Abstract
The linear homopolymer poly-β-1,6-N-acetyl-d-glucosamine (β-1,6-GlcNAc; PGA) serves as an adhesin for the maintenance of biofilm structural stability in diverse eubacteria. Its function in Escherichia coli K-12 requires the gene products of the pgaABCD operon, all of which are necessary for biofilm formation. PgaC is an apparent glycosyltransferase that is required for PGA synthesis. Using a monoclonal antibody directed against E. coli PGA, we now demonstrate that PgaD is also needed for PGA formation. The deletion of genes for the predicted outer membrane proteins PgaA and PgaB did not prevent PGA synthesis but did block its export, as shown by the results of immunoelectron microscopy (IEM) and antibody adsorption assays. IEM also revealed a conditional localization of PGA at the cell poles, the initial attachment site for biofilm formation. PgaA contains a predicted β-barrel porin and a superhelical domain containing tetratricopeptide repeats, which may mediate protein-protein interactions, implying that it forms the outer membrane secretin for PGA. PgaB contains predicted carbohydrate binding and polysaccharide N-deacetylase domains. The overexpression of pgaB increased the primary amine content (glucosamine) of PGA. Site-directed mutations targeting the N-deacetylase catalytic activity of PgaB blocked PGA export and biofilm formation, implying that N-deacetylation promotes PGA export through the PgaA porin. The results of previous studies indicated that N-deacetylation of β-1,6-GlcNAc in Staphylococcus epidermidis by the PgaB homolog, IcaB, anchors it to the cell surface. The deletion of icaB resulted in release of β-1,6-GlcNAc into the growth medium. Thus, covalent modification of β-1,6-GlcNAc by N-deacetylation serves distinct biological functions in gram-negative and gram-positive species, dictated by cell envelope differences.
Bacterial biofilms are multicellular communities that are formed in many natural habitats and provide protection against hostile conditions, including immunological defenses (10, 13). A distinguishing feature of biofilms is that cells are entrapped within an extracellular polymeric matrix (6, 44). While this matrix may contain a variety of components that affect its architecture, ion selectivity, and other properties, the structural integrity of biofilm often depends upon polysaccharides that are known or predicted to be neutral or basic in charge, e.g., Pel and Psl of Pseudomonas aeruginosa, cellulose, or poly-β-1,6-N-acetyl-d-glucosamine (poly-β-1,6-GlcNAc; PGA) (see references 16, 32, 50, and 52).
Poly-β-1,6-GlcNAc is a homopolymer that was originally described from Staphylococcus epidermidis and is referred to as polysaccharide intracellular adhesin or PIA (33). Staphylococcus aureus was reported to produce a similar succinylated polymer, poly-N-succinyl-β-1,6-glucosamine (34), although the succinyl moieties were later determined to be artifacts of purification (29). Poly-β-1,6-GlcNAc was subsequently isolated from Escherichia coli and shown to depend on the pgaABCD operon for its production (50). This operon is present in diverse bacterial species and appears to be part of a horizontally transferred locus (50). Other gram-negative species produce this polysaccharide, based on the presence of a pgaABCD-homologous locus in the genome, isolation and nuclear magnetic resonance analysis of the polymer, immunoreactivity of cell extracts with α-staphylococcal PIA, and/or the sensitivity of their biofilms to dispersin B, an enzyme that specifically hydrolyzes the β-1,6-glycosidic linkages of PGA (see references 23, 24, and 36). Besides mediating cell-to-cell and cell-to-surface adhesion in biofilms, poly-β-1,6-GlcNAc is essential for the formation of the nonrandom or periodic cellular architecture of E. coli biofilm microstructure and for conversion from temporary polar cell surface attachment to permanent lateral attachment during the initial stages of biofilm development (1, 2). Poly-β-1,6-GlcNAc has profound effects on host-microbe interactions. It apparently promotes the transmission of the plague bacillus Yersinia pestis from the flea vector to the mammalian host (22, 26) and affects colonization, virulence, and immune evasion in infections caused by gram-positive and gram-negative species (see references 4, 8, 9, 24, 41, and 48).
The synthesis of poly-β-1,6-GlcNAc requires the pgaABCD and icaADBC operons of E. coli and S. epidermidis, respectively (20, 50). PgaC and IcaA are homologous cytoplasmic membrane proteins of glycosyltransferase family 2 (GT-2) that are necessary for polymerization. In S. epidermidis bacteria, optimal poly-β-1,6-GlcNAc production also requires the IcaC and IcaD proteins (18), which are not related in sequence to the E. coli Pga proteins. PgaB and IcaB contain polysaccharide N-deacetylase domains belonging to carbohydrate esterase family 4. IcaB of S. epidermidis is a secreted, cell wall-associated protein that introduces the 15 to 20% deacetylated (glucosamine) residues found in the mature polymer (47). PgaB of E. coli is predicted to be an outer membrane lipoprotein (50), and its ortholog in Yersinia pestis, HmsF, copurifies with the outer membrane fraction of this bacterium (37). PgaB and its orthologs in gram-negative species are larger than IcaB and possess an additional domain of unknown function, which is predicted herein to be a PGA-binding domain. PgaA was not found by BLAST analyses to be related to any protein of defined function (50). It is predicted herein to form an N-terminal superhelical domain that is related to mitochondrial α-importin, human O-linked GlcNAc transferase and other eukaryotic transport proteins, and a C-terminal β-barrel porin for PGA secretion.
A csrA mutant of E. coli overproduces PGA and displays a dramatic increase in biofilm formation (25, 49). The CsrA protein posttranscriptionally represses PGA production by binding to six sites within the untranslated leader and proximal coding region of pgaA mRNA (49). This blocks ribosome access to the pgaA Shine-Dalgarno sequence and destabilizes the pgaABCD transcript. PGA synthesis in E. coli bacteria requires the LysR family DNA-binding protein NhaR, which activates pgaABCD transcription in response to high pH and high concentrations of sodium ions (19).
We have used molecular genetic approaches to show that PgaC and PgaD are necessary for the biosynthesis of PGA, while PgaB is an N-deacetylase whose catalytic activity, along with PgaA, is necessary for PGA export from the periplasm. Homologous proteins of other gram-negative bacteria are proposed to perform these same functions. A model for PGA synthesis and export by these proteins is presented.
MATERIALS AND METHODS
Bacterial strains and media.
All E. coli strains and plasmids used in the present study are listed in Table 1. For all experiments, cultures were started from a 1:100 dilution of an overnight culture and grown in Luria-Bertani medium (LB) (pH 7.4) (1% tryptone, 0.5% yeast extract, 1% NaCl), supplemented as required with the following antibiotics: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain and/or plasmid | Relevant description or genotype | Source or reference |
|---|---|---|
| E. coli K-12 strains and plasmids | ||
| DH5α | supE44 ΔlacU169 (φ80lacZ ΔM15) hsdR17 relA1 recA1 endA1 gyrA96 thi-1 | Laboratory strain |
| MG1655 | F− λ− | Michael Cashel |
| TRMG | MG1655 csrA::kan | 38 |
| TRMG (pPGA372) | cpsE::Tn10; overproduces PGA | 50 |
| XWMGΔC | MG1655 ΔpgaC | 50 |
| TRXWMGΔA | TRMG ΔpgaA | 50 |
| TRXWMGΔB | TRMG ΔpgaB | 50 |
| TRXWMGΔC | TRMG ΔpgaC | 50 |
| TRXWD146 | TRMG pgaD146::cam | 50 |
| pUC19 | Cloning vector, Apr | 40 |
| pCR 2.1-TOPO | Cloning vector, Apr and Kmr | Invitrogen |
| pCRpgaB | pgaB in pCR2.1-TOPO | This study |
| pD115A | pgaB D115A, from pCRPgaB | This study |
| pH184A | pgaB H184A, from pCRPgaB | This study |
| pCRPgaB 271 | pgaB truncation, from pCRPgaB | This study |
| pCRPgaB 410 | Same | This study |
| pCRPgaB 516 | Same | This study |
| Y. pestis strains | ||
| KIM6+ | Hms+ | Joe Hinnebusch |
| KIM6 | Hms− | Joe Hinnebusch |
Preparation of PGA.
For dot blot analysis, cell-associated PGA was prepared as described previously (19). Cultures were incubated for 24 h at 26°C or 37°C with shaking at 250 rpm or standing, harvested (5 ml) by centrifugation, and resuspended in 400 μl of a solution containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 1 μg lysozyme. This reaction mixture was incubated at room temperature for 30 min, and a solution (300 μl) containing 10 μg DNase I, 40 μg RNase, 200 μg α-amylase, and 40 mM MgCl2 was added. The mixture was incubated at room temperature for 1 h with occasional mixing before being heated to 37°C for 2 h. The resulting cell lysate was extracted once with 50 mM Tris-HCl (pH 8.0)-saturated phenol and once with chloroform. The aqueous phase (1 ml) was collected, and residual chloroform was allowed to evaporate overnight at room temperature. PGA in the spent LB medium was collected, concentrated using a YM-3 membrane (3,000 Da cutoff; Millipore, Billerica, MA), and assayed directly by immunoblotting.
For ninhydrin determination of free amino groups, PGA was prepared from batch cultures and purified by fast protein liquid chromatography using a previously described approach, which yields a highly purified polymer fraction (50). The strains for these assays were grown for 24 h at 26°C with shaking at 250 rpm. The purified polysaccharide fractions were acid hydrolyzed and assayed for their hexosamine content with 3-methyl-2-benzothiazolone hydrazone hydrochloride (42), and the neutral sugar content was determined by using a phenol-sulfuric acid assay (14). N-Acetyl-d-glucosamine and -d-glucose, respectively, were used as the standards for these assays.
MAb production.
Mouse immunizations, fusions, cloning line development, and antibody purification were performed in the ICBR Hybridoma Laboratory at the University of Florida. Eight-week-old female BALB/c mice were injected (two 50-μl subcutaneous and one 100-μl intraperitoneal injections per mouse) with 100 μg PGA (lyophilized and suspended in 0.2 ml phosphate-buffered saline (PBS; 10 mM sodium phosphate buffer, 0.15 mM NaCl, pH 7.2) containing RIBI adjuvant (RIBI Immunochem Research, Inc.) and were given a boost injection as described above at 2 weeks. Tail vein bleeds were obtained 12 days after the boost and screened by enzyme-linked immunosorbent assay (ELISA) for response against PGA (described below). Mice showing the greatest response received two further injections 2 weeks apart with 200 μg PGA in PBS plus adjuvant (injection site distribution as given above). Test bleeds were examined 2 weeks afterwards, and the mouse exhibiting the greatest titer was given a final intraperitoneal injection boost (without adjuvant) at 3 weeks after the test bleed. Four days after the final boost, mouse spleen cells were fused with the mouse myeloma cell line HP 4 (SP2/0) (University of Florida Hybridoma Laboratory deposit line) in Dulbecco's modified Eagle medium (DMEM) (Gibco) containing 25% HL 4 (SP2/0) conditioning medium and 20% horse serum (Atlanta Biologicals; designated DME-PC medium) plus 1× hypoxanthine-aminopterin-thymidine (HAT) medium supplement (Sigma-Aldrich). The HL 4 (SP2/0) conditioning medium component refers to the supernatant obtained from high-log-phase cultures of HL 4 (SP2/0) in DMEM plus 10% horse serum. These and subsequent cultures were incubated at 37°C in a 7% CO2 growth chamber. Positive samples from the ELISA screening of supernatants from 96-well-plate mass cultures were grown in DME-PC plus 1× HT medium supplement (Sigma-Aldrich) in 24-well plates and screened by ELISA. Finally, the lines with the strongest responses were cultured in six-well plates in DME-PC plus 1/2× HT medium supplement without conditioning medium. ELISA screening yielded the fusion line designated 2F3.MC. Expansive cultures of this line in DME-PC medium and ELISA screening of 96-well, then 24-well, and, finally, 6-well plates resulted in the clone HL 1993. ELISA screening throughout the process indicated that the antibody produced was of the immunoglobulin M (IgM) class. Monoclonal antibody (MAb) 2F3.1D4 was isolated from the filtered supernatants from six 10-ml cultures of 2F3.1D4 grown in DME-PC medium in CELLine CL 35 bioreactors (Wilson Wolf Manufacturing Corp.) by passage through a 25-ml mouse IgM affinity isolation column (American Qualex Antibodies). Following the removal of unbound material with 0.01 M PBS plus 0.02% NaN3, antibody was eluted with 0.1 M glycine (pH 2.8). The peak fractions, as determined by absorbance (A280), were pooled, desalted, and concentrated in 0.1 M PBS plus 0.02% NaN3 by centrifugation in a 10,000 NMWL Amicon Ultra-15 device (Millipore). The final MAb yield (approximately 22 mg, 20 mg ml−1) was estimated by measuring absorbance at 280 nm using the extinction coefficient 1.2 ml·mg−1·cm−1. The antibody exhibited a strong binding preference for the β-1,6-N-acetyl-glucosamine linkages of PGA compared to its binding to the β-1,4-N-acetyl-glucosamine linkages of fetuin (unpublished data).
ELISA screening.
Mouse bleeds (serially diluted up to 10,000 times in 0.01 M PBS plus 0.2% Tween 20 [PBST]), fusion lines (undiluted), clonal cell lines (serially diluted up to 1,000 times), and MAb (diluted to yield 0 to 10 μg/ml in PBST) were screened as primary antibodies (1°Ab) in 96-well Immulon 2 plates (Dynatech Laboratories, Inc.) for reactivity against PGA. Lyophilized PGA containing 200 μg glucosamine equivalents was solubilized with 5 N HCl (40 μl/cryovial), vortexed intermittently, neutralized after 1 h with 5 N NaOH (40 μl), and diluted to 10 μg/ml with coating buffer (CB) containing 15 mM Na2CO3 plus 33 mM NaHCO3 plus 0.02% NaN3. Diluted PGA or CB (100 μl) was presented to the wells, and the plates incubated at 4°C overnight. The remaining steps of the procedure were conducted at room temperature. The wells were aspirated, rinsed four times with 125 μl PBST, and blocked with 5% bovine serum albumin in PBST for 1 h. The wells were aspirated and rinsed as described above, and either 100 μl PBST or 1°Ab was added to triplicate wells containing CB (blanks) or PGA and incubated for 2 h. Normal mouse serum, fusion medium, or cloning medium served as the negative control for the appropriate screening experiment. Following aspiration and rinsing as described above, goat anti-mouse IgM (μ-chain specific)-alkaline phosphatase (AP) conjugate (Sigma-Aldrich) was diluted 1,000 times with PBST, 75 μl was added to all wells, and plates were incubated for 2 h. Goat anti-mouse IgG (γ-chain specific)-AP conjugate (Sigma-Aldrich) and goat anti-mouse IgG (whole molecule)-AP conjugate (Sigma-Aldrich) were also used to survey for IgG production and showed little response to the 1°Ab in all cases. Following aspiration and rinsing as described above, 75 μl of substrate (10 mg p-nitrophenyl phosphate per 10 ml of 50 mM NaHCO3 plus 50 mM Na2CO3 plus 0.5 mM MgCl2·6H2O) was dispensed in all wells and the absorbance at 405 nm determined over time with a model 680 microplate reader (Bio-Rad). The absorbance values were corrected for background to yield the net A405 values.
Western blotting of PgaB.
Cultures for Western blotting of PgaB proteins were grown at 26°C for 24 h with shaking, and 2.5 ml of cells was concentrated and resuspended in lysis buffer (90 mM Tris-HCl, 2% sodium dodecyl sulfate [SDS], pH 6.8). Samples were boiled for 5 min, cell debris was removed by centrifugation, and the supernatant solutions were assayed for total cell protein by using the bicinchoninic acid method with bovine serum albumin as the standard protein (Pierce) and saved at −80°C. A 96-well microplate reader was used to measure A562.
Four microliters of SDS-polyacrylamide gel electrophoresis loading buffer (135 mM Tris-HCl, 3% SDS, 0.03% bromphenol blue, 30% glycerol, pH 6.8) was added to 50 μg of total protein (∼8 μl), and proteins were separated by SDS-polyacrylamide gel electrophoresis (7.5% or 12.5%). Gels were equilibrated in 25 mM Tris-192 mM glycine plus 20% methanol (vol/vol) at pH 8.3 and electroblotted overnight to a nitrocellulose membrane (0.2 μm). Blots were blocked with PBS containing 0.05% Tween 20 (PBS-T05) and 5% nonfat dry milk and probed with rabbit polyclonal antiserum against HmsF of Yersinia pestis (from R. Perry) diluted 1:5,000 in PBS-T05 and 1% bovine serum albumin (PBS-TB). The blot was rinsed with PBS-T05 and treated with goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich) diluted 1:10,000 in PBS-TB. Signals were developed with Western Lightning chemiluminescent substrate (PerkinElmer) according to the manufacturer's recommendations.
Chemiluminescence was detected by using a Bio-Rad ChemiDoc system, with MagicMark XP polypeptides (Invitrogen) as molecular weight standards. The molecular weights of PgaB (HmsF) proteins were estimated by using the QuantityOne software package.
Immunoblotting.
For cell-bound PGA, 2 μl of sample, corresponding to 1.4 μl, 14 μl, 140 μl, or 700 μl of a 24-h culture, was applied to a nitrocellulose membrane. For PGA in spent medium, 2 μl of sample, corresponding to 2 μl, 20 μl, or 100 μl of spent medium, was applied directly onto a nitrocellulose membrane. After application of the sample, the membrane was allowed to air dry overnight at room temperature. It was blocked for 1 h in PBS-T05 plus 5% dry skim milk and treated for 1 h at room temperature with the murine IgM MAb 2F3.1D4 (diluted 1:10,000 in PBS-TB) that was raised against E. coli PGA. The membrane was washed twice for 5 min and twice for 10 min with PBS-T05, and treated with horseradish peroxidase-conjugated anti-murine Ig antibody (1:10,000; Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature. The membrane was then washed, and the signal was detected by chemiluminescence (Western Lightning Chemiluminescence Plus protocol; PerkinElmer). Membranes were photographed using a Bio-Rad ChemiDoc system. For each immunoblotting analysis, the complete experiment was conducted at least twice, with essentially identical results.
Immunoelectron microscopy (IEM). (i) Processing and embedding.
Cultures (1 ml) were grown with shaking at 250 rpm or standing for 24 h at 26°C and harvested, and cells were washed twice with 1 ml of 20 mM sodium PBS, pH 7.4 (PBS20). Fixation was performed by the method of Li et al. (31). Samples were fixed for 2 h at 4°C with a solution containing 2% paraformaldehyde and 0.1% glutaraldehyde in 100 mM HEPES (pH 6.8). Samples were washed twice with HEPES buffer and once with distilled water, incubated in 2% (wt/vol) aqueous uranyl acetate for 45 min at 4°C, dehydrated successively in 70, 80, and 90% ethanol for 15 min each at 4°C, and treated twice with 100% ethanol for 15 min at 4°C. After infiltration with 50% (vol/vol) LR White (Electron Microscopy Sciences) and ethanol for 4 h, 100% LR White overnight, and 100% LR White for 4 h, samples were embedded in 100% LR White and polymerized at 50°C for 24 h. The samples were sectioned (70 to 80 nm) by using a diamond knife and an RMC MT-7000 ultramicrotome, and the sections were collected onto Formvar-coated nickel grids.
(ii) Immunogold labeling.
The sample grids were blocked with a solution containing 0.5% glycine and 1% bovine serum albumin in PBS20 for 30 min, incubated for 30 min with goat serum (1:20), and incubated with the anti-PGA murine MAb (1:200) for 1 h. The grids were washed five times with distilled water and incubated for 1 h with 10-nm-diameter colloidal gold-conjugated secondary antibody (1:20; Ted Pella, Inc.), washed five times with distilled water, and poststained with 5% aqueous uranyl acetate for 30 s to 1 min. The specimens were examined with a JEOL JEM-1210 transmission electron microscope at 80 kV with the anticontamination device in place.
Antibody adsorption assay.
Standing cultures (24 ml) were incubated for 24 h at 26°C, harvested by centrifugation, and resuspended in 200 μ1 of LB medium containing 20 mM MgCl2. The adsorption assay mixture (40 μ1) contained cells, 1:200-diluted anti-PGA murine MAb, LB medium, and 20 mM MgCl2. The mixture was incubated for 2 h at room temperature and centrifuged, and the supernatant solution was collected. Antibody remaining in the supernatant solution was detected by applying it to a nitrocellulose membrane and processing the membrane as described above for immunoblotting. The complete experiment was conducted twice, with essentially identical results.
Cloning and mutagenesis of pgaB.
The wild-type E. coli K-12 pgaB gene was PCR amplified from chromosomal DNA using KOD DNA polymerase (Novagen, San Diego, CA) and the oligonucleotide primers PgaB-SD FWD and PgaB REV (Table 2) under reaction conditions described by the manufacturer. The resulting PCR product was cloned into the HincII site of pUC19 (40). The pgaB-containing BamHI-XbaI fragment was excised and subcloned into the corresponding restriction sites of pCR2.1-TOPO (Invitrogen, Carlsbad, CA) to produce pCRpgaB. The resulting plasmid, pCRpgaB, was found to complement the ΔpgaB biofilm phenotype. Site-directed mutagenesis of pgaB was performed using pCRpgaB and PfuUltra high-fidelity DNA polymerase (Stratagene) for PCR. The primers for mutagenesis are listed in Table 2. Site-directed mutant BamHI-SacII fragments (888 bp) were excised and individually subcloned into the corresponding restriction sites of pCRpgaB to produce pD115A and pH184A. The pgaB truncation mutants were constructed by preparing BamHI-XhoI PCR fragments, which were cloned into pCR2.1-TOPO. The pgaB constructs of this study were analyzed by sequencing (SeqWright DNA Technology Service, Houston, TX) to confirm that they contained no unwanted mutations.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer name | Sequence 5′ to 3′ | Comments |
|---|---|---|
| PgaB-SD FWD | GGATCCATAGTGGAGTAATACAGGATGTTACGTAATGGAAATAAATATCTCC | Contains pgaB SD, BamHI for cloning |
| PgaB REV | TCTAGATTAATCATTTTTCGGATACCAGGC | Contains pgaB stop codon, XbaI for cloning |
| D115A FWD | GTAGTGCTGACTTTTGATGCCGGCTACCAGAGTTTTTATAC | For site-directed mutagenesis |
| D115A REV | GTATAAAAACTCTGGTAGCCGGCATCAAAAGTCAGCACTAC | For site-directed mutagenesis |
| H184A FWD | CTCGTTGAGCTCGCTTCTGCTACATGGAATTCTCACTACGGTATT | For site-directed mutagenesis |
| H184A REV | AATACCGTAGTGAGAATTCCATGTAGCAGAAGCGAGCTCAACGAG | For site-directed mutagenesis |
| PgaB-BamHI-SD-FW-2 | GGATCCATAGTGGAGTAATACAGG | Contains pgaB SD, BamHI for cloning |
| PgaB 271 | TATTTCTAGATTAGAACATGTCATAACCGAGTTTTTTTAAT | Contains stop codon, XbaI for pgaB deletions |
| PgaB 410 | TATTTCTAGATTAGGGATCTAAATCCCAG | Contains stop codon, XbaI for pgaB deletions |
| CTTAATACC | ||
| PgaB 516 | TATTTCTAGATTAGTCAGTTAACGCACGA | Contains stop codon, XbaI for pgaB deletions |
| CTTTTAAAG |
All primers were purchased from Integrated DNA Technologies, Inc., Coralville, IA. Restriction sites are underlined. SD, Shine-Dalgarno.
Biofilm assay.
The amount of biofilm formation in microtiter plates was determined by crystal violet staining of adherent cells, as described previously (50). At least six replicates were conducted for each sample, and each experiment was performed at least twice. The results were calculated as the averages and standard errors of the data from two or more experiments.
Ninhydrin assay.
The primary amine content of purified PGA was determined by ninhydrin assay (30) using d-glucosamine as the standard, as previously described (23).
Bioinformatics.
The amino acid sequences of Pga proteins were submitted to the HHpred server for remote protein homology detection and structure prediction (43). An alignment of homologs for each sequence was built by multiple iterations of PSI-BLAST searches against the NCBI nonredundant database and annotated with predicted secondary structure and confidence values from PSIPRED (28). Next, a profile hidden Markov model (HMM) was constructed that included the information about predicted secondary structure. The query HMM was then compared with HMMs from the Protein Data Bank (PDB) by the HHsearch software (43). HHsearch used position-specific gap penalties and scored for secondary structure similarity to construct local alignments with the query sequence and the matching database sequences. The most significant hits for the domains exhibited extremely high probability scores, ranging from 99.9 to 100, and E values from 4.2 × 10−21 to 0 (43). The best-matched proteins were selected as templates to be used for comparative modeling with the MODELLER software (39). The resulting structure models were viewed with PyMOL version 1.0 (12).
RESULTS AND DISCUSSION
Examination of PGA production and localization by immunoblotting.
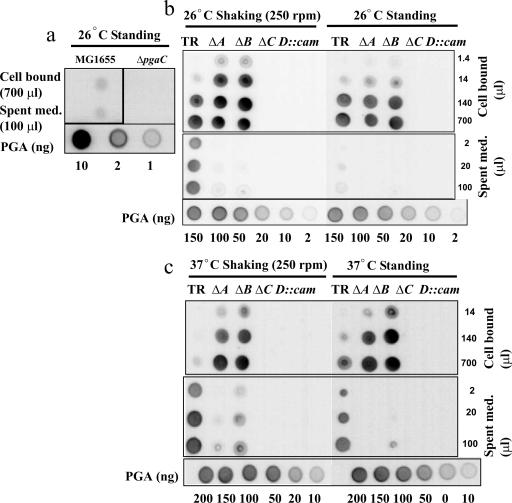
While PGA synthesis requires pgaC, its dependency on the other pga genes in E. coli or other gram-negative bacteria had not been previously determined. Therefore, polysaccharide was prepared from E. coli cells and spent medium and probed for PGA by immunoblotting using a murine anti-PGA IgM MAb (Fig. 1). In E. coli K-12 MG1655, PGA in both the cell-associated and spent-medium fractions was detectable but not abundant (Fig. 1a). Therefore, a csrA::kanR mutant (TRMG1655), which is derepressed for pgaABCD gene expression, was also examined for PGA accumulation using immunoblotting (Fig. 1b and c). The results of this experiment revealed a substantial effect of csrA on PGA accumulation, which was greater than that previously reported (50), apparently due to the greater specificity of the MAb than of the hexosamine assay that was previously employed. The examination of this strain and its isogenic nonpolar pga mutants showed that pgaC and pgaD were both needed for PGA accumulation (Fig. 1b and c). Because immunodetection requires the antigen to bind to nitrocellulose, this result cannot eliminate the possibility that small, inefficiently bound GlcNAc oligosaccharides might have been synthesized by the pgaD mutant.
FIG. 1.
Immunological detection of cell-bound and cell-free PGA. MG1655 and its isogenic ΔpgaC mutant (a) and TRMG1655 (csrA::kanR) (TR) and its isogenic pga mutants (b and c) were grown at 26°C or 37°C for 24 h under standing or shaking conditions as indicated. PGA was prepared from equal quantities of spent medium (Spent med.) or concentrated cells and analyzed by immunoblotting (Materials and Methods). A standard series of purified PGA is shown at the bottom of each panel for comparison. This experiment was repeated in its entirety twice, with essentially identical results. ΔA, ΔpgaA; ΔB, ΔpgaB; ΔC, ΔpgaC.
Under static growth conditions, PGA was primarily cell bound in the pga wild-type strain (TRMG1655), with very little PGA appearing in the spent medium (Fig. 1b). However, much of this polymer was released into the spent medium when the strain was grown with shaking. In contrast, the isogenic pgaA and pgaB mutants of this strain released little or no PGA into the medium under standing or shaking conditions. This observation suggested that PGA secretion is defective in the latter mutants or that PGA is secreted but remains tightly bound to cells.
The results for cells grown at 37°C, which is suboptimal for biofilm formation (50), were similar (Fig. 1c), with some notable exceptions. First, the pga wild-type strain (TRMG1655) retained a smaller fraction of PGA in the cell-bound fraction than in the spent medium at 37°C. This effect was observed under standing conditions and was even more striking under shaking conditions. This finding suggested that PGA export and/or its release from the cell surface is enhanced at the higher temperature (37°C). Second, some PGA was detectable in the spent medium from the ΔpgaB mutant under this growth condition. This suggests that the pgaB mutant possesses a weak transport activity at 37°C. However, we cannot entirely discount the possibility that PGA was released nonspecifically from this strain due to cell lysis or leakage from the cell envelope.
Imaging of PGA by IEM.
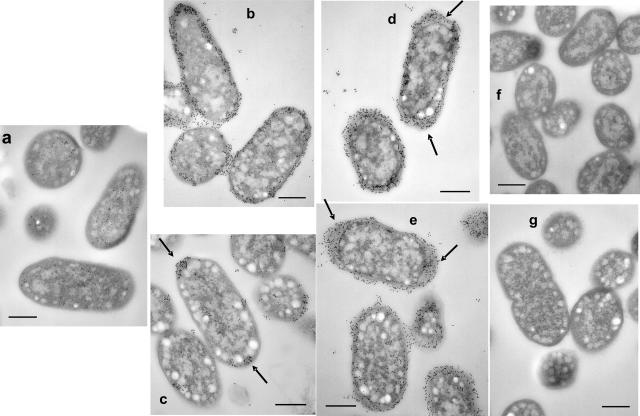
PGA localization was examined more precisely by transmission electron microscopy of thin sections with immunostaining using the anti-PGA MAb (Fig. 2). Consistent with the immunoblotting results (Fig. 1a), the parental E. coli K-12 strain MG1655 exhibited a very weak reaction with the antibody (Fig. 2a). In contrast, the isogenic csrA mutant, TRMG1655, exhibited strong labeling around the perimeter of cells that were grown under standing conditions (Fig. 2b). As predicted from the results of the blotting experiments (Fig. 1), growth under shaking conditions substantially decreased the total amount of PGA that was present on the cells (Fig. 2c). In addition, longitudinal sections of these cells revealed that PGA was predominantly localized at the poles (Fig. 2c). This pattern of polar localization was observed in 76% (125/165) of longitudinally sectioned cells from 17 different fields (data not shown), raising the possibility that PGA biosynthesis originates at the cell poles.
FIG. 2.
Localization of PGA shown by IEM of wild-type and mutant pga strains. Cultures were grown for 24 h at 26°C under standing (b) or shaking (a, c, d, e, f, and g) conditions. Samples were prepared, and PGA was detected with a primary anti-PGA murine MAb and a secondary, gold-labeled anti-murine Ig antiserum (Materials and Methods). Sample identities: MG1655 (a); TRMG1655 (csrA::kanR) (b and c); TRMG1655 ΔpgaA (d); TRMG1655 ΔpgaB (e); TRMG1655 ΔpgaC (f); and TRMG1655 pgaD::cam (g). Note the gold labeling around the perimeter and between cells of the wild-type, standing pga strain culture (b) and the decreased labeling of cells from the shaken culture (c), with label retention at the cell poles (arrows). Polar localization of PGA was observed in 76% (125/165) of longitudinally sectioned cells from 17 different fields. In contrast, the ΔpgaA and ΔpgaB strains from the shaken cultures retained strong circumferential labeling with shaking (d and e). Labeling patterns of ΔpgaA and ΔpgaB mutants under standing conditions (not shown) were essentially identical to those observed under shaking conditions. Size bars are 400 nm in length.
The PGA localization in the ΔpgaA and ΔpgaB mutant strains was essentially indistinguishable from that in the parent strain under standing conditions (Fig. 2 and data not shown). However, under shaking conditions, these mutants still retained the PGA at the cell surface (Fig. 2d and e). This confirmed the immunoblotting results (Fig. 1) and suggested that PgaA and PgaB may be involved in PGA export. One additional difference between ΔpgaA and ΔpgaB mutants and the parent strain was that the periplasm of the mutants was dramatically expanded and immunoreactive at the cell poles (Fig. 2d and e), indicative of PGA accumulation at this site. As expected from the immunoblotting results (Fig. 1), the ΔpgaC and pgaD mutants showed essentially no gold labeling (Fig. 2f and g).
pgaA and pgaB are necessary for PGA export.
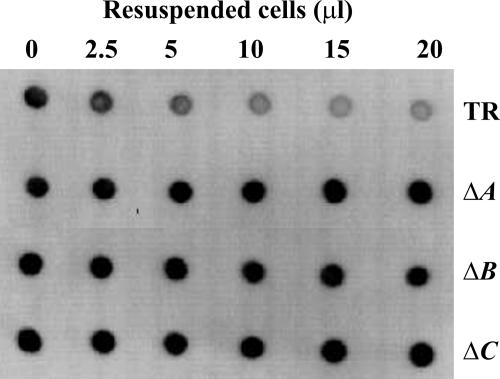
While the results of IEM studies revealed that PGA is retained at the perimeter of pgaA and pgaB mutant cells, this analysis was not precise enough to determine whether the polymer was exposed to the external milieu. To answer this question, we examined the ability of cells of the pga wild-type strain TRMG1655 and isogenic pgaA, pgaB, and pgaC nonpolar deletion mutants to adsorb the anti-PGA MAb from solution. Cells were mixed with the antibody, allowed to react for 2 h, and removed by centrifugation, and the remaining soluble (unbound) antibody was assayed by immunoblotting (Fig. 3). The results of the control reactions demonstrated that the wild-type strain adsorbed antibody from the solution, confirming that PGA is externally exposed in this strain, while the pgaC mutant, which cannot synthesize PGA, failed to adsorb this antibody. The pgaA and pgaB mutants also failed to adsorb the antibody. Thus, the PGA that is associated with pgaA and pgaB mutant cells (Fig. 1 and 2) is not externally exposed, confirming that PgaA and PgaB are needed for its export.
FIG. 3.
Adsorption of anti-PGA MAb to test for surface-exposed PGA on ΔpgaA and ΔpgaB mutant cells. Equal quantities of cells of TRMG1655 (TR) and its isogenic pga mutants were harvested, resuspended, and incubated at several concentrations with the anti-PGA MAb for 1 h. Antibody remaining in the supernatant solution after removal of cells was detected by immunoblotting (Materials and Methods). The reactions across the top and bottom rows depict the results for positive and negative experimental controls, respectively, for surface-exposed PGA. ΔA, ΔpgaA; ΔB, ΔpgaB; ΔC, ΔpgaC.
pgaB mutagenesis: evidence that N-deacetylation of PGA facilitates secretion.
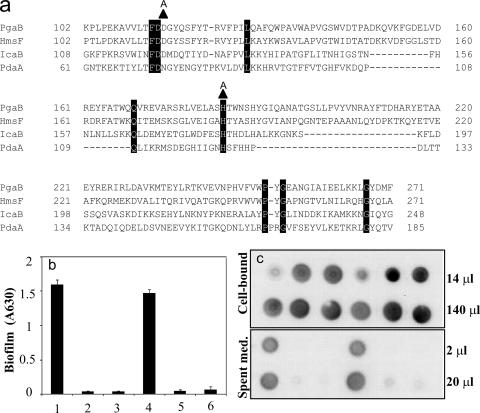
Genetic studies in S. epidermidis revealed that IcaB is a deacetylase that converts 15 to 20% of the GlcNAc residues of PIA to glucosamine. Furthermore, deletion of icaB led to shedding of this polymer from the cell surface and an inability to support biofilm formation (47). In contrast, deletion of pgaB of E. coli led to a defect in PGA export and retention of the polymer in the periplasm (Fig. 1 to 3). To determine whether deacetylation by PgaB is required for PGA export and biofilm formation, two amino acids of PgaB, which were predicted to be necessary for catalytic activity based on conservation with amino acids of Bacillus subtilis PdaA N-deacetylase and which were required for biofilm formation by Y. pestis (5, 15, 45), were changed to alanine (Fig. 4a). Alanine replacement is widely used to assess the functionality of amino acid residues in proteins, in part because it rarely disrupts higher-order protein structure (see reference 35). Plasmidic expression of the wild-type pgaB gene complemented both the biofilm defect (Fig. 4b) and the inability of a nonpolar ΔpgaB mutant to export PGA (Fig. 4c). However, the site-directed mutants were uniformly unable to restore either process (Fig. 4b and c), suggesting that the deacetylation of PGA by PgaB is a requirement for the export of this polysaccharide. To further assess the catalytic defect caused by one of the mutations, D115A, PGA was isolated and purified by fast protein liquid chromatography from a ΔpgaB strain containing the corresponding plasmid [TRXWMGΔB(pD115A)], the pgaB wild-type plasmid [TRXWMGΔB(pCRpgaB)]), or the vector control [TRXWMGΔB(pCR2.1-TOPO)], as well as a pgaB wild-type strain [TRMG(pPGA372)]. Ninhydrin analysis of the free amino groups in these four preparations revealed percent glucosamine contents of 1.4 ± 0.7 (mean ± standard deviation), 22 ± 0.3, 0.0 ± 0.4, and 5 ± 0.5, respectively. These results confirmed the predictions that pgaB is involved in the N-deacetylation of PGA and that the D115A substitution in pgaB would decrease this activity. Taken in context with the results of immunoblotting and biofilm analyses, this finding suggests that the deacetylation of PGA by PgaB promotes its export from the cell.
FIG. 4.
Sequence alignment of polysaccharide N-deacetylase domains and effects of pgaB site-directed mutations on biofilm formation and PGA synthesis and localization. (a) The E. coli PgaB deacetylase domain sequence is compared to that of HmsF of Yersinia pestis, IcaB of Staphylococcus epidermidis, and PdaA of Bacillus subtilis. Amino acid residues are numbered with respect to the deduced initiating methionine of each protein. Highly conserved amino acid residues are highlighted in black. Alanine replacements made in E. coli PgaB are shown as triangles located immediately above the PgaB sequence. The effects of these alanine replacements on biofilm formation (b) and PGA localization (c) are shown. Samples 1 to 6 represent the following strains: 1, TRMG; 2, TRMG ΔpgaB; 3, TRMG(pCR2.1-TOPO) ΔpgaB; 4, TRMG(pCRpgaB) ΔpgaB; 5, TRMG(pD115A) ΔpgaB; 6, TRMG(pH184A) ΔpgaB. Error bars depict standard errors relative to the means. This experiment was repeated in its entirety twice, with essentially identical results. A630, A630; Spent med., spent medium.
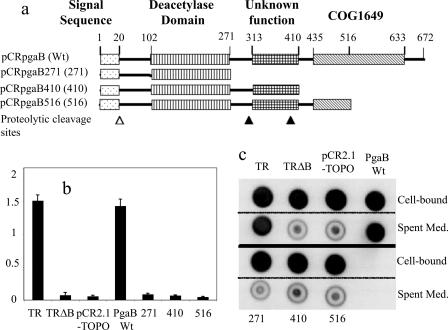
Truncation of the PgaB DUF187 domain.
To test the functionality of the C-terminal region of PgaB, we prepared three truncated pgaB constructs and examined their effects on biofilm formation and PGA secretion. The truncation mutants were uniformly dysfunctional in both processes (Fig. 5). PGA was purified from the strains containing the least- and most-severe truncations, TRXWMG(pCRpgaB516) and TRXWMG(pCRpgaB271), respectively. Ninhydrin analysis of these PGA preparations was conducted, and the results compared to those from the isogenic pgaB-overexpressing strain [TRXWMGΔB(pCRPgaB)], pgaB site-directed mutant [TRXWMGΔB(pD115A)], and vector control [TRXWMGΔB(pCR2.1-TOPO)] (data provided above). PGA from the pgaB516 and pgaB271 truncation mutants exhibited very low levels of amino groups, 1.1% ± 0.2% and 0.9% ± 0.4% relative to the level in the isogenic strain expressing the wild-type protein (22% ± 0.3%). These data raise the possibility that the DUF187 domain of PgaB promotes the deacetylase activity of this protein.
FIG. 5.
Effects of PgaB truncation on biofilm formation and PGA localization. (a) Plasmids expressing C-terminally truncated PgaB proteins are shown on the domain map of PgaB along with predictions for the prepeptide cleavage (open triangle) and palmitoylation sites, residues 20 and 21, respectively; polysaccharide N-deacetylase domain; and the domain of unknown function referred to as COG1649 or DUF187. Two putative proteolytic cleavage sites, identified by Western blotting of PgaB polypeptides (see Fig. 6), are depicted by closed triangles. (b) Biofilm formation by TRMG (csrA::kanR) (TR), TRΔB (pgaB deletion), TRΔB(pCR2.1-TOPO) (vector control), and TRΔB containing the pgaB plasmids depicted in panel a. (c) These strains were analyzed for PGA in cell-bound and cell-free fractions by immunoblotting (see Materials and Methods). Wt, wild type; Spent Med., spent medium.
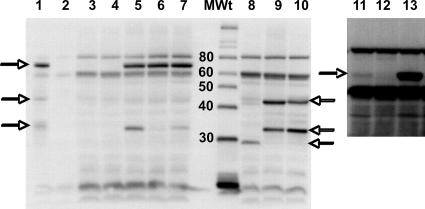
Western blotting of PgaB polypeptides.
Western blotting was used to determine whether the site-directed mutants and truncated PgaB proteins accumulate properly. Because antiserum against E. coli PgaB was unavailable, antiserum against the Y. pestis PgaB ortholog, HmsF, was used for these analyses. Y. pestis strains KIM6+, which possesses the hms locus, and KIM6, which lacks the hms genes, served as controls for the reactions (Fig. 6, lanes 1 and 2). The E. coli strain TRMG1655 showed a weak reaction with the antibody relative to that of its isogenic ΔpgaB mutant; nevertheless, a signal representing the full-length PgaB protein of the former strain was apparent at high exposures (Fig. 6, compare lanes 3 and 4 with lanes 11 and 12). In contrast, plasmidic expression of the wild-type and D115A and H184A site-directed mutant pgaB genes yielded strong signals corresponding to the full-length protein in all cases (Fig. 6, lanes 5 to 7). Because the levels of accumulation of the site-directed mutant PgaB proteins were essentially the same as that of the wild-type protein, their inability to support biofilm formation and PGA secretion (Fig. 5) cannot be attributed to differential instability or failure to accumulate.
FIG. 6.
Western blots of wild-type and mutant PgaB proteins. Cultures were grown, harvested, and analyzed for PgaB protein by Western blotting using anti-HmsF antibody (Materials and Methods). Strain identities were as follows: 1, Y. pestis KIM6+; 2, Y. pestis KIM6; 3, TRMG; 4, TRMG ΔpgaB(pCR2.1 TOPO); 5, TRMG ΔpgaB(pCRpgaB); 6, TRMG ΔpgaB(pD115A); 7, TRMG ΔpgaB(pH184A); 8, TRMG ΔpgaB(pCRpgaB271); 9, TRMG ΔpgaB(pCRpgaB410); 10, TRMG ΔpgaB(pCRpgaB516). MWt indicates the molecular mass standards and their corresponding sizes in kDa. The horizontal arrows mark the positions of PgaB (HmsF) full-length and partial polypeptides that were detected by the antiserum. Lanes 1 to 10 depict samples separated on a 12.5% gel. Lanes 11 to 13 depict the same samples as in lanes 3 to 5 separated on a 7.5% gel and overexposed to reveal the PgaB native protein (∼74 kDa) from TRMG1655. The Y. pestis and E. coli proteins were loaded at 5 μg and 50 μg per lane, respectively.
Examination of the pgaB truncation mutants yielded more-complex results. The protein from pCRpgaB271 (Fig. 5a) accumulated in its full-length form (Fig. 6, lane 8). However, the full-length proteins from pCRpgaB410 and pCRpgaB516 (Fig. 5a and Fig. 6, lanes 9 and 10) were not observed. Instead, two major smaller products appeared in both of these samples. These polypeptides most likely represent products of proteolysis, as pgaB-dependent products similar in size to one of these polypeptides were present in other E. coli extracts (Fig. 6, lanes 5 and 7) and proteins similar in size to both of these products were observed in extracts from Y. pestis KIM6+, but not KIM6 (lanes 1 and 2). In any case, the PgaB271 protein and both of the putative proteolytic fragments from pCRpgaB410 and pCRpgaB516 should contain the intact PgaB deacetylase domain (Fig. 5a). In context with the results of the biofilm formation (Fig. 5b), PGA localization (Fig. 5c), and ninhydrin assays, the results of these Western analyses indicate that the PgaB deacetylase domain is necessary, but not sufficient, for normal catalytic activity of this enzyme in the bacterial cell, which also requires the C terminus of PgaB.
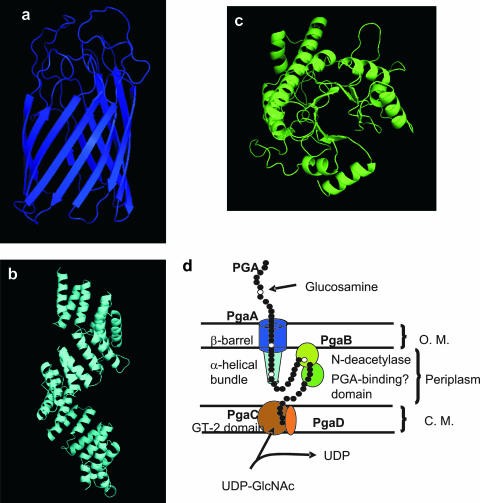
Domain predictions for Pga proteins.
Since PgaA and PgaB are necessary for PGA export, we sought to infer structural and functional information from homologous proteins. While conventional sequence search methods failed to identify homologous proteins of known structure or function, the HHpred server (43) detected and aligned remotely homologous sequences in the Protein Data Bank (PDB). The N-terminal domain of PgaA had significant hits to the tetratricopeptide repeat domains of human nucleoporin O-linked GlcNAc transferase, yeast mitochondrial outer membrane translocon protein Tom70p, and human peroxisomal targeting signal-1 receptor PEX5. These tetratricopeptide repeat domains contain pairs of antiparallel α-helices stacked in parallel to form an elongated superhelical structure with a spiraling groove that mediates protein-protein interactions (17, 27, 51). Human nucleoporin O-linked GlcNAc transferase represented the best match for the PgaA N-terminal domain. The C-terminal domain of PgaA had a significant hit to the E. coli long-chain fatty acid transporter protein FadL. This protein forms an outer membrane pore composed of 14 antiparallel β strands (46). These relationships suggest that PgaA contains a porin domain that facilitates the export of PGA across the outer membrane and a periplasmic domain that may mediate protein-protein interactions, perhaps with PgaB.
In addition to a predicted cleavable N-terminal signal peptide, palmitoylation site, and deacetylase domain, PgaB and its orthologs in gram-negative bacteria possess a C-terminal domain of unknown function (DUF187 or COG1649). Using HHpred, we found a series of matches to carbohydrate binding and modifying domains (data not shown), the most significant of which was the match to the N-terminal domain of Thermus thermophilus β-galactosidase. This domain forms a TIM barrel fold that harbors the galactose binding and catalytic sites (21). Based on this relationship and the observation that deletions lacking this domain are defective for PGA N-deacetylation, we predict that this domain binds to unmodified poly-β-1,6-GlcNAc and thereby assists catalysis by the deacetylase domain.
PgaC and PgaD are cytoplasmic membrane proteins (11). PgaC contains a single domain, which is a member of the GT-2 glycosyltransferase family (50). Neither the BLAST (50) nor the HHpred analysis permitted assignment of the small (137 amino acid) protein PgaD to a functional domain family. We propose that PgaD of E. coli is representative of proteins that assist glycosyltransferases (PgaC orthologs) of gram-negative bacteria in the polymerization of β-1,6-GlcNAc.
A model for Pga protein assembly in the cell envelope.
Figure 7 shows comparative structure models for PgaA and PgaB, as well as a model of the four Pga proteins in the cell envelope. While it is reasonable to expect that PGA synthesis and transport may require the Pga proteins to physically interact, our data do not address this issue. PgaC and PgaD are cytoplasmic membrane proteins that are required for PGA synthesis and are shown interacting based on this functional data. The model predicts that subsequent to polymerization, PGA is modified by the lipoprotein PgaB in the periplasm. This introduces a limited amount of glucosamine into the polymer, which facilitates its export through the outer membrane by the PgaA porin.
FIG. 7.
HHpred predictions of Pga protein domain structures (a to c) and a hypothetical model for Pga protein assembly in the cell envelope (d). Structural predictions were derived by a threading algorithm using the best domain matches for PgaA C-terminal domain (FadL protein of E. coli) (a), PgaA N-terminal domain (human nucleoporin O-linked GlcNAc transferase) (b), and PgaB C-terminal domain (LacZ of T. thermophilus) (c). The amino acid sequences of these domains are 518 to 807, 65 to 515, and 333 to 646, respectively. The N-terminal domain of PgaB and the sole domain of PgaC belong to carbohydrate esterase (N-deacetylase) family 4 and GT-2 glycosyltransferase family, respectively (50). No prediction was obtained for PgaD. (d) The model for PGA synthesis and Pga protein localization in the cell envelope is discussed in the text. O.M., outer membrane; C.M., cytoplasmic membrane.
Conclusions.
This study examines the differential functions of the Pga gene products in the synthesis, covalent modification, and export of the E. coli biofilm adhesin PGA. While it was not surprising to find that genes for the cytoplasmic membrane proteins (PgaC and PgaD) were necessary for PGA synthesis and the apparent outer membrane proteins (PgaA and PgaB) were needed for PGA export, a number of unanticipated findings of this study are worth noting. (i) PGA was primarily cell associated under standing conditions, yet much of it was released into the medium when cultures were grown under shaking conditions. The sloughing of PGA probably accounts for the relatively weak surface association of E. coli cells and biofilm under sheering conditions in comparison with that of species such as Pseudomonas aeruginosa, which form strong surface attachments under such conditions (1). (ii) Under shaking conditions, PGA was preferentially located at the cell poles. This is intriguing because during the initiation of biofilm formation, E. coli and certain other gram-negative rods form temporary attachments to abiotic surfaces via the cell poles prior to forming more-stable lateral attachments (see references 2 and 7). PGA-deficient E. coli cells still form the temporary polar attachments, indicating that another adhesin may also be present at these sites. However, PGA apparently is necessary for temporarily attached cells to undergo the transition to permanent lateral attachment (2). This localization raises the possibility that the PGA synthesis apparatus is located at the cell poles. The observation that the periplasm of pgaA and pgaB mutants is expanded at the cell poles is also consistent with this idea. (iii) While IcaB of S. epidermidis and PgaB of E. coli are needed for deacetylation of β-1,6-GlcNAc, the basis for this requirement differs in each case. In staphylococci, there is no outer membrane barrier to exopolysaccharide secretion and the relatively abundant (15 to 20%) glucosamine residues of PIA are thought to be needed for the polymer to remain anchored to the cell envelope by electrostatic interactions (47). In E. coli, it is apparent that the glucosamine residues of PGA facilitate PGA export through the outer membrane porin, PgaA. (iv) The observation that an elevated temperature (37°C) permitted a ΔpgaB mutant, but not a ΔpgaA mutant, to release some PGA into the medium suggests that higher temperature might improve PGA export through a pgaA channel in the absence of deacetylation. (v) The similarity of the PGA secretion system to the Tom system for transport of unfolded peptides across the mitochondrial outer membrane has not escaped our attention. Tom70p, which is structurally related to the PgaA N-terminal domain, contains stacked α-helices that form an N-terminal domain which interacts with the Hsp70 chaperone protein, as well as a C-terminal domain that forms a hydrophobic region believed to interact with unfolded cargo polypeptides (51). Tom70p apparently assists in the transfer of prepeptides through the mitochondrial outer membrane by the porin Tom40p (3). Additional studies will be required to determine whether the bacterial transport of PGA and mitochondrial prepeptide transport by the Tom system are mechanistically related processes.
Acknowledgments
These studies were funded in part by the National Institutes of Health (GM066794). Contributions from the University of Florida were supported by CRIS project R-101049 of the University of Florida Institute of Food and Agricultural Sciences.
We are grateful to Robert Perry and Alexander Bobrov for the generous gift of polyclonal antibody against Y. pestis HmsF (PgaB). Kane Biotech, Inc., may develop applications related to the findings herein. T. Romeo owns equity in and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Agladze, K., D. Jackson, and T. Romeo. 2003. Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 1855632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 1878237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, M. J., A. E. Frazier, J. M. Gulbis, and M. T. Ryan. 2007. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17456-464. [DOI] [PubMed] [Google Scholar]
- 4.Begun, J., J. M. Gaiani, H. Rohde, D. Mack, S. B. Calderwood, F. M. Ausubel, and C. D. Sifri. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair, D. E., and D. M. van Aalten. 2004. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 57013-19. [DOI] [PubMed] [Google Scholar]
- 6.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 7.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 1864476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerca, N., K. K. Jefferson, T. Maira-Litrán, D. B. Pier, C. Kelly-Quintos, D. A. Goldmann, J. Azeredo, and G. B. Pier. 2007. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-β-(1-6)-glucosamine. Infect. Immun. 753406-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerca, N., T. Maira-Litrán, K. K. Jefferson, M. Grout, D. A. Goldmann, and G. B. Pier. 2007. Protection against Escherichia coli infection by antibody to the Staphylococcus aureus poly-N-acetylglucosamine surface polysaccharide. Proc. Natl. Acad. Sci. USA 1047528-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49711-745. [DOI] [PubMed] [Google Scholar]
- 11.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 3081321-1323. [DOI] [PubMed] [Google Scholar]
- 12.DeLano, W. 2002. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 13.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28350-356. [DOI] [PubMed] [Google Scholar]
- 15.Forman, S., A. G. Bobrov, O. Kirillina, S. K. Craig, J. Abney, J. D. Fetherston, and R. D. Perry. 2006. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 1523399-3410. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 1864457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatto, G. J., Jr., B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 71091-1095. [DOI] [PubMed] [Google Scholar]
- 18.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 27318586-18593. [DOI] [PubMed] [Google Scholar]
- 19.Goller, C., X. Wang, Y. Itoh, and T. Romeo. 2006. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 1888022-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 431367-1378. [DOI] [PubMed] [Google Scholar]
- 21.Hidaka, M., S. Fushinobu, N. Ohtsu, H. Motoshima, H. Matsuzawa, H. Shoun, and T. Wakagi. 2002. Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 32279-91. [DOI] [PubMed] [Google Scholar]
- 22.Hinnebusch, B. J., and D. L. Erickson. 2008. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague, p. 229-248. In T. Romeo (ed.), Current Topics in Microbiology and Immunology, vol. 322. Bacterial biofilms, in press. Springer-Verlag, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izano, E. A., I. Sadovskaya, E. Vinogradov, M. H. Mulks, K. Velliyagounder, C. Ragunath, W. B. Kher, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190783-792. [DOI] [PubMed] [Google Scholar]
- 27.Jínek, M. J. Rehwinkel, B. D. Lazarus, E. Izaurralde, J. A. Hanover, and E. Conti. 2004. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 111001-1007. [DOI] [PubMed] [Google Scholar]
- 28.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292195-202. [DOI] [PubMed] [Google Scholar]
- 29.Joyce, J. G., C. Abeygunawardana, Q. Xu, J. C. Cook, R. Hepler, C. T. Przysiecki, K. M. Grimm, K. Roper, C. C. Ip, L. Cope, D. Montgomery, M. Chang, S. Campie, M. Brown, T. B. McNeely, J. Zorman, T. Maira-Litrán, G. B. Pier, P. M. Keller, K. U. Jansen, and G. E. Mark. 2003. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr. Res. 338903-922. [DOI] [PubMed] [Google Scholar]
- 30.Leane, M. M., R. Nankervis, A. Smith, and L. Illum. 2004. Use of the ninhydrin assay to measure the release of chitosan from oral solid dosage forms. Int. J. Pharm. 271241-249. [DOI] [PubMed] [Google Scholar]
- 31.Li, Z., A. J. Clarke, and T. J. Beveridge. 1996. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J. Bacteriol. 1782479-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, L., H. Lu, A. Sprinkle, M. R. Parsek, and D. Wozniak. 2007. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 1898353-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenney, D., K. L. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 2841523-1527. [DOI] [PubMed] [Google Scholar]
- 35.Mercante, J., K. Suzuki, X. Cheng, P. Babitzke, and T. Romeo. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 28131832-31842. [DOI] [PubMed] [Google Scholar]
- 36.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry, R. D., A. G. Bobrov, O. Kirillina, H. A. Jones, L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 1861638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234779-815. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sloan, G. P., C. F. Love, N. Sukumar, M. Mishra, and R. Deora. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 1898270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, R. L., and E. Gilkerson. 1979. Quantitation of glycosaminoglycan hexosamine using 3-methyl-2-benzothiazolone hydrazone hydrochloride. Anal. Biochem. 98478-480. [DOI] [PubMed] [Google Scholar]
- 43.Söding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutherland, I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 1473-9. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Berg, B., P. N. Black, W. M. Clemons, Jr., and T. A. Rapoport. 2004. Crystal structure of the long-chain fatty acid transporter FadL. Science 3041506-1509. [DOI] [PubMed] [Google Scholar]
- 47.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 27954881-54886. [DOI] [PubMed] [Google Scholar]
- 48.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6269-275. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 561648-1663. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 1862724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, Y., and B. Sha. 2006. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13589-593. [DOI] [PubMed] [Google Scholar]
- 52.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]