Abstract
Chlamydia pneumoniae is an important obligate intracellular pathogen that replicates within an inclusion in the eukaryotic cell. The initial event of a chlamydial infection is the adherence to and subsequent uptake of the infectious elementary bodies (EBs) by the human cell. These processes require yet-unidentified bacterial and eukaryotic surface proteins. The GroEL1 protein, which exhibits a very strong antigenicity and in vitro can activate various eukaryotic cells, is a potential pathogenicity factor. We localized the protein during the infection process and found it in the inclusion but outside the chlamydial particles. GroEL1 was also localized on the surface of EBs, and the protein could be washed off the EBs. Latex beads coated with recombinantly produced GroEL1 (rGroEL1) bound in a dose-dependent manner to HEp-2 cells. Likewise, GroEL1, when expressed and displayed on the yeast cell surface, mediated adhesion to HEp-2 cells. Interestingly, the homologous GroEL2 and GroEL3 proteins showed no adhesive properties. Incubation of primary umbilical vein endothelial cells with soluble GroEL1 and GroEL1-coated latex beads activated the translocation of the general transcription factor NF-κB into the nucleus. Finally, preincubation of HEp-2 cells with rGroEL1 significantly reduced subsequent infection with C. pneumoniae, although adhesion of infectious bacteria to eukaryotic cells was not affected. Taken together, these data support a role for extracellular GroEL1 in the establishment of the chlamydial infection.
The human pathogen Chlamydia pneumoniae is an obligate intracellular gram-negative bacterium characterized by a unique biphasic life cycle. Infectious but metabolically inactive elementary bodies (EBs) attach to and invade eukaryotic cells, evade fusion with lysosomes, and differentiate into metabolically active reticulate bodies within a host-derived vacuole (termed an inclusion) in which they replicate. After a period of growth, the reticulate bodies redifferentiate into EBs which are released from the infected cells (25, 30). Chlamydiae enter host cells by multiple routes, and this versatility is thought to account for species differences and tissue tropism (63).
C. pneumoniae causes several severe diseases of the respiratory tract (42, 46, 79). In addition, a link between C. pneumoniae infection, atherosclerosis, and coronary artery disease has been reported (64, 86). More recently the pathogen has also been associated with several other chronic diseases (23, 26, 52, 77, 89).
The molecular mechanisms of chlamydial pathogenesis are poorly understood, and many of the factors that contribute to the pathogenicity of this bacterium have not yet been delineated precisely. Infection by EBs requires both attachment to and uptake by target cells. Several reports have demonstrated that glycosaminoglycans (GAGs) on host cells are involved in attachment and subsequent infection, but controversy remains concerning the precise mechanisms involved (90; reviewed in reference 10). However, it has recently been shown that the bacterial surface-associated OmcB protein is the major GAG-binding adhesin (19, 55, 76). Other bacterial proteins, including the cysteine-rich membrane proteins Hsp70 and MOMP, have been analyzed for their potential contribution to the attachment of Chlamydia trachomatis and Chlamydia psittaci EBs to eukaryotic cells (47, 60, 70, 76, 78, 80), but none of these acts as a high-affinity chlamydial surface ligand. Interestingly, antibodies directed against PmpD from C. trachomatis or Pmp21 from C. pneumoniae significantly reduced subsequent infection, suggesting that they might represent chlamydial adhesins (13, 85). Apart from GAGs, the host components that participate in EB binding have so far eluded detection, although protein disulfide isomerase has been implicated in the process (11, 14). Thus, the molecular mechanisms involved in chlamydial attachment and entry remain largely unresolved and controversial.
One of the few proteins so far identified as being relevant for chlamydial pathogenesis is the GroEL1 protein, also referred to as heat shock protein 60 (Hsp60) (84). This protein belongs to the group I chaperonins produced by almost all prokaryotic and eukaryotic cells, which as intracellular proteins assist in the correct folding of nascent or denatured proteins under both normal and stress conditions (8, 91). However, molecular chaperones clearly have additional functions. Several reports indicate that molecular chaperones produced by pathogenic bacteria, such as Hsp60, can function as intracellular, cell surface, or extracellular signals in the course of infection processes (for a review, see reference 27). Conversely, human cells respond to infection or other forms of stress by secreting heat shock proteins (20, 58, 73).
Genome sequence analysis has revealed that all chlamydial species possess three groEL-like genes (groEL1, groEL2, and groEL3) (36, 62, 75). GroEL1 shows the highest homology to GroEL proteins from other organisms, and the chaperonin function of chlamydial GroEL1 has been demonstrated by complementation of a temperature-sensitive Escherichia coli groEL mutant (3, 37).
Recombinant C. pneumoniae GroEL1 (rGroEL1) can stimulate innate immune cells in vitro via Toll-like receptor 2 (TLR2) and TLR4 and thus may contribute to the proinflammatory responses seen during infection (12). Moreover, C. pneumoniae GroEL1 can activate the granulocyte-macrophage colony-stimulating factor in human bronchial epithelial cells almost as effectively as viable or UV-inactivated whole bacteria, giving rise to a sustained proinflammatory phenotype (43). Similarly, C. trachomatis GroEL1 induces endothelial cells, smooth muscle cells, and macrophages to produce adhesion factors and proinflammatory cytokines. In both cases, GroEL1 operates by activating NF-κB (9, 40, 51, 66, 81).
The immune responses to chlamydial GroEL1 correlate significantly with disease sequelae in humans, and 80 to 90% of patients infected with C. trachomatis have antibodies directed against GroEL1 (7, 17, 28, 65). But it is still not clear whether antibodies specific for chlamydial GroEL1 make a direct contribution to disease pathogenesis. It is assumed that, in the course of chlamydial infections, bacterial GroEL induces cross-reactive immune responses to endogenous GroEL, and this may account for much of the tissue damage observed during chronic infections (57, 91). However, in guinea pigs it has been shown that immunization with native GroEL1 actually reduces the degree of gross ocular pathology induced by subsequent infection with Chlamydophila caviae (GPIC), indicating that the anti-GroEL1 response confers a certain degree of protection against ocular disease (59).
The high degree of antigenicity of GroEL1 in patients implies that the protein is easily accessible to the immune system, perhaps because it is localized on the surface of the chlamydial particles. Early studies on isolated outer membrane complexes from C. trachomatis and C. psittaci EBs had indeed pointed to the possibility that GroEL1 might be associated with chlamydial membranes (3). In the present study, we show that C. pneumoniae GroEL1 is present on the surface of infectious EBs and binds to host cells. In addition, we demonstrate that preincubation of target cells with chlamydial rGroEL1 substantially inhibits the infection process.
MATERIALS AND METHODS
Eukaryotic cells.
HEp-2 cells (ATCC CCL-23) were propagated as described previously (33). For immunofluorescence assays, cells were seeded on glass coverslips (1-cm diameter) and incubated for 48 h at 37°C. Cells to be used for chlamydial infection were seeded onto 25-cm2 polystyrene flasks to form confluent monolayers. Attachment assays were performed using aliquots (1 × 105) of HEp-2 cells seeded onto glass coverslips 24 h prior to the experiments.
Primary human umbilical vein endothelial cells (HUVECs) (provided by J. Schrader, Institut fuer Kreislaufphysiologie, HHU Duesseldorf) were cultivated at 37°C (second to third passage) in Ham's F12K medium (Invitrogen) supplemented with 10% fetal bovine serum, 30 μg/ml endothelial cell growth supplement (Sigma), 10 U/ml heparin (Sigma), and 6% CO2.
Bacterial and yeast strains.
Chlamydia pneumoniae respiratory isolate GiD was cultivated as described previously (32). Escherichia coli XL-1 Blue (Stratagene) was grown in LB with or without 50 μg/ml ampicillin.
Saccharomyces cerevisiae CEN.PK2-1C (MATa leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2) (18) was grown in YP medium supplemented with 4 mg/liter adenine and 20 mg/liter tryptophan plus 2% glucose (YPD) or in synthetic medium plus 2% glucose (SD) (72). The S. cerevisiae strain EBY100 (MATa ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL), used for adhesion experiments, was grown in synthetic medium containing 2% raffinose (SR) or 2% galactose (SG) (72) (Invitrogen) (55). For expression of Aga2p or Aga2p fusion proteins in strain EBY100, cells were grown in accordance with the manual supplied by Invitrogen.
Plasmids.
Plasmid pMal-c2 (New England Biolabs) was modified to allow amplification in yeast by the addition of a 1,921-bp S. cerevisiae CEN6/ARSH4/URA3 DNA fragment, which was amplified from plasmid pRS316 (74) by PCR using oligonucleotides C-284 and C-285 (Table 1) and Pfx polymerase (Invitrogen). The PCR product was integrated into SwaI-cleaved plasmid pMal-c2 by homologous recombination in yeast using the transformation protocol described by Schiestl and Gietz (69). Total yeast DNA was then isolated (35) and used to transform E. coli cells to recover the recombinant plasmid pAC-2. The maltose-binding protein (MBP)-invasin (Inv) fusion protein was expressed from plasmid pRI284 (kindly provided by P. Dersch) (16).
Cloning and expression of groEL genes.
The C. pneumoniae groEL2 (Cpn0777) and groEL3 (Cpn0898) genes were amplified by PCR using the primer pairs C-123/C-124 and C-125/C-126, respectively (Table 1) and C. pneumoniae genomic DNA as the template. Both amplicons were cloned into the EcoRI-cleaved yeast-E. coli shuttle plasmid pAC-2 by homologous recombination in S. cerevisiae. Integration of the groEL2 or groEL3 gene into pAC-2 led to fusion with the malE gene for MBP. The groEL gene of E. coli K-12 and the gene fragment encoding the Inv adhesion domain of Yersinia pseudotuberculosis (bp 2235 to 2826) were amplified using oligonucleotides C-154/C-155 and C-578/C-579, respectively (Table 1). The amplicons were transformed into yeast together with the BglII/EcoRI-cleaved pAC-2 plasmid to yield a GroEL or Inv expression plasmid coding for N-terminally His6-tagged fusion proteins.
Protein expression was induced in E. coli grown in liquid culture by treatment with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h. Expression and purification of the MalE fusion proteins MBP-Inv, MBP-LacZ, MBP-GroEL2, and MBP-GroEL3 were performed as suggested by the supplier of the progenitor plasmid pMal-c2 (New England Biolabs). The His-tagged rGroEL1 from C. pneumoniae, the GroEL from E. coli, and the Inv protein from Y. pseudotuberculosis were expressed and purified as described previously (33). GroEL and GroEL1 were isolated in the presence of 20 μg/ml polymyxin B (Sigma) (33). Purified proteins were dialyzed three times against phosphate-buffered saline (PBS) and stored at −20°C. Residual lipopolysaccharide (LPS) was removed by passage over polymyxin B-agarose columns (Detoxi-Gel AffinityPak prepacked columns; catalog no. 20344, Pierce) according to the manufacturer's recommendations. The endotoxin concentration of these protein preparations was <0.05 endotoxin units/mg protein, as determined by the Limulus amoebocyte lysate assay (assays were performed according to FDA standards by ACILA AG, Mörfelden-Walldorf, Germany).
To express GroEL1 on the yeast cell surface, the full-length groEL1 coding sequence was amplified by PCR using oligonucleotides C-236 and C-237 (Table 1) with C. pneumoniae GiD genomic DNA and cloned into the EcoRI/NotI-digested vector pYD1 (Invitrogen) by homologous recombination in S. cerevisiae. Plasmids expressing Aga2-Inv and Aga2-OmcB have been described by Mölleken and Hegemann (55). All constructs used were verified by sequencing.
Immunolocalization of C. pneumoniae proteins.
HEp-2 cells grown on glass coverslips in 24-well plates for 48 h (1 × 105 per well) were infected with C. pneumoniae GiD at a multiplicity of infection (MOI) of 0.5. At 60 hours postinfection the cells were washed once with PBS, fixed with 3.7% formaldehyde for 1 h., and permeabilized with PBS containing 0.5% Triton X-100 or fixed with 96% methanol for 10 min. The monolayer was then washed three times with PBS (39). Polyclonal antibodies directed against OmpA (1:20) or C. trachomatis S1 protein (1:4), or monoclonal antibodies against DnaK (1:50) were kindly provided by S. Birkelund and F. Wuppermann (F. Wuppermann and J. Hegemann, unpublished) (39, 50). Polyclonal antibodies directed against C. pneumoniae OmcB were preabsorbed against rOmcB protein (1:25) (55). The monoclonal antibody against GroEL1 (A57B9) (1:200) is directed against a N-terminal epitope of GroEL1 and was kindly provided by R. Morrison (88). The fluorescein isothiocyanate-labeled secondary antibodies were used at a dilution of 1:20 (Dako). The Cy3-labeled secondary antibodies were used at a dilution of 1:200 (Sigma). Cells were viewed using a Zeiss Axioskop.
For immunolocalization of GroEL1 on chlamydial EBs, the chlamydiae were purified using a Percoll gradient (2). Viable or methanol-fixed EBs were processed for immunofluorescence as described above.
Fractionation of chlamydial GroEL1 protein from infectious EBs.
Chlamydial particles (EBs) were purified on Percoll gradients by centrifugation at 30,000 × g as described previously (2). The bacterial pellet (approximately 1 × 108 particles) was resuspended in 250 μl of PBS, incubated for 2 h at 4°C, pelleted again at 30,000 × g for 30 min, incubated for an additional 2 h at 4°C in 250 μl PBS, and recovered by centrifugation at 30,000 × g. The chlamydial EB pellet was then prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The two supernatants were lyophilized separately and dissolved in SDS-PAGE buffer. The pellet and the two supernatant fractions were then subjected to SDS-PAGE and Western blot analysis using OmpA, DnaK, and GroEL1 antibodies. The wash fractions loaded contained three times as much protein as the pellet samples.
SDS-PAGE and Western blot analysis.
Proteins were fractionated by SDS-PAGE and subjected to Western blot analysis. An antibody specific for the His tag (Qiagen) was used to detect His-tagged rGroEL1. As secondary antibodies, anti-mouse or anti-rabbit antibodies, each linked to alkaline phosphatase (Promega), were used.
Adhesion assays with protein-coated latex beads.
Latex beads (Sigma) (diameter, 1 μm) were coated with protein (16). The coating efficiency was analyzed by immunofluorescence microscopy and immunoblotting and found to be between 90 and 95%. Adhesion assays with protein-coated latex beads and HEp-2 cells were performed as reported earlier (16), and the average numbers of beads bound to cells were determined.
Yeast adhesion assays.
Yeast adhesion assays were performed with HEp-2 cells cultivated on glass coverslips as described previously (55). Briefly, yeast cells (1 × 106 in 1 ml of PBS) expressing Aga2p alone or a specific fusion protein on the cell surface were added to 1 × 105 HEp-2 cells. Coverslips were incubated for 1 h. at 4°C with gentle shaking, washed three times with 0.5 ml PBS, and fixed with 3.7% formaldehyde. The numbers of yeast cells attached to 1,000 HEp-2 cells were counted microscopically. Each experiment was repeated four times.
NF-κB activation assays.
HUVECs were seeded onto gelatin-coated 24-well plates at a density of 2 × 105 cells/well and allowed to adhere for 24 h at 37°C in the presence of 6% CO2. The cells were incubated either with protein or with protein-coated beads. As a control, HUVECs were infected with C. pneumoniae (MOI, ∼10) and analyzed 2 h later. The subcellular localization of NF-κB was analyzed in fixed cells using a polyclonal rabbit anti-human NF-κB p65 antibody (Santa Cruz Biotechnology) and a fluorescein isothiocyanate-labeled secondary antibody (Sigma), and the percentage of HUVECs exhibiting a strong nuclear signal was determined.
Infection competition assay.
Confluent HEp-2 cell monolayers grown on glass coverslips were washed with PBS and incubated with 20 or 200 μg/ml rGroEL1 or bovine serum albumin (BSA) for 2 h at 37°C under gentle agitation. Percoll gradient-purified C. pneumoniae EBs were added and incubated for an additional 2 h at 37°C under light agitation. The HEp-2 cells were washed three times, covered with chlamydial growth medium, and incubated for 3 days at 37°C. Cells were fixed, and chlamydial inclusions were labeled using a chlamydia-specific antibody directed against LPS (Pathfinder; Bio-Rad). The number of inclusions in 80 microscope fields (20/coverslip) was determined and expressed as a percentage of the number found in PBS-treated samples.
Flow cytometric assay for adhesion of CFSE-labeled C. pneumoniae EBs to human cells.
Purified C. pneumoniae EBs (2 × 108) were labeled for 1 h at 37°C with 25 μmol carboxyfluorescein succinimidyl ester (CFSE) (Molecular Probes) and washed twice with PBS containing 1% BSA as previously described (71). Confluent monolayers of HEp-2 cells or HUVECs in 24-well plates were incubated for 2 h at 37°C with 20 μg/ml or 200 μg/ml recombinant protein or 500 μg/ml heparin, and CFSE-labeled C. pneumoniae EBs (MOI, 10) were then added and left for 1 h. Cells were washed with PBS, trypsinized, and fixed with formaldehyde, and the degree of bacterial adhesion was measured by flow cytometry using a FACSAria (BD Biosciences).
RESULTS
C. pneumoniae GroEL1 protein is localized within the chlamydial inclusion but outside the bacterial cell.
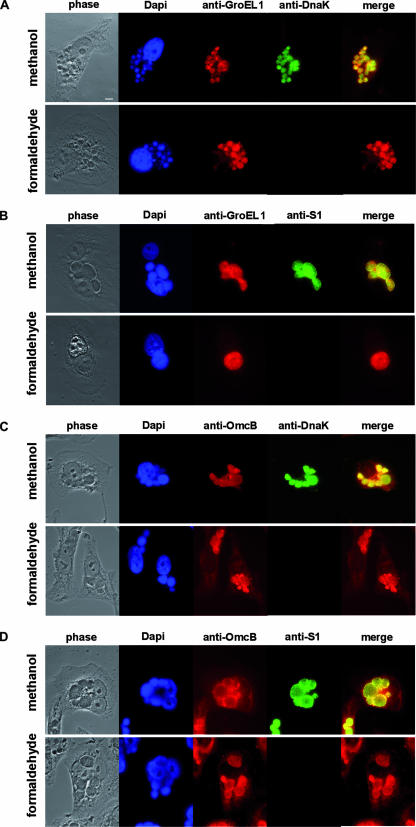
To determine the localization of GroEL1 protein during chlamydial infection, human epithelial HEp-2 cells were infected with C. pneumoniae. At 60 h postinfection, the infected cells were fixed with either methanol (rendering all chlamydial antigens accessible to antibodies) or formaldehyde plus Triton X-100 (intrachlamydial antigens remain inaccessible) and processed for indirect immunofluorescence (39). In methanol-fixed cells, both chlamydial outer membrane proteins (OmcB and OmpA) and the intracellular chlamydial proteins DnaK and S1 could readily be detected in the chlamydial inclusion (Fig. 1A to D; see Fig. S1B and S1J in the supplemental material). Staining of inclusions with a monoclonal antibody directed against the GroEL1 protein likewise revealed that the antigen was localized in the chlamydial inclusion (Fig. 1A and B; see Fig. S1F in the supplemental material). In formaldehyde-fixed cells the extrachlamydial OmcB and OmpA proteins could still be detected within the inclusion (Fig. 1C and D; see Fig. S1L in the supplemental material), while the intracellular chlamydial DnaK and S1 proteins were not accessible to the antibody under these fixation conditions (Fig. 1A and B; see Fig. S1D in the supplemental material). Note that formaldehyde does not damage the S1 and DnaK epitopes, as revealed by the fact that subsequent fixation with methanol allows antibodies to recognize both proteins (data not shown). Interestingly, staining of the chlamydial inclusions with the monoclonal antibody directed against GroEL1 revealed that GroEL1 could be detected within the inclusion but outside the bacterial cells (Fig. 1A and B; see Fig. S1H in the supplemental material). The fact that the intrachlamydial DnaK and S1 proteins are not detectable in formaldehyde-fixed cells indicates that bacterial lysis within the inclusion is negligible. The same localization pattern was observed with a polyclonal antibody directed against GroEL1 (data not shown). Collectively, these data indicate that some of the chlamydial GroEL1 protein is secreted from the chlamydial particles present in the inclusion.
FIG. 1.
Localization of chlamydial GroEL1 within the chlamydial inclusion. HEp-2 cells infected with C. pneumoniae at an MOI of 0.5 for 60 h were fixed either with methanol to visualize both intra- and extrachlamydial proteins or with formaldehyde plus 0.5% Triton X-100 to visualize extrachlamydial proteins (5). Indirect immunofluorescence microscopy was performed using antibodies against intrachlamydial DnaK (A and C) or C. trachomatis ribosomal S1 protein (B and D) and surface-localized chlamydial OmcB (C and D) and chlamydial GroEL1 (A and B). Bar, 10 μm.
C. pneumoniae GroEL1 protein is associated with the surface of infectious EBs.
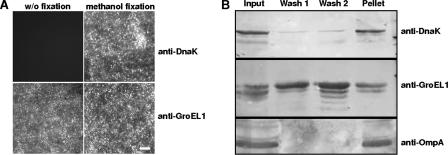
The GroEL1 localization pattern described above was detectable throughout the entire infection process (24 to 72 h postinfection) (data not shown). Because most of the reticulate bodies within the inclusions have redifferentiated into EBs by 72 h postinfection, this result suggested that infectious EBs released from the infected eukaryotic cells might also carry extracellularly exposed GroEL1 protein. To address this issue directly, infectious EBs were purified on Percoll gradients and either stained without fixation or first fixed with methanol and then stained with the GroEL1-specific monoclonal antibody. In both cases a strong fluorescence signal was detected on the EBs, indicating that GroEL1 protein is indeed present on the surface of infectious EBs (Fig. 2A). In contrast, staining of unfixed EBs using an antibody directed against the chlamydial DnaK protein produced no signals, while the DnaK protein was readily detectable after fixation with methanol (Fig. 2A).
FIG. 2.
Surface localization of C. pneumoniae GroEL1. (A) Chlamydial GroEL1 protein could be detected on Percoll gradient-purified EBs without (w/o) fixation as well as after methanol fixation using indirect immunofluorescence microscopy. Antibodies directed against the intrachlamydial DnaK protein could detect the antigen only after methanol fixation. Bar, 1 μm. (B) Percoll gradient-purified EBs were washed twice with PBS; the input, wash fractions 1 and 2, and pellet were separated by SDS-PAGE and analyzed by Western blot analysis using antibodies against chlamydial OmpA, DnaK, and GroEL1. The wash fractions loaded contained three times as much protein as the pellet samples. Data are representative of several separate experiments.
We next asked what fraction of the C. pneumoniae GroEL1 protein is associated with the EB surface. In fractionation experiments, Percoll gradient-purified EBs were washed with PBS and the amounts of GroEL1 present in the pellet and supernatant were determined by Western blot analysis (Fig. 2B). Less than 10% of the input DnaK protein and less than 5% of the OmpA control protein were detected in the wash solutions, while approximately 50% of the total GroEL1 protein detected in the input was found in the wash solutions (Fig. 2B; compare lane 1 with lanes 2 and 3). Approximately 50% of the GroEL1 protein remained associated with the pelleted EBs (Fig. 2B, lane 4). This result suggests that at least half of the chlamydial GroEL1 protein is associated with the bacterial surface.
GroEL1-coated latex beads bind HEp-2 cells.
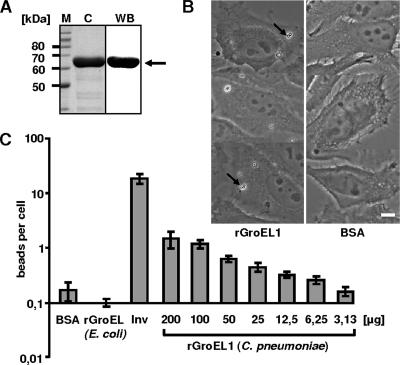
The localization of C. pneumoniae GroEL1 at the surface of infectious EBs suggested that the protein might interact with eukaryotic cells during the infection process. To test this possibility directly, HEp-2 cells were exposed to latex beads coated with purified His-tagged chlamydial rGroEL1 protein (Fig. 3A and B).
FIG. 3.
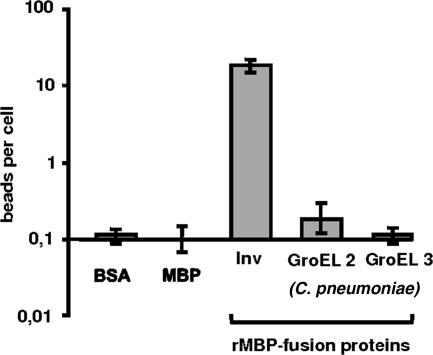
Adhesion of rGroEL1-coated latex beads to HEp-2 cells. (A) Coomassie blue-stained SDS-PAGE of the affinity-purified rGroEL1 protein after expression in E. coli (lane C) and analysis of purified rGroEL1 by Western blot analysis using an antibody against the N-terminal His tag (lane WB; the arrow marks the rGroEL1 protein band). Lane M, molecular mass markers. (B) Latex beads were coated with BSA or His-tagged recombinant proteins as indicated (protein concentration during coating, 200 μg/ml). Coated beads were incubated with HEp-2 cells. Adhesion of protein-coated latex beads was visualized by phase-contrast microscopy (magnification, ×63). Arrows indicate latex beads attached to HEp-2 cells. Bar, 10 μm. (C) Dose-dependent adhesion of rGroEL1-coated beads to HEp-2 cells. Beads were coated with different rGroEL1 protein concentrations. The control beads (BSA, Inv, and E. coli GroEL) were always coated with a protein concentration of 200 μg/ml. Results of the adhesion experiments are given as bound beads per HEp-2 cell (n = 1,000 HEp-2 cells; number of experiments = 4). Data shown are means ± standard deviations.
Light microscopy revealed that control BSA-coated latex beads rarely associated with HEp-2 cells (Fig. 3B and C). As a positive control, beads were coated with a known bacterial adhesin, invasin (InV) from Yersinia pseudotuberculosis (16). Beads coated with His-tagged rInv bound in large numbers to the eukaryotic cells (Fig. 3C). Interestingly, His-tagged rGroEL1-coated beads also showed a significant association with HEp-2 cells (Fig. 3C). In contrast His-tagged E. coli rGroEL protein did not mediate adhesion of beads to HEp-2 cells (Fig. 3C). The binding of C. pneumoniae rGroEL1-coated beads was dose dependent; when beads were coated with rGroEL1 at a concentration of 200 μg/ml, an average of 1 to 2 beads per cell was observed (Fig. 3C). These experiments indicate that the rGroEL1 protein from C. pneumoniae is able to adhere to eukaryotic cells.
GroEL1 mediates attachment of yeast cells to HEp-2 cells.
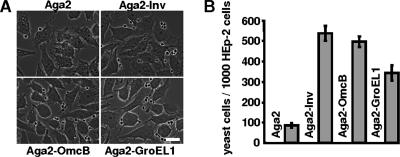
In order to confirm the results obtained with the latex bead assay, we employed the yeast display system, which allows one to study the interaction of bacterial proteins with human cells (55). To this end, full-length GroEL1 protein was expressed as a fusion with the yeast a-agglutinin receptor subunit Aga2p, which is anchored in the yeast cell wall via the Aga1p subunit. The plasmid-borne gene for the Aga2-GroEL1 fusion protein is expressed via the galactose-inducible GAL1 promoter. Aga2-GroEL1 could be detected on the yeast cell surface with anti-GroEL1 antibodies (data not shown). Yeast cells expressing Aga2-GroEL1 were incubated with HEp-2 cells seeded on glass coverslips. In contrast to yeast cells expressing Aga2p alone, yeast cells presenting the Aga2-GroEL1 fusion protein on the cell surface showed a significant affinity for HEp-2 cells, similar to that of yeast cells expressing either of two positive control proteins, the Inv protein and the C. pneumoniae OmcB protein (Fig. 4). Thus, the yeast display experiments verify that C. pneumoniae GroEL1 can confer the ability to adhere to human epithelial cells.
FIG. 4.
Yeast cells expressing Aga2-GroEL1 on the yeast cell surface adhere to human cells. (A) Untransformed yeast cells or yeast cells expressing Aga2, Aga2-Inv, Aga2-OmcB, or Aga2-GroEL1 from a plasmid were incubated with 1 × 105 HEp-2 cells, and the number of yeast cells associated with HEp-2 cells was determined microscopically. Shown are typical photomicrographs. Bar, 10 μm. (B) Diagrammatic representation of the number of yeast cells expressing Aga2, Aga2-Inv, Aga2-OmcB, and Aga2-GroEL-1 adhering to 1,000 HEp-2 cells (number of experiments = 4). Data shown are means ± standard deviations.
GroEL1, but not GroEL2 or GroEL3, can mediate adhesion to human cells.
We tested whether the ability of C. pneumoniae GroEL1 to adhere to HEp-2 cells was a general property of GroEL and other GroEL-like proteins. All chlamydial genomes so far sequenced have three groEL-like genes (groEL1, groEL2, and groEL3), which encode proteins that are between 20% and 60% identical to the prototype GroEL protein from E. coli (36, 62, 75). GroEL1 from C. pneumoniae strain GiD shows 33% and 24% identity to GroEL2 and GroEL3, respectively (see Fig. S2 in the supplemental material). To test whether these GroEL-like proteins all have the ability to adhere to HEp-2 cells, we expressed and purified MBP-tagged C. pneumoniae rGroEL2 and rGroEL3 and performed bead assays. None of these proteins exhibited binding to HEp-2 cells at levels significantly above those found for the negative controls MBP and BSA (Fig. 5). In contrast, the positive control, MBP-tagged Y. pseudotuberculosis Inv protein, mediated significant adhesion of beads to the human cells (Fig. 5). Together these experiments show that of the GroEL/GroEL-like proteins tested, GroEL1 is the only one capable of mediating binding to eukaryotic cells.
FIG. 5.
Assay for binding of C. pneumoniae rGroEL2- or rGroEL3-coated latex beads to HEp-2 cells. Latex beads were coated with the proteins indicated (protein concentration during coating, 200 μg/ml) and incubated with HEp-2 cells as described in Materials and Methods. Results of the binding experiments are given as bound beads per single HEp-2 cell (n = 1,000 HEp-2 cells; number of experiments = 4). Data shown are means ± standard deviations.
rGroEL1 protein from C. pneumoniae activates NF-κB in HUVECs.
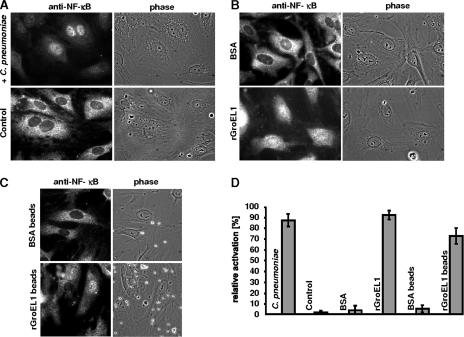
Infection of cultured endothelial cells by C. pneumoniae activates the ubiquitous nuclear factor NF-κB, which, upon translocation from the cytosol to the nucleus, up-regulates the expression of a variety of genes involved in inflammatory and proliferative responses (1, 15, 44). Chlamydia-induced activation of NF-κB leads to the up-regulation of genes for tissue factor, E-selectin, and the adhesion molecules ICAM-1 and VCAM-1 (22, 40, 44, 56). We asked whether the surface-localized GroEL1 from C. pneumoniae was the constituent activating NF-κB in endothelial cells. To test this hypothesis, primary HUVECs were cultivated and incubated with C. pneumoniae, with buffer alone, or with rGroEL1, and the number of cells showing translocation of NF-κB from the cytosol into the nucleus was determined. In all experiments using recombinant protein, the concentration of LPS endotoxin (binding of which also results in nuclear uptake of NF-κB) in the final GroEL1 preparations was <0.05 endotoxin unit/mg protein, as determined by the Limulus amoebocyte lysate assay. Incubation of HUVECs with infectious C. pneumoniae EBs induced translocation of NF-κB in 87% of the eukaryotic cells. In contrast, incubation with buffer resulted in translocation in fewer than 2% of the HUVECs (Fig. 6A and D). Likewise, when HUVECs were treated with BSA, only about 3% of the cells exhibited translocation of NF-κB. Interestingly, incubation of endothelial cells with soluble rGroEL1 protein resulted in translocation of NF-κB in up to 92% of HUVECs (Fig. 6B and D). Similarly, addition of rGroEL1-coated latex beads to HUVECs resulted in translocation of the transcription factor in about 72% of eukaryotic cells; BSA-coated beads induced translocation in only 5% (Fig. 6C and D). These data clearly imply that C. pneumoniae rGroEL1 on its own is capable of activating endothelial cells by stimulating translocation of NF-κB.
FIG. 6.
C. pneumoniae rGroEL1 activates NF-κB translocation in HUVECs. (A) HUVECs were infected with C. pneumoniae or as control with PBS for 2 h. NF-κB localization was determined by indirect immunofluorescence microscopy as described in Materials and Methods. (B) HUVECs were stimulated with 20 μg/ml rGroEL1 or with BSA for 5 h. NF-κB localization was determined as described above. (C) HUVECs were stimulated with rGroEL1-coated latex beads or BSA-coated latex beads for 5 h. NF-κB localization was determined as described above. (D) Quantification of NF-κB activation in the experiments shown in panels A, B, and C. Bars represent the ratio of NF-κB translocation into the nucleus to the total number of HUVECs cells. Each value represents the mean from three experiments ± the standard deviation.
Preincubation of target cells with C. pneumoniae rGroEL1 protein inhibits infection by C. pneumoniae.
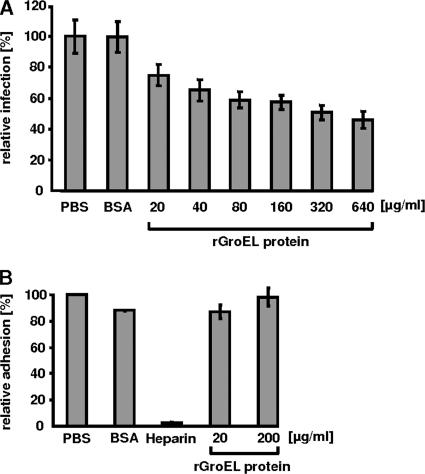
To characterize more directly the possible biological role of surface-exposed GroEL1 during the initial phase of infection, we performed neutralization experiments with the purified recombinant protein. Preincubation of HEp-2 cells with 20 μg/ml rGroEL1 prior to exposure to C. pneumoniae EBs was associated with a significant (24%) reduction in the number of inclusions formed, relative to the controls pretreated with PBS alone (Fig. 7A). The negative control BSA had no effect on the infection rate. The inhibitory effect of rGroEL1 on infection was dose dependent, and maximum inhibition was observed at an rGroEL1 concentration of 640 μg/ml (Fig. 7A). This dose-dependent reduction in infectivity suggests that GroEL1 protein plays a role in establishing C. pneumoniae infections.
FIG. 7.
Soluble chlamydial rGroEL1 protein does not interfere with EB attachment to human cells but reduces subsequent infection by C. pneumoniae. (A) HEp-2 cells were incubated with increasing amounts (20 μg/ml to 640 μg/ml) of rGroEL1 for 2 h prior to infection with purified C. pneumoniae EBs. At 60 h postinfection, cells were fixed and analyzed by indirect immunofluorescence microscopy as described in Materials and Methods. HEp-2 cells pretreated with PBS or 200 μg/ml BSA prior to infection with C. pneumoniae served as controls. The number of chlamydial inclusions was determined, and the value found for the PBS control was set to 100% (n = 20 microscopic fields; number of experiments = 4). (B) Binding of purified, viable, CFSE-stained C. pneumoniae EBs to HEp-2 cells was detected by flow cytometric analysis. In control experiments, the attachment of C. pneumoniae EBs to HEp-2 cells incubated with PBS, 200 μg/ml BSA, or 500 μg/ml heparin was monitored and compared with binding rates in samples to which 20 μg/ml or 200 μg/ml rGroEL1 protein had been added (number of experiments = 4). Error bars indicate standard deviations.
Preincubation of target cells with rGroEL1 protein does not interfere with the adhesion of C. pneumoniae EBs.
The reduction of infectivity mediated by rGroEL1 could be brought about in two ways, which are not mutually exclusive: addition of soluble rGroEL1 either could directly block receptors on the HEp-2 cell surface that are required for attachment and/or internalization of EBs or could induce the premature activation of pathways that negatively influence the establishment of infection (e.g., via the NF-κB activation observed in HUVECs [Fig. 6]). In order to test whether surface-localized chlamydial GroEL1 is a prerequisite for adhesion of EBs to host cells, flow cytometric analyses of binding of viable CFSE-labeled EBs to HEp-2 cells were performed. The inclusion of heparin led to a 95% decrease in EB attachment, whereas the addition of rGroEL1 at 20 μg/ml or 200 μg/ml had no inhibitory effect on EB binding to Hep-2 cells (Fig. 7B). This result suggests that GroEL1 seems not to play a role in EB attachment.
DISCUSSION
This study demonstrates that C. pneumoniae GroEL1 is associated with the surface of infectious EBs and can be found in the inclusion throughout the later course of the infection. Fractionation experiments revealed that approximately half of the total GroEL1 found in EBs is localized on their surfaces. Latex beads coated with rGroEL1 adhered to HEp-2 cells in a dose-dependent way, while beads treated with the other two GroEL homologues from C. pneumoniae and GroEL from E. coli failed to do so. The binding properties of GroEL1 were confirmed using yeast display assays. Incubation of HUVECs with soluble rGroEL1 or with rGroEL1-coated beads resulted in translocation of NF-κB into the nucleus. Finally, infection by C. pneumoniae was inhibited in a dose-dependent manner by preincubation of HEp-2 cells with soluble rGroEL1 protein. However, direct adhesion experiments revealed that EB binding to HEp-2 cells seems not to be mediated by GroEL1.
GroEL1 protein is associated with the EB surface and the chlamydial inclusion during the developmental cycle.
Our results support and extend earlier findings reported on GroEL1 localization. Early biochemical experiments had already suggested that GroEL1 is associated with outer membrane fractions of infectious EBs from C. trachomatis and C. psittaci, from which it could be released using mild detergents (3). More recently, electron microscopy studies detected C. trachomatis GroEL1 antigens in inclusions not only in the bacterial cytoplasm but also at the chlamydial envelope and in the lumen of the inclusion (61). This has recently been described for C. pneumoniae GroEL1 also. In this case the protein was detected by electron microscopy in inclusions of replication-permissive epithelial HEp-2 cells as well as in the extracellular space (12). Our immunolocalization data also prove that C. pneumoniae GroEL1 protein is the inclusion but outside the chlamydial particles as well as on the surface of infectious EBs. Furthermore, our data provide evidence that at least 50% of the GroEL1 in isolated infectious EBs is surface associated, as it could be recovered by washing the EBs with PBS. Bacterial cell lysis was excluded as a possible source for this fraction of the protein by the finding that an intrachlamydial protein, DnaK, was not enriched in the soluble phase under these conditions. Most probably the relative amount of GroEL1 associated with the EB surface is higher in vivo, since the EBs used in the fractionation experiments had been purified using Percoll gradients, which might have stripped some of the GroEL1 from their surfaces. The fraction of GroEL1 that remains associated with the EBs is presumably located in the bacterial cytoplasm, where it probably serves as a molecular chaperonin. The fact that chlamydial GroEL1 (but not GroEL2 and GroEL3) can functionally substitute for the temperature-sensitive endogenous GroEL in an E. coli mutant strongly suggests that it can function as a chaperonin (37). Consequently, the differential compartmentalization of GroEL1 would suggest a dual role for this protein, and thus GroEL1 might belong to the growing group of “moonlighting proteins” (34).
Extracellular localization of and a role for GroEL1 in the molecular cross talk between a pathogenic microorganism and its host cell are unusual features for a heat shock protein, but several precedents for such multifunctionality now exist. Thus, an extracellular localization for GroEL has been reported in Bartonella bacilliformis, Helicobacter pylori, Legionella pneumophila, and Francisella tularensis (reviewed in references 21, 27, 29, 48, and 54). H. pylori, Clostridium difficile, Haemophilus ducreyi, and Salmonella enterica serovar Typhimurium all use surface GroEL-like proteins to promote attachment to human cells (reviewed in reference 27). In the lactic acid bacterium Lactobacillus johnsonii, a natural inhabitant of the gastrointestinal tracts of mammals, GroEL protein was found at the bacterial surface, where it may promote attachment to mucus and epithelial cells, stimulate cytokine secretion, and mediate aggregation of the gastric pathogen H. pylori (4). Likewise, the GroEL-like protein found on the surface of virulent L. pneumophila strains acts as an adhesin and mediates internalization and specific trafficking of latex particles into HeLa cells (21). In the eukaryotic dimorphic fungal pathogen Histoplasma capsulatum, a surface-localized GroEL-like protein mediates attachment of the facultatively intracellular fungus to β2 integrin receptors (CD11/CD18) on macrophages (49). Thus, it is feasible that in various, mainly pathogenic, organisms, GroEL and GroEL-like proteins have been diversified during evolution and have evolved new functions associated with different subcellular localization patterns. For example, in the endosymbiont Enterobacter aerogenes, the GroEL-like protein differs at 11 positions from E. coli GroEL and is found both intra- and extracellularly, acting as a paralyzing toxin on insects (87).
The GroEL proteins, which are thought to be prototypes of cytosolic proteins, are probably not the only cytosolic proteins that can also be diverted to the bacterial cell surface. Recently, the translation elongation factor EF-Tu was found to be present on the surface of L. johnsonii cells, mediating their attachment to human cells (24). Characterization of the cell wall subproteome of Listeria monocytogenes has revealed the presence of GroEL, enolase, and EF-Tu, among other typical “cytoplasmic” proteins (68). Like C. pneumoniae GroEL1, these proteins do not carry any obvious secretion signals or known surface-binding domains. The mechanism by which they reach the surface of the bacteria therefore remains unclear. It has been speculated, however, that these “moonlighting” proteins might be exported by ABC transporters or by passive release upon osmotic stress (68). Alternatively, these proteins might be exported by the so-called type VI secretion pathway described for gram-negative bacteria. Many of them release vesicles from their outer membrane as secretory vehicles for components relevant for bacterial adhesion and invasion, cytotoxicity, and host responses (45).
rGroEL1 activates primary human endothelial cells.
In the present study we have demonstrated that C. pneumoniae rGroEL1 mediates the activation of NF-κB in HUVECs and that soluble GroEL1 and latex beads coated with the protein are almost equally efficient in doing so. Activation and translocation of NF-κB to the nucleus in eukaryotic cells infected by C. pneumoniae have been demonstrated in different studies (6, 40, 44). Activation of NF-κB is an important step in the pathogen-induced eukaryotic signaling observed for a variety of pathogens, such as H. pylori, enterohemorrhagic E. coli, or Campylobacter jejuni (38, 53, 67).
Activation of NF-κB during infection by C. pneumoniae has been described for a variety of different cell types (e.g., epithelial cells, HUVECs, human monocytes, and Mono Mac cells) (22, 44, 82, 83). The activation of NF-κB in HUVECs is very fast, as it could be detected within 15 min of infection with C. pneumoniae (44). Similarly, in Mono Mac cells NF-κB activation peaked within the first 60 min after infection (83, 82). This strongly suggests that the intracellular signaling events leading to the translocation of NF-κB are induced upon initial contact of the chlamydial particles with the eukaryotic cells. In this respect the activation of NF-κB by GroEL1-coated latex beads may mimic the contact of C. pneumoniae EBs with eukaryotic cells.
Infection with C. pneumoniae triggers additional host cell responses, such as phosphorylation of the mitogen-activated kinase p40/42; secretion of interleukin-1 (IL-1), IL-8, IL-10, and monocyte chemoattractant protein 1; and the expression of cell surface receptors such as ICAM, VCAM, E-selectin, and tissue factor (6, 9, 15, 41, 44, 56, 66, 83). Several of these host cell responses were also observed when eukaryotic cells were incubated with C. trachomatis GroEL1 (41, 66). This suggests that such activation might be a general property of chlamydial GroEL1 proteins. Indeed C. pneumoniae rGroEL1 initiates secretion of IL-12 and tumor necrosis factor via TLRs in dendritic cells and activates vascular endothelial cells to secrete the cytokines IL-1β, IL-6, and IL-8 and to express cell surface adhesion proteins (12, 51). Therefore, a feasible model is one in which surface localization of C. pneumoniae GroEL1 contributes to the pathogen-provoked activation of eukaryotic host cells upon chlamydial attachment and subsequently triggers the intracellular signal transduction pathways within the eukaryotic cells, as described by others (31, 40, 44, 56, 66, 82, 83). The nature of the GroEL1 receptor on the host cell is still unclear. However, it has been reported that GroEL1 from C. pneumoniae binds to CD14, TLR4, and/or TLR2 (9, 12, 41, 51).
The surface localization of C. pneumoniae GroEL1 on EBs and the activation of HUVECs by rGroEL1 strongly suggest that GroEL1 plays an important role in the initial phase of the pathogen-host interaction, and indeed our data directly support this idea, as pretreatment of HEp-2 cells with rGroEL1 significantly reduces subsequent infectivity. However, pretreatment of the epithelial cells with rGroEL1 did not inhibit adhesion of infectious EBs, suggesting that bacterial cell surface-localized GroEL1 is not required for attachment of EBs to HEp-2 cells. It can be speculated, however, that GroEL1 binding to eukaryotic receptors might have an influence on later steps in the establishment of the infection; e.g., the protein might be relevant for the internalization of EBs or for the modification of the early inclusion. Thus, a premature and/or hyperactive response induced in the HEp-2 cells by rGroEL1 prior to addition of the EBs might well be deleterious for the host and/or for a successful infection. The fact that rGroEL1 can activate NF-κB translocation in HUVECs with kinetics similar to those found during a normal C. pneumoniae infection supports this signaling hypothesis.
In conclusion, our data provide evidence that a fraction of the C. pneumoniae GroEL1 protein is localized on the bacterial cell surface and plays an important role in the establishment of the chlamydial infection, possibly by inducing early signaling events that help to convert the eukaryotic cell into a hospitable environment suitable for the establishment of a successful infection.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 590, project number C5 (to J.H.H.) and the Era-NET PathoGenoMics network ECIBUG.
We thank Ursula Fleig for critical reading of the manuscript. The assistance of Aleksandra Culjak in constructing plasmid pAC-2 is greatly appreciated.
The authors do not have a commercial or other association that might pose a conflict of interest.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baldwin, A. S., Jr. 2001. Series introduction: the transcription factor NF-kappaB and human disease. J. Clin. Investig. 1073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batteiger, B. E., W. J. T. Newhall, and R. B. Jones. 1985. Differences in outer membrane proteins of the lymphogranuloma venereum and trachoma biovars of Chlamydia trachomatis. Infect. Immun. 50488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavoil, P., R. S. Stephens, and S. Falkow. 1990. A soluble 60 kilodalton antigen of Chlamydia spp. is a homologue of Escherichia coli GroEL. Mol. Microbiol. 4461-469. [DOI] [PubMed] [Google Scholar]
- 4.Bergonzelli, G. E., D. Granato, R. D. Pridmore, L. F. Marvin-Guy, D. Donnicola, and I. E. Corthesy-Theulaz. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkelund, S., A. G. Lundemose, and G. Christiansen. 1990. The 75-kilodalton cytoplasmic Chlamydia trachomatis L2 polypeptide is a DnaK-like protein. Infect. Immun. 582098-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blessing, E., C. C. Kuo, T. M. Lin, L. A. Campbell, F. Bea, B. Chesebro, and M. E. Rosenfeld. 2002. Foam cell formation inhibits growth of Chlamydia pneumoniae but does not attenuate Chlamydia pneumoniae-induced secretion of proinflammatory cytokines. Circulation 1051976-1982. [DOI] [PubMed] [Google Scholar]
- 7.Brunham, R. C., and R. W. Peeling. 1994. Chlamydia trachomatis antigens: role in immunity and pathogenesis. Infect. Agents Dis. 3218-233. [PubMed] [Google Scholar]
- 8.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92351-366. [DOI] [PubMed] [Google Scholar]
- 9.Bulut, Y., E. Faure, L. Thomas, H. Karahashi, K. S. Michelsen, O. Equils, S. G. Morrison, R. P. Morrison, and M. Arditi. 2002. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J. Immunol. 1681435-1440. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, L. A., and C. C. Kuo. 2006. Interactions of Chlamydia with the host cells that mediate attachment and uptake, p. 505-522. In P. M. Bavoil and P. B. Wyrick (ed.), Chlamydia genomics and pathogenesis. Horizon Bioscience, Norfolk, United Kingdom.
- 11.Conant, C. G., and R. S. Stephens. 2007. Chlamydia attachment to mammalian cells requires protein disulfide isomerase. Cell. Microbiol. 9222-232. [DOI] [PubMed] [Google Scholar]
- 12.Costa, C. P., C. J. Kirschning, D. Busch, S. Durr, L. Jennen, U. Heinzmann, S. Prebeck, H. Wagner, and T. Miethke. 2002. Role of chlamydial heat shock protein 60 in the stimulation of innate immune cells by Chlamydia pneumoniae. Eur. J. Immunol. 322460-2470. [DOI] [PubMed] [Google Scholar]
- 13.Crane, D. D., J. H. Carlson, E. R. Fischer, P. Bavoil, R. C. Hsia, C. Tan, C. C. Kuo, and H. D. Caldwell. 2006. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc. Natl. Acad. Sci. USA 1031894-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, C. H., J. E. Raulston, and P. B. Wyrick. 2002. Protein disulfide isomerase, a component of the estrogen receptor complex, is associated with Chlamydia trachomatis serovar E attached to human endometrial epithelial cells. Infect. Immun. 703413-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dechend, R., M. Maass, J. Gieffers, R. Dietz, C. Scheidereit, A. Leutz, and D. C. Gulba. 1999. Chlamydia pneumoniae infection of vascular smooth muscle and endothelial cells activates NF-kappaB and induces tissue factor and PAI-1 expression: a potential link to accelerated arteriosclerosis. Circulation 1001369-1373. [DOI] [PubMed] [Google Scholar]
- 16.Dersch, P., and R. R. Isberg. 1999. A region of the Yersinia pseudotuberculosis invasin protein enhances integrin-mediated uptake into mammalian cells and promotes self-association. EMBO J. 181199-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckert, L. O., S. E. Hawes, P. Wolner Hanssen, D. M. Money, R. W. Peeling, R. C. Brunham, C. E. Stevens, D. A. Eschenbach, and W. E. Stamm. 1997. Prevalence and correlates of antibody to chlamydial heat shock protein in women attending sexually transmitted disease clinics and women with confirmed pelvic inflammatory disease. J. Infect. Dis. 1751453-1458. [DOI] [PubMed] [Google Scholar]
- 18.Entian, K. D., T. Schuster, J. H. Hegemann, D. Becher, H. Feldmann, U. Guldener, R. Gotz, M. Hansen, C. P. Hollenberg, G. Jansen, W. Kramer, S. Klein, P. Kotter, J. Kricke, H. Launhardt, G. Mannhaupt, A. Maierl, P. Meyer, W. Mewes, T. Munder, R. K. Niedenthal, M. Ramezani Rad, A. Rohmer, A. Romer, A. Hinnen, et al. 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262683-702. [DOI] [PubMed] [Google Scholar]
- 19.Fadel, S., and A. Eley. 2007. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J. Med. Microbiol. 5615-22. [DOI] [PubMed] [Google Scholar]
- 20.Fleshner, M., and J. D. Johnson. 2005. Endogenous extra-cellular heat shock protein 72: releasing signal(s) and function. Int. J. Hyperthermia 21457-471. [DOI] [PubMed] [Google Scholar]
- 21.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 664602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gencay, M. M., M. Tamm, A. Glanville, A. P. Perruchoud, and M. Roth. 2003. Chlamydia pneumoniae activates epithelial cell proliferation via NF-κB and the glucocorticoid receptor. Infect. Immun. 715814-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerard, H. C., K. L. Wildt, J. A. Whittum-Hudson, Z. Lai, J. Ager, and A. P. Hudson. 2005. The load of Chlamydia pneumoniae in the Alzheimer's brain varies with APOE genotype. Microb. Pathog. 3919-26. [DOI] [PubMed] [Google Scholar]
- 24.Granato, D., G. E. Bergonzelli, R. D. Pridmore, L. Marvin, M. Rouvet, and I. E. Corthesy-Theulaz. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 722160-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grayston, J. T. 1989. Chlamydia pneumoniae, strain TWAR. Chest 95664-669. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, D. L., and R. McDonald. 1998. Can acute Chlamydia pneumoniae respiratory tract infection initiate chronic asthma? Ann. Allergy Asthma Immunol. 81339-344. [DOI] [PubMed] [Google Scholar]
- 27.Henderson, B., E. Allan, and A. R. Coates. 2006. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 743693-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horner, P. J., D. Cain, M. McClure, B. J. Thomas, C. Gilroy, M. Ali, J. N. Weber, and D. Taylor-Robinson. 1997. Association of antibodies to Chlamydia trachomatis heat-shock protein 60 kD with chronic nongonococcal urethritis. Clin. Infect. Dis. 24653-660. [DOI] [PubMed] [Google Scholar]
- 29.Huesca, M., S. Borgia, P. Hoffman, and C. A. Lingwood. 1996. Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect. Immun. 642643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 10411430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn, H. U., M. Krull, F. N. Wuppermann, A. C. Klucken, S. Rosseau, J. Seybold, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 2000. Infection and activation of airway epithelial cells by Chlamydia pneumoniae. J. Infect. Dis. 1821678-1687. [DOI] [PubMed] [Google Scholar]
- 32.Jantos, C. A., S. Heck, R. Roggendorf, M. Sen-Gupta, and J. H. Hegemann. 1997. Antigenic and molecular analyses of different Chlamydia pneumoniae strains. J. Clin. Microbiol. 35620-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jantos, C. A., C. Krombach, F. N. Wuppermann, A. Gardemann, S. Bepler, H. Asslan, J. H. Hegemann, and W. Haberbosch. 2000. Antibody response to the 60-kDa heat-shock protein of Chlamydia pneumoniae in patients with coronary artery disease. J. Infect. Dis. 1811700-1705. [DOI] [PubMed] [Google Scholar]
- 34.Jeffery, C. J. 1999. Moonlighting proteins. Trends Biochem. Sci. 248-11. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 37.Karunakaran, K. P., Y. Noguchi, T. D. Read, A. Cherkasov, J. Kwee, C. Shen, C. C. Nelson, and R. C. Brunham. 2003. Molecular analysis of the multiple GroEL proteins of chlamydiae. J. Bacteriol. 1851958-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keates, S., Y. S. Hitti, M. Upton, and C. P. Kelly. 1997. Helicobacter pylori infection activates NF-kappa B in gastric epithelial cells. Gastroenterology 1131099-1109. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen, K., A. S. Madsen, P. Mygind, G. Christiansen, and S. Birkelund. 1999. Identification of two novel genes encoding 97- to 99-kilodalton outer membrane proteins of Chlamydia pneumoniae. Infect. Immun. 67375-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kol, A., T. Bourcier, A. H. Lichtman, and P. Libby. 1999. Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J. Clin. Investig. 103571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kol, A., A. H. Lichtman, R. W. Finberg, P. Libby, and E. A. Kurt-Jones. 2000. Heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. J. Immunol. 16413-17. [DOI] [PubMed] [Google Scholar]
- 42.Korppi, M., T. Heiskanen Kosma, E. Jalonen, P. Saikku, M. Leinonen, P. Halonen, and P. H. Makela. 1993. Aetiology of community-acquired pneumonia in children treated in hospital. Eur. J. Pediatr. 15224-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krull, M., P. Bockstaller, F. N. Wuppermann, A. C. Klucken, J. Muhling, B. Schmeck, J. Seybold, C. Walter, M. Maass, S. Rosseau, J. H. Hegemann, N. Suttorp, and S. Hippenstiel. 2006. Mechanisms of Chlamydophila pneumoniae-mediated GM-CSF release in human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 34375-382. [DOI] [PubMed] [Google Scholar]
- 44.Krüll, M., A. C. Klucken, F. N. Wuppermann, O. Fuhrmann, C. Magerl, J. Seybold, S. Hippenstiel, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 1624834-4841. [PubMed] [Google Scholar]
- 45.Kuehn, M. J., and N. C. Kesty. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 192645-2655. [DOI] [PubMed] [Google Scholar]
- 46.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuo, C. C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S. I. Hakomori. 1996. An N linked high mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to Hela cells. J. Clin. Investig. 982813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, B. Y., M. A. Horwitz, and D. L. Clemens. 2006. Identification, recombinant expression, immunolocalization in macrophages, and T-cell responsiveness of the major extracellular proteins of Francisella tularensis. Infect. Immun. 744002-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long, K. H., F. J. Gomez, R. E. Morris, and S. L. Newman. 2003. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J. Immunol. 170487-494. [DOI] [PubMed] [Google Scholar]
- 50.Lundemose, A. G., S. Birkelund, P. M. Larsen, S. J. Fey, and G. Christiansen. 1990. Characterization and identification of early proteins in Chlamydia trachomatis serovar L2 by two-dimensional gel electrophoresis. Infect. Immun. 582478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maguire, M., S. Poole, A. R. Coates, P. Tormay, C. Wheeler-Jones, and B. Henderson. 2005. Comparative cell signalling activity of ultrapure recombinant chaperonin 60 proteins from prokaryotes and eukaryotes. Immunology 115231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melby, K. K., T. K. Kvien, A. Glennas, and G. Anestad. 1999. Chlamydia pneumoniae as a trigger of reactive arthritis. Scand. J. Infect. Dis. 31327-328. [DOI] [PubMed] [Google Scholar]
- 53.Mellits, K. H., J. Mullen, M. Wand, G. Armbruster, A. Patel, P. L. Connerton, M. Skelly, and I. F. Connerton. 2002. Activation of the transcription factor NF-kappaB by Campylobacter jejuni. Microbiology 1482753-2763. [DOI] [PubMed] [Google Scholar]
- 54.Minnick, M. F., L. S. Smitherman, and D. S. Samuels. 2003. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of GroEL. Infect. Immun. 716933-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moelleken, K., and J. H. Hegemann. 2008. The Chlamydia outer membrane protein OmcB is required for adhesion and exhibits biovar-specific differences in glycosaminoglycan binding. Mol. Microbiol. 67403-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molestina, R. E., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1999. Infection of human endothelial cells with Chlamydia pneumoniae stimulates transendothelial migration of neutrophils and monocytes. Infect. Immun. 671323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrison, R. P. 1991. Chlamydial hsp60 and the immunopathogenesis of chlamydial disease. Semin. Immunol. 325-33. [PubMed] [Google Scholar]
- 58.Pfister, G., C. M. Stroh, H. Perschinka, M. Kind, M. Knoflach, P. Hinterdorfer, and G. Wick. 2005. Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J. Cell Sci. 1181587-1594. [DOI] [PubMed] [Google Scholar]
- 59.Rank, R. G., C. Dascher, A. K. Bowlin, and P. M. Bavoil. 1995. Systemic immunization with Hsp60 alters the development of chlamydial ocular disease. Investig. Ophthalmol. Vis. Sci. 361344-1351. [PubMed] [Google Scholar]
- 60.Raulston, J. E., C. H. Davis, T. R. Paul, J. D. Hobbs, and P. B. Wyrick. 2002. Surface accessibility of the 70-kilodalton Chlamydia trachomatis heat shock protein following reduction of outer membrane protein disulfide bonds. Infect. Immun. 70535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raulston, J. E., T. R. Paul, S. T. Knight, and P. B. Wyrick. 1998. Localization of Chlamydia trachomatis heat shock proteins 60 and 70 during infection of a human endometrial epithelial cell line in vitro. Infect. Immun. 662323-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reynolds, D. J., and J. H. Pearce. 1991. Endocytic mechanisms utilized by chlamydiae and their influence on induction of productive infection. Infect. Immun. 593033-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii983-986. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Campillo, M., L. Bini, M. Comanducci, R. Raggiaschi, B. Marzocchi, V. Pallini, and G. Ratti. 1999. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis 202269-2279. [DOI] [PubMed] [Google Scholar]
- 66.Sasu, S., D. LaVerda, N. Qureshi, D. T. Golenbock, and D. Beasley. 2001. Chlamydia pneumoniae and chlamydial heat shock protein 60 stimulate proliferation of human vascular smooth muscle cells via Toll-like receptor 4 and p44/p42 mitogen-activated protein kinase activation. Circ. Res. 89244-250. [DOI] [PubMed] [Google Scholar]
- 67.Savkovic, S. D., A. Koutsouris, and G. Hecht. 1997. Activation of NF-kappaB in intestinal epithelial cells by enteropathogenic Escherichia coli. Am. J. Physiol. 273C1160-1167. [DOI] [PubMed] [Google Scholar]
- 68.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 42991-3006. [DOI] [PubMed] [Google Scholar]
- 69.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16339-346. [DOI] [PubMed] [Google Scholar]
- 70.Schmiel, D. H., S. T. Knight, J. E. Raulston, J. Choong, C. H. Davis, and P. B. Wyrick. 1991. Recombinant Escherichia coli clones expressing Chlamydia trachomatis gene products attach to human endometrial epithelial cells. Infect. Immun. 594001-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnitger, K., F. Njau, U. Wittkop, A. Liese, J. G. Kuipers, A. Thiel, M. A. Morgan, H. Zeidler, and A. D. Wagner. 2007. Staining of Chlamydia trachomatis elementary bodies: a suitable method for identifying infected human monocytes by flow cytometry. J. Microbiol. Methods 69116-121. [DOI] [PubMed] [Google Scholar]
- 72.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 73.Shin, B. K., H. Wang, A. M. Yim, F. Le Naour, F. Brichory, J. H. Jang, R. Zhao, E. Puravs, J. Tra, C. W. Michael, D. E. Misek, and S. M. Hanash. 2003. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 2787607-7616. [DOI] [PubMed] [Google Scholar]
- 74.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282754-759. [DOI] [PubMed] [Google Scholar]
- 76.Stephens, R. S., K. Koshiyama, E. Lewis, and A. Kubo. 2001. Heparin-binding outer membrane protein of chlamydiae. Mol. Microbiol. 40691-699. [DOI] [PubMed] [Google Scholar]
- 77.Stratton, C. W., and S. Sriram. 2003. Association of Chlamydia pneumoniae with central nervous system disease. Microbes Infect. 51249-1253. [DOI] [PubMed] [Google Scholar]
- 78.Su, H., L. Raymond, D. D. Rockey, E. Fischer, T. Hackstadt, and H. D. Caldwell. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. USA 9311143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thom, D. H., J. T. Grayston, L. A. Campbell, C. C. Kuo, V. K. Diwan, and S. P. Wang. 1994. Respiratory infection with Chlamydia pneumoniae in middle-aged and older adult outpatients. Eur. J. Clin. Microbiol. Infect. Dis. 13785-792. [DOI] [PubMed] [Google Scholar]
- 80.Ting, L. M., R. C. Hsia, C. G. Haidaris, and P. M. Bavoil. 1995. Interaction of outer envelope proteins of Chlamydia psittaci GPIC with the HeLa cell surface. Infect. Immun. 633600-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vabulas, R. M., P. Ahmad-Nejad, C. da Costa, T. Miethke, C. J. Kirschning, H. Hacker, and H. Wagner. 2001. Endocytosed HSP60s use Toll-like receptor 2 (TLR2) and TLR4 to activate the Toll/interleukin-1 receptor signaling pathway in innate immune cells. J. Biol. Chem. 27631332-31339. [DOI] [PubMed] [Google Scholar]
- 82.Wahl, C., S. Maier, R. Marre, and A. Essig. 2003. Chlamydia pneumoniae induces the expression of inhibitor of apoptosis 2 (c-IAP2) in a human monocytic cell line by an NF-kappaB-dependent pathway. Int. J. Med. Microbiol. 293377-381. [DOI] [PubMed] [Google Scholar]
- 83.Wahl, C., F. Oswald, U. Simnacher, S. Weiss, R. Marre, and A. Essig. 2001. Survival of Chlamydia pneumoniae-infected Mono Mac 6 cells is dependent on NF-κB binding activity. Infect. Immun. 697039-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward, M. E. 1999. Mechanisms of chlamydia-induced disease, p. 171-210. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 85.Wehrl, W., V. Brinkmann, P. R. Jungblut, T. F. Meyer, and A. J. Szczepek. 2004. From the inside out—processing of the chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51319-334. [DOI] [PubMed] [Google Scholar]
- 86.Xu, Q. 2003. Infections, heat shock proteins, and atherosclerosis. Curr. Opin. Cardiol. 18245-252. [DOI] [PubMed] [Google Scholar]
- 87.Yoshida, N., K. Oeda, E. Watanabe, T. Mikami, Y. Fukita, K. Nishimura, K. Komai, and K. Matsuda. 2001. Chaperonin turned insect toxin. Nature 41144. [DOI] [PubMed] [Google Scholar]
- 88.Yuan, Y., K. Lyng, Y. X. Zhang, D. D. Rockey, and R. P. Morrison. 1992. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect. Immun. 602288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yucesan, C., and S. Sriram. 2001. Chlamydia pneumoniae infection of the central nervous system. Curr. Opin. Neurol. 14355-359. [DOI] [PubMed] [Google Scholar]
- 90.Zhang, J. P., and R. S. Stephens. 1992. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell 69861-869. [DOI] [PubMed] [Google Scholar]
- 91.Zügel, U., and S. H. E. Kaufmann. 1999. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1219-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.