Abstract
The system I cytochrome c biogenesis pathway requires CcmD, a small polypeptide of 69 residues in Escherichia coli. Here it is shown that CcmD is a component of the CcmABC ATP-binding cassette transporter complex. CcmD is not necessary for the CcmC-dependent transfer of heme to CcmE in the periplasm or for interaction of CcmE with CcmABC. CcmD is absolutely required for the release of holo-CcmE from the CcmABCD complex. Evidence is presented that the topology of CcmD in the cytoplasmic membrane is the N terminus outside and the C terminus inside with one transmembrane domain.
Cytochromes c are electron transport proteins involved in respiration and photosynthesis. Typical c-type cytochromes are characterized by the covalent attachment of the two heme vinyl groups via thioether linkages to the cysteine residues of the conserved CXXCH signature motif, where the histidine residue serves as an axial ligand to the heme iron. Cytochromes c are synthesized at their site of action (e.g., outside the cytoplasmic membrane of bacteria) by one of three systems (designated systems I, II, and III [for reviews, see references 16, 24, and 26]). The system I pathway is the most complex of the three and is found in plant and some protozoan mitochondria (5, 14), archaea (3, 11), and α- and γ-proteobacteria (4, 7, 18, 19). The system I pathway of Escherichia coli is composed of eight integral membrane proteins encoded by the ccmABCDEFGH operon. The functions of some of these proteins have been established. CcmE takes heme from CcmC and acts as a periplasmic heme chaperone (9, 22, 23), and CcmC-bound holo-CcmE can function as a heme reservoir (10). Because heme is bound with high affinity to CcmC (9), an ATP-dependent CcmABC release complex (stoichiometry, A2B1C1) is required to release heme (as holo-CcmE). Holo-CcmE is putatively the substrate for the cytochrome c synthetase (CcmF/H) for subsequent heme ligation to apocytochrome c (2, 9). Using recombinant systems I (E. coli) and II (Helicobacter hepaticus CcsBA) in an E. coli strain with the ccm operon deleted (Δccm) and endogenous heme synthesis deleted (ΔhemA), we have shown that system I has at least a fivefold-higher affinity for heme than system II (21). This suggests that the heme reservoir holo-CcmE and the CcmA2B1C1 release complex allow system I to use heme at endogenous levels lower than the levels used by system II, thus potentially giving an organism using the system I pathway a distinct physiological advantage over an organism using the system II pathway (21).
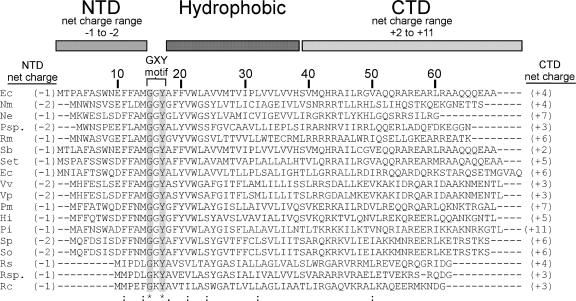
The function of the short CcmD protein has been a question of considerable interest (1, 13, 20). While CcmD shows little conservation of the primary sequence (Fig. 1), the predicted domain structure consists of a negatively charged N-terminal domain (NTD), a hydrophobic region, and a positively charged C-terminal domain (CTD). In Rhodobacter capsulatus a functional CcmD (HelD)-LacZ fusion coimmunoprecipitated with CcmA, suggesting that it is part of a CcmABC complex and that the CTD is in the cytoplasm (13). In E. coli, CcmD has been shown by coimmunoprecipitation to interact with CcmC and to a lesser extent with CcmE (1). Ahuja and Thony-Meyer concluded that CcmD is an “assembly factor of membrane proteins” involved in “increased incorporation (of CcmE) into the membrane” (1). Recently, we have shown (9) that CcmC, but not CcmD, is necessary for interaction with CcmE (see below). In the present report we characterize the E. coli CcmA2B1C1 release complex and define an essential function for CcmD in the system I pathway. We show that CcmD is not required for the transfer of heme to CcmE but that, as part of the CcmABC release complex, CcmD is required for the release of holo-CcmE from this complex. We also reevaluate the membrane topology of the CcmD protein in E. coli and provide evidence that supports the topology proposed for R. capsulatus CcmD (13), with the NTD oriented in the periplasm and the CTD oriented in the cytoplasm.
FIG. 1.
Alignment of the E. coli CcmD protein with representative bacterial CcmD sequences. The amino acid sequences of CcmD proteins of E. coli K-12 strain MG1655 (accession no. POABM5) (Ec), Nitrospira multiformis ATCC 2516 (YP_411906) (Nm), Nitrosomonas eutropha C71 (YP_747848) (Ne), Polynucleobacter sp. strain QLW-PIDMWA-1 (Psp.), Ralstonia metallidurans CH34 (YP_584534) (Rm), Shigella boydii Sb227 (YP_408515) (Sb), Salmonella enterica subsp. enterica serovar Typhi CT1 (NP_456796) (Set), Erwinia carotovora subsp. atroseptica SCRI1043 (YP_049982) (Ec), Vibrio vulnificus YJ016 (NP_935245) (Vv), Vibrio parahaemolyticus RIMD 2210633 (NP_798599) (Vp), Pasteurella multocida subsp. multocida PM70 (NP_244945) (Pm), Haemophilus influenzae Rd KW20 (NP_439249) (Hi), Psychromonas ingrahamii 37 (ZP_01349302) (Pi), Shewanella putrefaciens CN-32 (ZP_00815699) (Sp), Shewanella oneidensis MR-1 (NP_715900) (So), Rhodobacter sphaeroides 2.4.1 (YP_351852) (Rs), Roseovarius sp. strain 214 (ZP_01034520) (Rsp.), and R. capsulatus SB1003 (P29963) (Rc) were aligned with ClustalW (25). Amino acid similarities are indicated as follows: asterisk and gray shading, identical residues in all sequences; colons, conserved substitution; and period, semiconserved substitution. The predicted domain structure of side chains, the net charges ([R + K] − [D + E]) of the NTD and CTD, and the location of the GXY motif are also indicated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain RGK103 (Δccm) has a deletion of all eight ccm genes (ccmABCDEFGH) and has been previously described (10). All E. coli strains were grown aerobically (with shaking at 300 rpm) at 37°C in Luria-Bertani media (Difco). Antibiotics were obtained from Sigma-Aldrich (St. Louis, MO) and were used at the following concentrations: carbenicillin, 100 μg ml−1; and chloramphenicol, 20.4 μg ml−1. Isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma-Aldrich) and arabinose (Sigma-Aldrich) were used at final concentrations of 1 mM and 0.2% (wt/vol), respectively.
Construction of plasmids.
E. coli strain TB1 was used as the initial host for all cloning experiments. All oligonucleotide sequences are given in Table 1. A C terminally FLAG-tagged (DYKDDDDK; Sigma-Aldrich) version of CcmD was generated by PCR using pRGK333 (pGEXsystemI) as a template and the oligonucleotide primers ccmDrFLAG and ccmDf-EcoRI. The amplified product was digested with EcoRI and SalI and ligated into pRGK359 (pGEXCcmABC:6XHis [9]) to generate pRGK369 (pGEXCcmABC:6XHisCcmDFLAG). This pGEX4T-1 (GE Healthcare, Piscataway, NJ)-based IPTG-inducible plasmid encodes a glutathione S-transferase (GST) fusion to the N terminus of CcmA, a six-His tag on the C terminus of CcmC, and a FLAG tag on the C terminus of CcmD. A pGEXsystemI plasmid without ccmD and ccmE was engineered in a two-step process. First, ccmABC were amplified from pRGK333 (10) using oligonucleotide primers ccmNterm (10) and ΔccmD-left (9), and the PCR product was digested with BglII/EcoRI and ligated into BamHI/EcoRI-digested pGEX4T-1. Second, ccmFGH were amplified from pRGK333 (10) using oligonucleotide primers delccmErt (9) and ccmCterm (10). The PCR product was digested with NdeI/EcoRI and ligated into the intermediate plasmid mentioned above digested with NdeI/EcoRI, resulting in pRGK370 (pGEXsystemI- ΔccmDE). To construct a pBAD-derived plasmid expressing CcmE, the ccmE gene was amplified from pRGK333 (10) using oligonucleotide primers ccmEf-NcoI and ccmE*-right (9), digested with NcoI/KpnI, and ligated into NcoI/KpnI-digested pRGK330 (10). In the resulting plasmid, pRGK371 (pBADCcmE:6XHis), the CcmE gene is overexpressed from the araB promoter and is arabinose inducible (not shown). A pBADc4-derived plasmid that contains a gene encoding an N-terminally six-His-tagged CcmD was engineered in a two-step process. First, the CcmD gene was amplified from pRGK333 (10) with oligonucleotide primers ccmDf-NdeI and ccmDr-NdeI, and the PCR product was digested with NdeI and ligated into NdeI-digested pET15b (Novagen-EMD Biosciences, Gibbstown, NJ). Second, this intermediate plasmid was digested with XbaI/XhoI, and the 0.3-kb band containing the His6:CcmD gene was gel purified and ligated into XbaI/SalI-digested pRGK332 (10), yielding pRGK372 (pBADc4-6XHis:CcmD). This plasmid contains a transcriptional fusion of the pET15b-derived E. coli His6:CcmD gene positioned after the Bordetella pertussis cytochrome c4 gene, which is under the control of the araB promoter. A control plasmid containing a gene encoding a protein lacking the N-terminal six-His tag on CcmD was also engineered in a two-step process. First, ccmD was amplified with oligonucleotide primers ccmDf-NcoI and ccmDr-NdeI, and the PCR product was digested with NcoI/NdeI and ligated into NcoI/NdeI-digested pET15b. This intermediate plasmid was digested with XbaI/XhoI, and the 0.25-kb fragment containing the CcmD gene was ligated into XbaI/SalI-digested pRGK332 (10), yielding pRGK373 (pBADc4-CcmD). pRGK373 contains a transcriptional fusion of the pET15b-derived wild-type E. coli CcmD gene after the B. pertussis cytochrome c4 gene, which is under the control of the araB promoter.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequence (5′-3′)a | Purpose |
|---|---|---|
| ccmDr-FLAG | CGGTCGACTCACTTGTCGTCATCGTCCTTGTAGTCTGCAGCCTCTGCTGTGC | pRGK369 (pGEXABCHisDFLAG) |
| ccmDf-EcoRI | CGGAATTCATGACCCCTGCATTTGC | pRGK369 (pGEXABCHisDFLAG) |
| ccmNterm | TTGCAGATCTATGCTTGAAGCCAGAGAGTTAC | pRGK370 (pGEXSystemI-ΔDE) |
| ΔccmD-left | GGAAGCGAATTCAGGGGTCATATGCGGCCTCTT | pRGK370 (pGEXSystemI-ΔDE) |
| delCcmE-rt | GGCGAATTCTCATATGGCCTCCTGCTGTTG | pRGK370 (pGEXSystemI-ΔDE) |
| ccmCterm | CGGAATTCTTTTTATTTACTCCTCCTGCGGCGAC | pRGK370 (pGEXSystemI-ΔDE) |
| ccmEf-NcoI | ATCCATGGATATTCGCCGTAAAAACCG | pRGK371 (pBADCcmE:6XHis) |
| ccmE*-right | ACGGGTACCTCAGTGGTGGTGGTGGTGGTGTGATGCCTGGGTCCTTATA | pRGK371 (pBADCcmE:6XHis) |
| ccmDf-NdeI | GAGGCCGCATATGACCCCTGCATTTGCTTC | pRGK372 (pBADc4-6XHis:CcmD) |
| ccmDr-NdeI | CGGCGAATATTCATATGGCCTCCTGCTGTTG | pRGK372 (pBADc4-6XHis:CcmD) |
| ccmDf-NcoI | GCCGCCATGGCCCCTGCATTTGCTTCCTGG | pRGK373 (pBADc4-CcmD) |
Restriction sites are underlined.
Protein induction, preparation, and solubilization.
E. coli cultures containing protein overexpression plasmid constructs were grown at 37°C in Luria-Bertani media containing the appropriate antibiotics to an optical density at 600 nm of approximately 0.5. Protein expression was induced for 3 h with 1 mM IPTG (pGEX4T derived) or 0.2% (wt/vol) arabinose (pBAD derived), unless otherwise noted. The bacterial cells were harvested by centrifugation (16,000 × g for 10 min) and were disrupted by sonication, except when B-Per (Pierce, Rockford, IL) was used for crude preparations (see below). Membrane proteins were isolated by ultracentrifugation (1 h at 207,000 × g), resuspended, and solubilized in 10 mM Tris buffer (pH 8.0) containing 1% n-dodecyl-β-d-maltoside (Sigma-Aldrich). Periplasmic proteins were isolated as previously described (9). B. pertussis cytochrome c4 was isolated using B-Per (Pierce) (8).
Protein purification.
To purify CcmC:His6, solubilized extracts were passed over a column containing His Bind (Novagen-EMD Biosciences) nickel affinity resin and washed according to the manufacturer's suggestions. FLAG-tagged CcmD was purified from solubilized membrane extracts using FLAG antibody resin (Sigma-Aldrich) according to the manufacturer's specifications. GST: CcmA was purified from solubilized extracts as previously described (9).
Other methods.
Protein concentrations were obtained using the bicinchoninic acid assay (Pierce) with bovine serum albumin as a standard. For heme staining and Western blotting, proteins were first separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or, for extracts containing CcmD:FLAG, by using 12.5% Next Gel (Amresco, Solon, OH) and transferred onto Hybond-C nitrocellulose membranes (Hybond; GE Healthcare, Piscataway, NJ). Chemiluminescence from heme or Western blots was detected using the SuperSignal Femto chemiluminescent substrate (Pierce) with a LAS1000plus luminescent image analyzer charge-coupled device camera system (Fujifilm, Valhalla, NY). Heme staining was quantified using a LOLITA II (low light test array; Raytest USA, Wilmington, NC) for standardization of light intensity detection.
RESULTS AND DISCUSSION
CcmD is part of the CcmABC release complex.
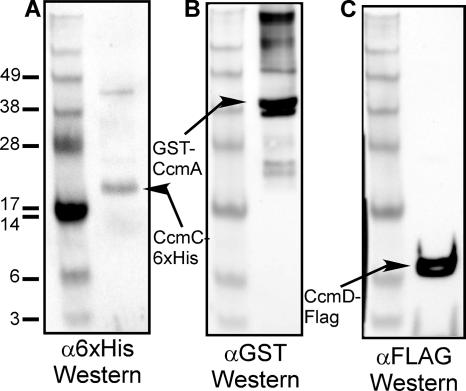
To determine whether E. coli CcmD is part of the CcmABC complex, a FLAG tag was added to the C terminus of CcmD. FLAG-tagged CcmD was functional and absolutely required for cytochrome c maturation (not shown) and allowed the detection of CcmD, expressed from pRGK369 (pGEXCcmABCHisDFLAG), with Western blots using FLAG monoclonal antibody (Sigma-Aldrich). As expected, GST:CcmA copurifies with CcmC:His6 (Fig. 2A shows a 25-kDa CcmC:His6 polypeptide, and Fig. 2B shows the 43-kDa GST:CcmA), confirming previously reported coimmunoprecipitation results (9). CcmD:FLAG also copurifies with CcmC:His6 (Fig. 2C shows a band at 7.8 kDa, the predicted molecular mass of CcmD). We have used the FLAG tag for immunoprecipitation with a FLAG antibody, in which case both GST:CcmA and CcmC:His6 coimmunoprecipitated (not shown). Additionally, affinity purification of GST:CcmA on a glutathione column brought down the CcmD:FLAG (not shown). These results are consistent with the hypothesis that CcmD is a component of the CcmABCD release complex in E. coli.
FIG. 2.
Protein-protein interactions with CcmC:His6 determined by analysis of proteins brought down with a nickel column. E. coli RK103 cultures containing pRGK369 (pGEXCcmABC:6XHisD:FLAG) were grown, membrane proteins were isolated, and nickel affinity chromatography was performed as described in the text. Purified proteins were separated by SDS-PAGE and probed with (A) six-His antisera (α6xHis), (B) GST antisera (αGST), or (C) FLAG antisera (αFLAG). The positions of standard proteins with the indicated molecular masses (103) are shown on the left, and the positions of CcmC:His6, GST:CcmA, and CcmD:FLAG are indicated by arrows.
CcmD is not essential for heme transfer to CcmE.
Ahuja and Thony-Meyer (1) suggested that CcmD plays a role (e.g., by protein-protein interaction) in formation of a complex between CcmC and CcmE. More recently, Feissner et al. (9) showed that GST:CcmA coprecipitated (using GST antisera) the same amount of CcmE whether CcmD was present or not, suggesting that CcmD is not required for the interaction between CcmE and the CcmABCD release complex. Two possible functions for CcmD are a role in heme transfer between CcmC and CcmE and a role in the ATP-dependent release of holo-CcmE from CcmABCD.
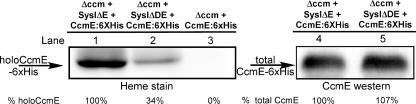
In order to test whether CcmD affects heme transfer to CcmE, system I plasmids without ccmD and ccmE or without only ccmE were used in conjunction with a plasmid expressing CcmE:His6. To establish that CcmD is required for cytochrome c maturation in this system, we coexpressed the B. pertussis cytochrome c4 and ccmE genes on another plasmid, pRGK349 (10). Only constructs containing ccmD (and other ccm genes) synthesized cytochrome c4 (not shown). When no system I is present with pRGK371 (pBADCcmE:6XHis), no holo-CcmE is detected (Fig. 3, lane 3), consistent with a requirement for CcmC for heme transfer to CcmE and with the hypothesis that there are no background levels of holo-CcmE, as reported previously (9, 20, 22). Holo-CcmE is present in the strain without only CcmD (Fig. 3, lane 2). The same levels of total CcmE (apo-CcmE and holo-CcmE) were detected using antisera to CcmE whether CcmD was present or not (Fig. 3, lanes 4 and 5). Although more holo-CcmE (approximately three times more) is present when CcmD is part of the CcmABCD complex than when CcmD is absent (Fig. 3, compare lanes 1 and 2), we conclude that CcmD is not essential for heme transfer to CcmE or for the stability or membrane insertion of CcmE. Below we address why less holo-CcmE is observed in the CcmD mutant.
FIG. 3.
Heme is transferred to CcmE with or without CcmD. E. coli RK103 harboring pRGK345 and pRGK371 (lanes 1 and 4), pRGK370 and pRGK371 (lanes 2 and 5), or pRGK371 (lane 3) was grown and induced as described in the text. Ten micrograms of crude n-dodecyl-β-d-maltoside-solubilized membrane proteins was separated by SDS-PAGE, the presence of holo-CcmE was detected by heme staining, and total CcmE was detected with CcmE antibodies. A typical gel with heme staining (lanes 1 to 3) and a CcmE Western blot (lanes 4 and 5) are shown, and the percentages of holo-CcmE, corrected for the amount of total CcmE present in each lane, are indicated at the bottom. Abbreviations: Δccm, E. coli RK103; SysIΔE, pRGK345; SysIΔDE, pRGK370; CcmE:6XHis, pRGK371.
CcmD is required for the release of holo-CcmE.
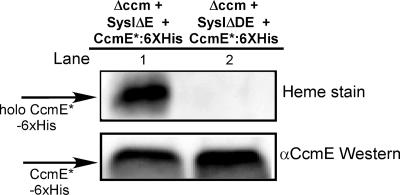
To determine if CcmD is involved in the release of holo-CcmE from CcmC, we employed the same assay that was used to show that the ATP hydrolysis features of CcmA and the presence of CcmB are required to release holo-CcmE from the CcmABC complex (9). The “release” assay uses a version of CcmE (designated CcmE*) in which the endogenous membrane tether is replaced with the cleavable B. pertussis cytochrome c4 signal sequence, resulting in CcmE* expressed as a soluble protein in the E. coli periplasm. With an intact system I present, holo-CcmE* is released, as expected, to the periplasmic fraction (Fig. 4, top panel, lane 1). When CcmD is absent, no holo-CcmE* is detected in the periplasmic fraction (Fig. 4, top panel, lane 2). The levels of total CcmE* detected by Western blotting were similar (Fig. 4, bottom panel, lanes 1 and 2) with and without CcmD. The results indicate that CcmD does not affect the stability or accumulation of CcmE* but CcmD is essential for the release of holo-CcmE* from the CcmABCD complex.
FIG. 4.
Holo-CcmE* is not released into the periplasm without CcmD. Periplasmic protein fractions were prepared from E. coli RK103 coexpressing pRGK345 and pRGK362 (lane 1) or pRGK370 and pRGK362 (lane 2). Twenty micrograms of periplasmic proteins was separated by SDS-PAGE and transferred to nitrocellulose. (Top panel) Heme staining showing the levels of holo-CcmE*. (Bottom panel) Immunoblot with CcmE antisera. Abbreviations: Δccm, E. coli RK103; SysIΔE, pRGK345; SysIΔDE, pRGK370; CcmE*:6XHis, pRGK362; αCcmE, CcmE antisera.
CcmD has a topology with the N terminus outside the cytoplasmic membrane and the C terminus inside the cytoplasmic membrane.
Using results obtained with a LacZ fusion to the C terminus of the R. capsulatus CcmD protein, Goldman et al. (13) proposed that the likely topology was the NTD located in the periplasm and the CTD located in the cytoplasm, with one transmembrane domain. However, because of the short NTD, the N terminus can be particularly refractive to experimental topological analysis (6, 15). The NTD and topology of E. coli CcmD were recently investigated further by using an N-terminal six-His tag, a C-terminal strep tag (SAWSHPQFEK), and the differential proteinase K sensitivity of spheroplasts and inverted membrane vesicles (1). Based on the results, Ahuja and Thony-Meyer (1) reported a topology for the E. coli CcmD protein where the NTD and the CTD were both in the cytoplasm, with the hydrophobic core in the inner leaflet of the cytoplasmic membrane. Given a periplasmic holo-CcmE release function for CcmD, we wanted to reevaluate the data that suggest that the E. coli CcmD NTD is cytoplasmic. The N-terminal amino acid sequence of purified E. coli CcmD, MTPAF, was determined, confirming that the translation start shown in Fig. 1 is correct.
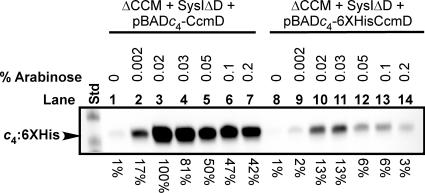
Ahuja and Thony-Meyer (1) did not show whether the addition of a six-His tag and other residues (see below) to the NTD might interfere with the optimal function of CcmD. Evaluation of the CcmD NTD and CTD (Fig. 1) for the inside-out rule (6, 15, 17) suggested that the addition of the six-His tag plus the adjoining 14 amino acids of vector-derived sequence (MGSSHHHHHHSSGLVPRGSH) added by Ahuja and Thony-Meyer (1) would affect the inside-out rule via the charge of the NTD (from −1 to at least +2). This could possibly lead to a change in the topology of most CcmD molecules in the membrane. To determine whether the addition of the pET15b-derived six-His tag to the N terminus of CcmD affects its activity, we constructed two “activity” reporter plasmids. We engineered the B. pertussis cytochrome c4 reporter plasmid (pRGK332) to include either His6:CcmD or untagged wild-type CcmD. The plasmids were constructed so that the ribosome-binding site (RBS) sequence and spacing to the ATG start codon, the six-His tag, and other vector-derived sequence were identical to those used by Ahuja and Thony-Meyer (1). A plasmid with the identical RBS but lacking the six-His tag and additional pET15b-derived amino acid sequence was constructed and served as the wild-type CcmD control plasmid for cytochrome c4 expression studies. Note also that the DNA upstream of the RBS and the spacing and sequence to the ATG of the six-His tag and wild-type CcmD are identical. Cytochrome c4 levels were used to estimate the “activity” of His6:CcmD and CcmD. The maximum level (defined as 100%) of cytochrome c4 from pRGK373 (wild-type CcmD) was detected with 0.02% arabinose (Fig. 5, lane 3). The maximum level of cytochrome c4 from pRGK372 (pBADc4-6XHis:CcmD) was also detected with 0.02% arabinose (Fig. 5, lane 10), but it was only 13% of the wild-type level. In general, quantification of “activity” with different arabinose concentrations indicates that the six-His tag on the NTD reduces function by approximately 90%. These results suggest that the addition of the six-His tag plus the 14 amino acids of pET15b sequence to the NTD of CcmD (MGSSHHHHHHSSGLVPRGSH) leads to highly reduced function of His6:CcmD when data are directly compared (Fig. 5, compare lanes 2 to 7 to lanes 9 to 14) to data for wild-type CcmD with the natural NTD sequence (MTPAFASWNE) (Fig. 1). Although other explanations are possible, the six-His tag could change the topology of the CcmD and hence reduce the function. The fact that some function remains could be because a small percentage of molecules have the NTD outside or because only proteolysed (i.e., proteolysis of the six-His tag) CcmD can assume the proper conformation. Our results suggest that the protease sensitivity data (1) and hence conclusions concerning CcmD NTD topology are based on nonfunctional CcmD molecules. Other lines of evidence also suggest that the topology of E. coli CcmD is the same as that suggested for R. capsulatus CcmD (12, 13). As mentioned above, the positive-inside/negative-outside rule (6, 15, 17) for membrane protein topology predicts that the CcmD NTD (charge, at least −1) is on the outside of the cytoplasmic membrane in all CcmD homologs (Fig. 1). The CTD is highly positively charged, consistent with the LacZ fusion results that place it in the cytoplasm. Because CcmD is required for the periplasmic release of holo-CcmE*, it is possible that a domain of CcmD in the periplasm is involved in protein-protein (or protein-heme) interactions with the CcmABCD release complex and/or holo-CcmE. After aligning the N termini of many CcmD proteins, we noted a completely conserved GXY motif (Fig. 1) that may be involved in such release interactions.
FIG. 5.
Heme staining of holocytochrome c4:His6 induced with increasing concentrations of arabinose shows that an N-terminal six-His tag on CcmD reduces its function. E. coli RK103 coexpressing pRGK353 and either pRGK373 (lanes 1 to 7) or pRGK372 (lanes 8 to 14) was grown and treated as described in the text. A typical blot to determine the levels of holocytochrome c4 (indicated at the bottom) compared to 100% (lane 3) is shown. The concentrations of arabinose are indicated above the lanes. The molecular masses of the standards (lane Std) are 29.2 kDa (top band) and 20.2 kDa (bottom band). Abbreviations: ΔCCM, E. coli RK103; SysIΔD, pRGK353; pBADc4-CcmD, pRGK373; pBADc4-6XHis:CcmD, pRGK372; c4, B. pertussis cytochrome c4.
In summary, we have shown that CcmD is part of the ATP-dependent CcmABCD holo-CcmE release complex and is not essential for heme transfer and attachment from CcmC to CcmE. We have shown that CcmD is absolutely required for the release of holo-CcmE* from the complex, and we suggest that the topology of CcmD is consistent with this function (i.e., the NTD in the periplasm and the CTD in the cytoplasm, with one transmembrane domain). We speculate that the lower levels of holo-CcmE formed without CcmD (threefold lower [Fig. 3]) could be due to decreased “recycling” through the heme attachment and release pathway. Future goals are to understand the details of the CcmABCD holo-CcmE release complex and the involvement of key residues of these proteins in the heme attachment and release pathway.
Acknowledgments
This study was funded by National Institutes of Health grant GM47909 to R.G.K.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Ahuja, U., and L. Thony-Meyer. 2005. CcmD is involved in complex formation between CcmC and the heme chaperone CcmE during cytochrome c maturation. J. Biol. Chem. 280236-243. [DOI] [PubMed] [Google Scholar]
- 2.Ahuja, U., and L. Thony-Meyer. 2003. Dynamic features of a heme delivery system for cytochrome C maturation. J. Biol. Chem. 27852061-52070. [DOI] [PubMed] [Google Scholar]
- 3.Allen, J. W., E. M. Harvat, J. M. Stevens, and S. J. Ferguson. 2006. A variant system I for cytochrome c biogenesis in archaea and some bacteria has a novel CcmE and no CcmH. FEBS Lett. 5804827-4834. [DOI] [PubMed] [Google Scholar]
- 4.Beckman, D. L., D. R. Trawick, and R. G. Kranz. 1992. Bacterial cytochromes c biogenesis. Genes Dev. 6268-283. [DOI] [PubMed] [Google Scholar]
- 5.Bonnard, G., and J. M. Grienenberger. 1995. A gene proposed to encode a transmembrane domain of an ABC transporter is expressed in wheat mitochondria. Mol. Gen. Genet. 24691-99. [DOI] [PubMed] [Google Scholar]
- 6.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 3081321-1323. [DOI] [PubMed] [Google Scholar]
- 7.Deshmukh, M., G. Brasseur, and F. Daldal. 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35123-138. [DOI] [PubMed] [Google Scholar]
- 8.Feissner, R. E., C. S. Beckett, J. A. Loughman, and R. G. Kranz. 2005. Mutations in cytochrome assembly and periplasmic redox pathways in Bordetella pertussis. J. Bacteriol. 1873941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, and R. G. Kranz. 2006. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61219-231. [DOI] [PubMed] [Google Scholar]
- 10.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, J. A. Loughman, K. W. Earley, and R. G. Kranz. 2006. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol. Microbiol. 60563-577. [DOI] [PubMed] [Google Scholar]
- 11.Fitz-Gibbon, S. T., H. Ladner, U. J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman, B. S., D. L. Beck, E. M. Monika, and R. G. Kranz. 1998. Transmembrane heme delivery systems. Proc. Natl. Acad. Sci. USA 955003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman, B. S., D. L. Beckman, A. Bali, E. M. Monika, K. K. Gabbert, and R. G. Kranz. 1997. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J. Mol. Biol. 268724-738. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, D. H., G. Bonnard, and J. M. Grienenberger. 1993. A gene involved in the biogenesis of c-type cytochromes is co-transcribed with a ribosomal protein gene in wheat mitochondria [corrected]. Curr. Genet. 24248-255. [DOI] [PubMed] [Google Scholar]
- 15.Heijne, G. V. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 53021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29383-396. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson, J., B. Persson, and G. von Heijne. 2005. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins 60606-616. [DOI] [PubMed] [Google Scholar]
- 18.Page, M. D., and S. J. Ferguson. 1997. Paracoccus denitrificans CcmG is a periplasmic protein-disulphide oxidoreductase required for c- and aa3-type cytochrome biogenesis; evidence for a reductase role in vivo. Mol. Microbiol. 24977-990. [DOI] [PubMed] [Google Scholar]
- 19.Ramseier, T. M., H. V. Winteler, and H. Hennecke. 1991. Discovery and sequence analysis of bacterial genes involved in the biogenesis of c-type cytochromes. J. Biol. Chem. 2667793-7803. [PubMed] [Google Scholar]
- 20.Ren, Q., and L. Thony-Meyer. 2001. Physical interaction of CcmC with heme and the heme chaperone CcmE during cytochrome c maturation. J. Biol. Chem. 27632591-32596. [DOI] [PubMed] [Google Scholar]
- 21.Richard-Fogal, C. L., E. R. Frawley, R. E. Feissner, and R. G. Kranz. 2007. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J. Bacteriol. 189455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz, H., R. A. Fabianek, E. C. Pellicioli, H. Hennecke, and L. Thony-Meyer. 1999. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. USA 966462-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz, H., H. Hennecke, and L. Thony-Meyer. 1998. Prototype of a heme chaperone essential for cytochrome c maturation. Science 2811197-1200. [DOI] [PubMed] [Google Scholar]
- 24.Stevens, J. M., O. Daltrop, J. W. Allen, and S. J. Ferguson. 2004. C-type cytochrome formation: chemical and biological enigmas. Acc. Chem. Res. 37999-1007. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thony-Meyer, L. 2002. Cytochrome c maturation: a complex pathway for a simple task? Biochem. Soc. Trans. 30633-638. [DOI] [PubMed] [Google Scholar]