOI] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402689-692. [DOI] [PubMed] [Google Scholar]
- 3.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33274-283. [DOI] [PubMed] [Google Scholar]
- 4.Burnie, J. P., T. L. Carter, S. J. Hodgetts, and R. C. Matthews. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 3053-88. [DOI] [PubMed] [Google Scholar]
- 5.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlin, L. K., and H. C. Nelson. 2007. The natural osmolyte trehalose is a positive regulator of the heat-induced activity of yeast heat shock transcription factor. Mol. Cell. Biol. 271505-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eastmond, D. L., and H. C. Nelson. 2006. Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J. Biol. Chem. 28132909-32921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24469-486. [DOI] [PubMed] [Google Scholar]
- 9.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germaniuk, A., K. Liberek, and J. Marszalek. 2002. A bichaperone (Hsp70-Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J. Biol. Chem. 27727801-27808. [DOI] [PubMed] [Google Scholar]
- 11.Giardina, C., and J. T. Lis. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 152737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg, A. L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426895-899. [DOI] [PubMed] [Google Scholar]
- 13.Görner, W., E. Durchschlag, M. T. Martínez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grably, M. R., A. Stanhill, O. Tell, and D. Engelberg. 2002. HSF and Msn2/4p can exclusively or cooperatively activate the yeast HSP104 gene. Mol. Microbiol. 4421-35. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, J. S., and D. J. Thiele. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 2795169-5176. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, J. S., D. W. Neef, and D. J. Thiele. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60240-245. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 245249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashikawa, N., and H. Sakurai. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 243648-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashikawa, N., N. Yamamoto, and H. Sakurai. 2007. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J. Biol. Chem. 28210333-10340. [DOI] [PubMed] [Google Scholar]
- 20.Hashikawa, N., Y. Mizukami, H. Imazu, and H. Sakurai. 2006. Mutated yeast heat shock transcription factor activates transcription independently of hyperphosphorylation. J. Biol. Chem. 2813936-3942. [DOI] [PubMed] [Google Scholar]
- 21.Hirata, Y., T. Andoh, T. Asahara, and A. Kikuchi. 2003. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 14302-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito-Harashima, S., K. Kuroha, T. Tatematsu, and T. Inada. 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 91559-1571. [DOI] [PubMed] [Google Scholar]
- 24.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1830-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindquist, S., and G. Kim. 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. USA 935301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, X. D., and D. J. Thiele. 1996. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 10592-603. [DOI] [PubMed] [Google Scholar]
- 27.Martínez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 152227-2235. [PMC free article] [PubMed] [Google Scholar]
- 28.Mori, K., A. Sant, K. Kohno, K. Normington, M. J. Gething, and J. F. Sambrook. 1992. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 112583-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, K., N. Ogawa, T. Kawahara, H. Yanagi, and T. Yura. 1998. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 2739912-9920. [DOI] [PubMed] [Google Scholar]
- 30.Nomura, M., S. Nakamori, and H. Takagi. 2003. Characterization of novel acetyltransferases found in budding and fission yeasts that detoxify a proline analogue, azetidine-2-carboxylic acid. J. Biochem. (Tokyo) 13367-74. [DOI] [PubMed] [Google Scholar]
- 31.Parsell, D. A., and S. Lindquist. 1993. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27437-496. [DOI] [PubMed] [Google Scholar]
- 32.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13349-356. [DOI] [PubMed] [Google Scholar]
- 33.Piper, P. W. 1993. Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 11339-355. [DOI] [PubMed] [Google Scholar]
- 34.Sakurai, H., and Y. Takemori. 2007. Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: binding specificities of yeast HSFs and human HSF1. J. Biol. Chem. 28213334-13341. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez, Y., and S. L. Lindquist. 1990. HSP104 required for induced thermotolerance. Science 2481112-1115. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, Y., D. A. Parsell, J. Taulien, J. L. Vogel, E. A. Craig, and S. Lindquist. 1993. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J. Bacteriol. 1756484-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 935777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt, M., W. Neupert, and T. Langer. 1996. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell Biol. 1341375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seppä, L., and M. Makarow. 2005. Regulation and recovery of functions of Saccharomyces cerevisiae chaperone BiP/Kar2p after thermal insult. Eukaryot. Cell 42008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seppä, L., A. L. Hanninen, and M. Makarow. 2004. Upregulation of the Hsp104 chaperone at physiological temperature during recovery from thermal insult. Mol. Microbiol. 52217-225. [DOI] [PubMed] [Google Scholar]
- 41.Singer, M. A., and S. Lindquist. 1998. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 16460-468. [DOI] [PubMed] [Google Scholar]
- 42.Smith, B. J., and M. P. Yaffe. 1991. Uncoupling thermotolerance from the induction of heat shock proteins. Proc. Natl. Acad. Sci. USA 8811091-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorger, P. K., and H. R. Pelham. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54855-864. [DOI] [PubMed] [Google Scholar]
- 44.Takemori, Y., A. Sakaguchi, S. Matsuda, Y. Mizukami, and H. Sakurai. 2005. Stress-induced transcription of the endoplasmic reticulum oxidoreductin gene ERO1 in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 27589-96. [DOI] [PubMed] [Google Scholar]
- 45.Treger, J. M., A. P. Schmitt, J. R. Simon, and K. McEntee. 1998. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem. 27326875-26879. [DOI] [PubMed] [Google Scholar]
- 46.Trott, A., and K. A. Morano. 2003. The yeast response to heat shock, p. 71-119. In S. Hohmann and P. W. H. Mager (ed.), Yeast stress responses. Springer-Verlag, Heidelberg, Germany.
- 47.Trotter, E. W., C. M. Kao, L. Berenfeld, D. Botstein, G. A. Petsko, and J. V. Gray. 2002. Misfolded proteins are components to mediate a subset of the response to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 27744817-44825. [DOI] [PubMed] [Google Scholar]
- 48.Westerheide, S. D., and R. I. Morimoto. 2005. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 28033097-33100. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, A., and H. Sakurai. 2006. The DNA-binding domain of yeast Hsf1 regulates both DNA-binding and transcriptional activities. Biochem. Biophys. Res. Commun. 3461324-1329. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, A., J. Ueda, N. Yamamoto, N. Hashikawa, and H. Sakurai. 2007. Role of heat shock transcription factor in Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 61373-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 28011911-11919. [DOI] [PubMed] [Google Scholar]
- 52.Young, J. C., V. R. Agashe, K. Siegers, and F. U. Hartl. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5781-791. [DOI] [PubMed] [Google Scholar]