Abstract
Legionella pneumophila expresses two peroxide-scavenging alkyl hydroperoxide reductase systems (AhpC1 and AhpC2D) that are expressed differentially during the bacterial growth cycle. Functional loss of the postexponentially expressed AhpC1 system is compensated for by increased expression of the exponentially expressed AhpC2D system. In this study, we used an acrylamide capture of DNA-bound complexes (ACDC) technique and mass spectrometry to identify proteins that bind to the promoter region of the ahpC2D operon. The major protein captured was an ortholog of OxyR (OxyRLp). Genetic studies indicated that oxyRLp was an essential gene expressed postexponentially and only partially complemented an Escherichia coli oxyR mutant (GS077). Gel shift assays confirmed specific binding of OxyRLp to ahpC2D promoter sequences, but not to promoters of ahpC1 or oxyRLp; however, OxyRLp weakly bound to E. coli OxyR-regulated promoters (katG, oxyR, and ahpCF). DNase I protection studies showed that the OxyRLp binding motif spanned the promoter and transcriptional start sequences of ahpC2 and that the protected region was unchanged by treatments with reducing agents or hydrogen peroxide (H2O2). Moreover, the OxyRLp (pBADLpoxyR)-mediated repression of an ahpC2-gfp reporter construct in E. coli GS077 (the oxyR mutant) was not reversed by H2O2 challenge. Alignments with other OxyR proteins revealed several amino acid substitutions predicted to ablate thiol oxidation or conformational changes required for activation. We suggest these mutations have locked OxyRLp in an active DNA-binding conformation, which has permitted a divergence of function from a regulator of oxidative stress to a cell cycle regulator, perhaps controlling gene expression during postexponential differentiation.
Legionella pneumophila is found in aquatic and moist soil environments as an intracellular parasite of protozoa (12, 14, 15). Legionnaires' disease results from aerosol transmission of bacteria from the environment to susceptible humans. In natural hosts (and some mammalian cell lines), L. pneumophila displays a dimorphic lifestyle in which vegetative replicating bacilli differentiate into metabolically dormant cyst-like planktonic forms (17). Analogous to the small-cell variants of Coxiella burnetii or the elementary bodies of Chlamydia spp., these highly resilient cyst-like forms of L. pneumophila enable the organism to survive extracellularly for extended periods of time in a highly infectious state. The cyst form likely contributes to the noted resilience of L. pneumophila to the actions of oxidizing agents (chlorine), chloramines, and biocides, used to control infection of man-made aquatic systems (17). Several lines of evidence suggest that antioxidant enzymes play crucial roles during the early stages of infection of protozoa or human phagocytes (3, 6, 8, 19, 27) and may also be important for environmental persistence (25).
We previously characterized two peroxide-scavenging alkyl hydroperoxide reductase systems of L. pneumophila: AhpC1, similar to the AhpC-thioredoxin system of Helicobacter pylori (30, 49), and AhpC2D, similar to the AhpC AhpD system found in mycobacteria (30, 42). These studies demonstrated that both systems contributed to efficient scavenging of H2O2 and organic peroxides and that at least one system was required for viability. Moreover, these genes were inversely expressed during the growth cycle; ahpC2D induced early in exponential phase and repressed postexponentially, concomitant with induction of ahpC1. Finally, a compensatory increase in expression of ahpC2D was observed in an ahpC1 mutant, suggesting that the ahpC2D operon might be upregulated in response to changes in cellular redox status or by increases in intracellular peroxides. However, previous studies of the antioxidant defense genes of L. pneumophila (sodB, sodC, katA, katB, ahpC1, or ahpC2D) had concluded that none of these genes were activated in response to oxidative stress (4, 5, 30, 39, 43). In contrast, these studies revealed that all of these antioxidant defense genes, with the exception of sodB (constitutive), were growth stage regulated.
The emerging pattern of growth stage-regulated antioxidant defense genes in L. pneumophila together with the postexponential expression of transmission (virulence) traits is consistent with the dimorphic lifestyle of this organism (17). In support of this view, microarray analyses comparing sessile and planktonic cells of L. pneumophila under biofilm conditions revealed ahpC2 and ahpD to be the most highly induced genes in sessile cells (25). Related studies have similarly shown that expression of the ahpC2D operon increases significantly during intracellular growth in protozoa (8). Despite these findings, little is known of the regulatory systems controlling ahpC2D expression or other antioxidant defense genes in L. pneumophila. With the exception of an OxyR homolog (described herein), typical regulators of oxidative stress defenses (such as SoxRS, OhrR, and PerR) are absent in the L. pneumophila genome. Growth phase-dependent regulation of genes associated with virulence and transmission traits has been attributed in part to global regulators, such as the stationary-phase sigma factor (RpoS) (2, 20), the carbon storage regulator (CsrA) (13, 33), and the LetAS, CpxRA, and PmrBA two-component regulatory systems (13, 16, 22, 32, 52); however, the predicted hierarchal regulatory cascade is incomplete, and none of these regulators has been linked to oxidative stress defenses.
Here we report the use of an acrylamide capture of DNA-bound complexes (ACDC) technique (35) and tandem mass spectrometry of ahpC2D promoter-bound proteins to identify a previously uncharacterized putative regulator of oxidative stress in L. pneumophila, designated OxyRLp. These studies will show that while oxyRLp was able to partly complement an oxyR mutant of Escherichia coli and purified OxyRLp exhibited weak binding to OxyR-regulated promoters of E. coli by gel shift experiments, the protein was no longer redox active and its repression of ahpC2D could not be reversed by H2O2. Finally, the apparent essentiality of oxyRLp together with its postexponential expression, concomitant with repression of ahpC2D, suggests regulatory roles that are unrelated to oxidative stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1 and oligonucleotides in Table 2. L. pneumophila strains were grown aerobically at 37°C on buffered charcoal yeast extract agar or in buffered yeast extract (BYE) broth, supplemented with α-ketoglutaric acid (1 mg/ml), ferric pyrophosphate (250 μg/ml), l-cysteine (40 μg/ml), thymidine (100 μg/ml), and antibiotics when required. E. coli strains DH5α, MG1655, and GS077 were grown at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with the appropriate antibiotics. For some studies, E. coli GS077 was grown at 37°C under anaerobic conditions using the BD GasPak EZ (Becton Dickinson, Oakville, Ontario) and AnaeroGen anaerobic atmosphere generation system (Oxoid, Ltd., Nepean, Ontario). Antibiotics (Sigma-Aldrich Canada Ltd., Oakville, Ontario) were added to media at the following concentrations, when appropriate: streptomycin (100 μg/ml), kanamycin (40 μg/ml), chloramphenicol (20 μg/ml), and ampicillin (100 μg/ml). All strains were stored at −85°C in nutrient broth containing 10% dimethyl sulfoxide.
TABLE 1.
Strains, genotypes, and plasmids used in this study
| Bacterial strain, genotype, or plasmid | Relevant property(ies) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Clontech |
| BL21 Codon Plus | F−ompT hsdS (rB− mB−) dcm+ Tetrgalλ (DE3) endA Hte [argU ileY leuW Camr] | Stratagene |
| MG1655 | K-12, F− λ− | ATCC 47076 |
| GS077 | MG1655-derived oxyR::kan | 51 |
| L. pneumophila | ||
| Lp02 | Philadelphia-1 derivative, rpsL hsdR thyA (Smr) mutant | 7 |
| ahpC1::kan | Lp02 ahpC1::kan mutant | 30 |
| ahpC2D::kan | Lp02 ahpC2D::kan mutant | 30 |
| rpoS::kan | Lp02 strain MB 379::kan | 2 |
| letA::kan | Lp02 strain MB 414::kan | 22 |
| himA::gm himB::kan | Lp02 integration host factor double mutant (lacking both α and β subunits); Kanr Gmr | This laboratory |
| Plasmids | ||
| pET29b | IPTG-inducible T7 expression vector | Novagen |
| petLpoxyR | pET29b-derived overexpression vector for His6-OxyRLp | This study |
| pBH6119 | RSF1010 ori, promoterless gfpmut3 tdΔi (Ampr) | 21 |
| pC2gfp | PahpC2 region (−199 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | 30 |
| pC2(−179/+27)gfp | PahpC2 region (−179 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| pC2(−156/+27)gfp | PahpC2 region (−156 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| pC2(−132/+27)gfp | PahpC2 region (−132 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| pC2(−109/+27)gfp | PahpC2 region (−109 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| pC2(−91/+27)gfp | PahpC2 region (−91 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| pC2(−66/+27)gfp | PahpC2 region (−66 to +27 bp upstream of ahpC2) cloned upstream of gfpmut3 of pBH6119 | This study |
| poxyRgfp | oxyR promoter region cloned upstream of gfpmut3 of pBH6119 | This study |
| pBAD22 | Arabinose-inducible expression vector (Ampr) | 18 |
| pBADEcoxyR | pBAD22-derived expression vector for E. coli oxyR | This study |
| pBADLpoxyR | pBAD22-derived expression vector for L. pneumophila oxyR | This study |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| Ac-FC2PROM | GGATCCACCAATATTCCTCCACAGGC |
| RC2PROM | GAATTCCTGGAAATTTGTTGCCTACCG |
| FECOXYRPBAD | GGCGAATTCATTATGAATCGTGATCTTGAG |
| RECOXYRPBAD | GGCAAGCTTAAACCGCCTGTTTTAAAACTT |
| FLPOXYRPBAD | GGCGAATTCATTATGAATTTAAGAGATTTACATTAT |
| RLPOXYRPBAD | GGCAAGCTTCTATGATAATTTGGATTGAACATT |
| FBLA | TCCGTGTCGCCCTTATTCC |
| RBLA | AACTACGATACGGGAGGGC |
| FLPOXYRPET | CATATGAATTTAAGAGATTTACATTAT |
| RLPOXYRPET | CTCGAGTGCTAATTTGGATTGAACATTTT |
| FC1PROM | GAATTCGTGCCATTACTGAGCGATTC |
| RC1PROM | GGATCCGAGTGCCTTTCATGTAGAGC |
| FLPOXYRPROM | GAATTCGATGGCACAAAGAGTTGCA |
| RLPOXYRPROM | CTGGGATCCTGCTTTACATCTGCCAGG |
| FECAHPCPROM | GGCACTGAAGATACCAAAGG |
| RECAHPCPROM | TCGATGAGATGTAAGGTAACC |
| FECKATGPROM | CGAAATGAGGGCGGGAAAAT |
| RECKATGPROM | CGATACACAGCGTTAGAGAG |
| FECOXYRPROM | GGTGCCGCTCCGTTTCTG |
| RECOXYRPROM | GCTGGCTAACGTGGCAGG |
| FC2PROM179 | GGCGAATTCCCTTCTTTAAAAGTAGTTTATAAAA |
| FC2PROM156 | GGCGAATTCAAACATAAAAGCCAAATATTTTGC |
| FC2PROM132 | GGCGAATTCAAAATGGATTAATAATTCTTATATTG |
| FC2PROM109 | GGCGAATTCTTGATTGACTTTATTTATCGTCTT |
| FC2PROM91 | GGCGAATTCCGTCTTTATAAAAACAATTGATTTT |
| FC2PROM66 | GGCGAATTCATTTATTAAATGATTTCACCTAAATT |
| RGFP | ACCATAACCGAAAGTAGTGAC |
Standard molecular techniques.
Preparation of DNA, restriction enzyme digestions, and other molecular techniques have been described elsewhere (40). Plasmid isolation was performed with the QIAprep spin miniprep kit (Qiagen Inc., Mississauga, Ontario) or using a standard alkaline lysis method (40). Purification of DNA fragments from agarose gels for subcloning was carried out with a QIAquick gel purification kit (Qiagen). PCR was performed under conditions described by LeBlanc et al. (30) using oligonucleotides synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). DNA and protein sequencing was carried out by the DalGen Microbial Genetics Center (Halifax, Nova Scotia, Canada). Protein concentrations were estimated using a Bio-Rad protein assay (Bio-Rad, Hercules, CA).
ACDC.
To identify transcription factors that bind to the promoter region of ahpC2D (PahpC2), an ACDC was performed according to a modified version of the methods described by Nelson et al. (35). Briefly, PCRs were performed using forward primers containing 5′ Acrydite (Ac) moiety, Ac-FC2PROM, and unmodified reverse primer RC2PROM. Amplicons were purified using a MinElute gel extraction kit (Qiagen). Protein lysates were obtained as follows: bacteria were harvested (4,800 × g, 6 min, 4°C) from a 500-ml, 24-h culture of L. pneumophila. The pellet was washed with cold 50 mM Tris-HCl, pH 7.5, and resuspended in buffer consisting of 50 mM Tris-HCl, pH 7.5; 1 mM EDTA, pH 8.0; 50 mM NaCl; 10% glycerol with 0.1 mM phenylmethylsulfonyl fluoride (PMSF); and 1 mM dithiothreitol (DTT). Cells were lysed using a French press and subjected to ultracentrifugation at 100,000 × g. Protein lysates were aliquoted and stored at −85°C. Before use, protein lysates were diluted in a buffer consisting of 100 mM Tris-HCl, pH 7.5, and 10 mM MgCl2 and treated for 15 min at room temperature with 100 mM of acetyl phosphate (phosphorylate DNA-binding proteins). Binding reactions for the ACDC assay were performed in a 25-μl volume and consisted of 0 (no DNA for control), 2, 4 or 8 μg of Ac-DNA, 1 μg of protein lysate, 7.5 μl buffer D (20 mM HEPES, pH 7.8; 100 mM KCl; 10% glycerol; and 1 mM DTT), 1 μg poly(dI-dC), and 3.5 μl of 30% acrylamide. Reactions were incubated for 15 min at room temperature before addition of 1.25 μl of 5% ammonium persulfate and 1.25 μl of diluted N,N,N′,N′′-tetramethylethylenediamine (1:30 in buffer D). The reactions were immediately loaded into the wells of a precast nondenaturing 5% polyacrylamide gel (0.5× Tris-borate-EDTA [TBE], Bio-Rad Laboratories). After polymerization (approximately 5 min), electrophoresis was performed in 0.5× TBE for 1 h at 125 V to remove nonspecific proteins. Each well was excised from the gel, transferred to a microtube containing 100 μl of 10% sodium dodecyl sulfate (SDS) sample buffer (NEB) with 5% β-mercaptoethanol and heated for 3 min at 94°C to remove captured proteins. After a brief centrifugation, 50-μl aliquots were loaded onto a precast 4 to 15% Tris-HCl polyacrylamide gel (Bio-Rad Laboratories) and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After silver staining, proteins distinct from the no-DNA controls were excised and submitted for mass spectrometry analysis (DalGen Microbial Genomics Center).
Cloning and overexpression of OxyRLp in E. coli GS077.
For complementation of the E. coli strain GS077 (oxyR::kan) (generously donated by Gisela Storz, National Institutes of Health, Bethesda, MD), E. coli strain K-12 oxyR (oxyREc) or oxyRLp was expressed using the arabinose-inducible expression systems of pBAD22 (18). Amplicons were generated from PCRs using primer pairs FECOXYRPBAD and RECOXYRPBAD for oxyREc and FLPOXYRPBAD and RLPOXYRPBAD for oxyRLp, digested with EcoRI and HindIII and subcloned into pBAD22. The resulting constructs (pBADEcoxyR and pBADLpoxyR) were transformed into E. coli DH5α and verified by PCR and DNA sequencing. Plasmids were extracted and transformed into electrocompetent E. coli GS077. Ampicillin-resistant GS077 transformants obtained anaerobically were confirmed by PCR analysis and termed GS077 pBADEcoxyR and GS077 pBADLpoxyR. The pBAD22 empty vector was transformed into wild-type MG1655 and GS077 strains, verified by PCR analysis using primers for the ampicillin resistance cassette FBLA and RBLA, and termed MG1655 pBAD22 and GS077 pBAD22, respectively.
Disk diffusion assays.
Strains were grown under anaerobic conditions to mid-exponential phase (optical density at 620 nm [OD620] of 0.4), and expression of oxyREc or oxyRLp from respective pBAD22-derived plasmids was induced with various concentrations of arabinose (0, 0.02, and 0.2%) or repressed using 0.2% glucose. Samples were analyzed by SDS-PAGE to confirm the expression of OxyREc and OxyRLp. Cells corresponding to 0.1 OD620 were inoculated in 4 ml of prewarmed LB 0.75% top agar, mixed by inversion, and poured onto LB media with proper antibiotics and in the presence of arabinose or glucose. Once solidified, 10 μl of 30% H2O2 was placed on sterile one-fourth-inch antibiotic disks in the center of the plate. Following overnight incubation at 37°C under anaerobic conditions, the diameters of the zones of inhibition were measured and reported for three independent experiments.
Overproduction and purification of OxyRLp.
The primer pair FLPOXYRPET and RLPOXYRPET was used to generate an amplicon containing NdeI and XhoI restriction sites, respectively. Following digestion, the NdeI-XhoI fragment was subcloned into the corresponding sites of pET29b, generating pETLpoxyR. The in-frame fusion of L. pneumophila oxyR with the C-terminal hexameric histidine (His6) tag was confirmed by DNA sequencing. For overexpression, an overnight culture of E. coli BL21(DE3) Codon Plus harboring pETLpoxyR was inoculated 1% (vol/vol) into 200 ml of fresh LB broth and grown to an OD620 of 0.4. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mM, and incubation was continued for another 60 min at 37°C. Bacteria were then harvested by centrifugation (5,800 × g for 6 min), resuspended (5 ml/g [wet weight]) in suspension buffer (20 mM Tris-HCl, pH 7.9; 500 mM NaCl; 5 mM imidazole; and 1 mM PMSF), and lysed on ice by sonication (six bursts of 10 s at 200 W). Following clarification by centrifugation (10,000 × g for 15 min, 4°C), the His6-OxyRLp was purified by nickel interaction chromatography as previously described (41). Samples taken from each step of purification were analyzed by SDS-PAGE, and the purified protein fractions were pooled, aliquoted, and stored at −85°C. A single band was excised and submitted for mass spectrometry analysis. Purified OxyRLp protein used in this study is considered to be wholly active.
EMSA.
For electrophoretic mobility shift assay (EMSA), amplicons spanning the promoter regions of ahpC1, ahpC2, and oxyR (PahpC1, PahpC2, and PoxyR) were generated by PCR amplification using primers sets FC1PROM and RC1PROM, FC2PROM and RC2PROM, and FLPOXYRPROM and RLPOXYRPROM, respectively. For binding assays to the E. coli ahpCF, katG, and oxyREc promoters, primer pairs FECAHPCPROM and RECAHPCPROM, FECKATGPROM and RECKATGPROM, and FECOXYRPROM and RECOXYRPROM were used to generate respective amplicons. Fifty nanograms of the DNA probes was mixed with various amounts of His6-OxyRLp (0, 3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, and 500 ng) in a 20-μl reaction mixture containing 20 mM Tris-HCl, pH 8.0; 5 mM MgCl; 20 mM KCl; 50 μg bovine serum albumin; 20% glycerol; 1 mM PMSF; and 0 or 200 mM DTT (Promega, Madison, WI) for oxidizing or reducing conditions, respectively. After 30 min of incubation at room temperature, protein-DNA complexes were separated by electrophoresis for approximately 1 h at 125 V on a 5% nondenaturing polyacrylamide gel in 0.5× TBE buffer. Gels were then soaked in 10,000-fold diluted Sybr green I nucleic acid stain (Invitrogen, La Jolla, CA) and washed twice in distilled water, and DNA was visualized using a Typhoon 9410 system (GE Healthcare) under blue light excitation at 488 nm and using a 520-nm emission filter. Densitometry was performed using ImageQuant version 5.2 (Molecular Dynamics, Sunnyvale, CA).
Mapping of PahpC2.
An EMSA was performed as described above using 500 ng of His6-OxyRLp and 50 ng of variously sized 5′-end deletion DNA fragments generated by PCR using a common reverse primer, RC2PROM, with different forward primers: FC2PROM, FC2PROM179, FC2PROM156, FC2PROM132, FC2PROM109, FC2PROM91, and FC2PROM66. All promoter fragments were also subcloned into pBH6119 and electroporated into L. pneumophila Lp02 and the ahpC1::kan mutant (30) to evaluate promoter activity using the green fluorescence protein (GFP) reporter assay.
DNase I footprinting.
DNase I protection experiments were performed using a 226-bp DNA fragment of PahpC2 generated from PCRs using primer pair FC2PROM and RC2PROM (containing EcoRI and BamHI restriction sites, respectively). The digested DNA (EcoRI for the top strand and BamHI for the bottom strand) was then treated with calf intestinal phosphatase (New England BioLabs, Ipswich, MA), and reactions were purified using a QIAquick gel extraction kit (Qiagen). End-labeled DNA was prepared using [γ-32P]dATP and Ready-To-Go T4 polynucleotide kinase (GE Healthcare) as described by the manufacturer and purified with a QIAquick nucleotide removal kit (Qiagen). Binding reactions were performed by incubating approximately 20,000 cpm of labeled probe with 10 μg of OxyRLp protein in an assay buffer consisting of 20 mM Tris-HCl, pH 8.0; 10 mM MgCl2; 100 mM KCl; 1 mM CaCl2; 10% glycerol; 0.05 mg/ml bovine serum albumin; 4 μg/ml poly(dI-dC); and H2O2 (0 to 50 mM) for oxidizing or 200 mM DTT for reducing conditions. In cases where peroxidation abolished DNA binding, replicates were treated with 200 mM DTT to restore DNA binding activity. After 20 min of incubation, 0.5 to 1 U of DNase I (Sigma) was added. The reaction was continued for 90 s before 100 μl of a stop solution (200 mM EDTA, pH 8.0; 5 M ammonium acetate; and 100 μg of salmon sperm DNA) was added. DNA was ethanol precipitated following phenol-chloroform extraction. Dry pellets were mixed with a formamide loading dye and loaded onto a 6% sequencing gel. The DNA ladder was performed by cycle sequencing using an α-35S-dATP label according to the SequiTherm Excel II DNA sequencing kit (Epicenter Technologies).
Primer extension.
Total RNA was obtained from exponentially grown L. pneumophila by an automated Maxwell-16 protocol (Promega), and primer extension reactions were performed as follows. Briefly, 2 pmol of [γ-32P]ATP end-labeled RC2PROM primer was added to 25 μg total RNA and allowed to anneal by dropping the temperature from 80°C to 41°C over a period of 30 min. cDNA extension products were generated at 41°C for 1 h in a reaction mixture containing 1 μl of 200 U/μl SuperScript reverse transcriptase (Invitrogen) and subsequently treated with RNase A (Sigma) to eliminate residual RNA. Samples were loaded onto a 6% polyacrylamide urea sequencing gel along with the corresponding DNA sequence ladder obtained by cycle sequencing using an α-35S-dATP label with the appropriate primer in accordance with the SequiTherm Excel II DNA sequencing kit protocol.
GFP reporter assay.
A GFP-transcriptional fusion of PoxyR was constructed using primer pair FLPOXYRPROM and RLPOXYRPROM and cloned into the EcoRI and BamHI sites of pBH6119 (21). Plasmid constructs were first transformed into E. coli DH5α strains, and correct orientation was determined by PCR using the respective combination of FLPOXYRPROM and RGFP primers. The pBH6119 plasmid or its derivatives were electroporated into wild-type, ahpC1, ahpC2D, rpoS, letA, or himAB mutants of L. pneumophila. Transformants capable of growth without thymidine were verified for respective plasmids by PCR analysis using primer pairs FLPOXYRPROM and RLPOXYRPROM or FBLA and RBLA for the empty vector controls. Fluorometric detection of samples taken from BYE-grown bacteria was performed as described previously (30). Values are expressed as relative fluorescence units (RFU) per OD620 and represent triplicate values obtained from three independent experiments.
OxyRLp regulation of ahpC2-gfp in E. coli GS077.
DNA fragments containing gfp (no promoter) or PahpC2gfp were excised with EcoRI and PstI from plasmids pBH6119 and pC2gfp, respectively, and were subcloned into pMMB206. The resulting constructs, pgfp and pahpC2gfp, respectively, were electroporated into either GS077 pBAD22 or GS077 pBADLpoxyR. Ampicillin- and chloramphenicol-resistant transformants were confirmed by PCR analysis and subjected to the GFP reporter assay (30). Overnight cultures, grown at 37°C in LB broth under anaerobic conditions, were inoculated 1% into fresh media until an OD620 of 0.4 was obtained. Then, glucose or arabinose was added to a final concentration of 0.02%, and cultures were incubated for an additional hour. In some cases, strains were then challenged for 30 min with 100 μM of H2O2. Fluorometric detection of samples taken from LB-grown bacteria was performed as described previously (30). Values are expressed as RFU per OD620 and represent triplicate values obtained from three independent experiments.
RESULTS
Acrylamide capture of ahpC2D DNA-bound complexes.
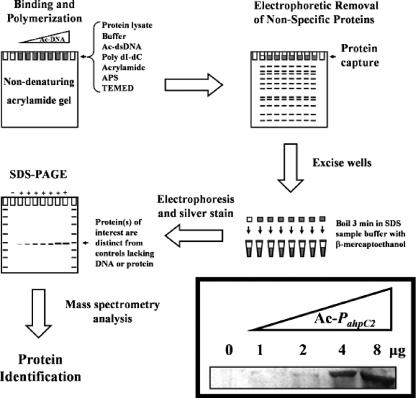
We employed the ACDC technique (Fig. 1) to screen for transcription factors that bound the promoter region of ahpC2D. The principle of this method relies on capture of DNA-binding proteins from L. pneumophila lysates by interaction with ahpC2D promoter DNA sequences immobilized in the polyacrylamide matrix by covalent linkage to its 5′ Ac moiety. Proteins that do not bind the Ac-DNA can be removed by standard nondenaturing electrophoresis. Following SDS-PAGE, silver staining, and mass spectrometry analysis of proteins captured in the wells of the native acrylamide gel, proteins able to bind target DNA sequences can be identified. Using increasing concentrations of Ac-labeled ahpC2D promoter DNA (Ac-PahpC2) in the ACDC assay (Fig. 1 inset), a distinct protein was captured by the immobilized Ac-DNA, whereas no bands could be seen in the absence of Ac-DNA. Peptides identified by tandem mass spectroscopy analysis corresponded to 181 of 296 amino acids (61% sequence coverage) of a previously uncharacterized protein annotated as the hydrogen peroxide-inducible gene activator, OxyR, of L. pneumophila Philadelphia-1. This protein, designated OxyRLp, shared 45 to 50% identity and 63 to 69% similarity to well-characterized OxyR regulators of Xanthomonas campestris, E. coli, Salmonella enterica serovar Typhimurium, Haemophilus influenzae, and C. burnetii by BLAST analysis.
FIG. 1.
Acrylamide capture of proteins interacting with PahpC2. The ACDC assay was performed by adding 1 μg of protein (cell extract) prepared from BYE-grown L. pneumophila to various amounts of Ac-DNA (0, 2, 4, and 8 μg). Nonspecific proteins were removed by nondenaturing electrophoresis on a 0.5× TBE-5% polyacrylamide gel. Captured proteins (inset) were excised from wells, heated in SDS sample buffer containing β-mercaptoethanol, and following SDS-PAGE, visualized by silver staining. dsDNA, double-stranded DNA; APS, ammonium persulfate; TEMED, N,N,N′,N′′-tetramethylethylenediamine.
The identification of OxyRLp as a putative regulator of ahpC2D was not unexpected, since OxyR mediates oxidative stress induction of alkyl hydroperoxide reductase and catalase genes in many organisms (1, 26, 31, 36). Moreover, in those organisms expressing the ahpC2D operon, including close relative C. burnetii, oxyR is usually located immediately upstream and shares promoter and regulatory sequences. However, the oxyRLp gene was not located upstream of ahpC2D, but rather shares a divergent promoter region with a gene annotated as a major facilitator transporter, and downstream is a gene of unknown function oriented in the opposite direction. Our attempts to knock out oxyRLp by allelic replacement with a drug resistance cassette (kanamycin) by methods previously used to obtain ahpC1 and ahpC2D mutants (30) proved unsuccessful. Since catalase supplementation to media enabled the recovery of oxyR mutants in Pseudomonas aeruginosa (23), buffered charcoal yeast extract agar was supplemented with catalase (10,000 units); however, no oxyRLp mutants were recovered. The apparent essentiality of oxyRLp was unexpected, since the genomic organization was uncomplicated by overlapping regulatory elements or downstream polarity effects. Possible functional divergence was considered, since previous studies had indicated that genes typically regulated in response to oxidative stress in other bacteria (ahpC1, ahpC2D, katA, katB, sodB, and sodC) were not similarly regulated in L. pneumophila (4, 5, 30, 38, 39, 43). Rather, most of these genes exhibited growth stage-dependent expression (4, 5, 30, 43).
Complementation of E. coli oxyR mutant.
Functional complementation was assessed by cloning (pBAD22) and expressing the oxyRLp gene in E. coli oxyR mutant strain GS077. Studies were conducted under anaerobic conditions because this strain is highly susceptible to peroxide killing (compared with wild-type strain MG1655) due to a loss of OxyR regulatory functions. Under inducing conditions (arabinose) or repressing conditions (glucose), the presence of pBAD22 did not alter peroxide susceptibility (Table 3). Complementation of GS077 with pBADEcoxyR fully restored peroxide resistance to wild-type E. coli MG1655 levels under inducing conditions, but not under repressing conditions. In contrast, overexpression of pBADLpoxyR partially rescued the peroxide sensitivity of E. coli GS077, suggesting that OxyRLp recognizes OxyR-regulated promoters. The noted partial complementation by OxyRLp might be due to DNA sequence differences in OxyR binding motifs between these organisms.
TABLE 3.
Zones of inhibition (mm) in a disk diffusion assay using 10 μl of 3% H2O2
| Straina | Zones of inhibition (mm) with:
|
|||
|---|---|---|---|---|
| Glucose (%)
|
Arabinose (%)b
|
|||
| 0 | 0.2 | 0.02 | 0.2 | |
| K-12† | 21 ± 1 | 23 ± 1 | 21 ± 1 | 21 ± 1 |
| K-12 pBAD22‡ | 21 ± 1 | 22 ± 1 | 22 ± 1 | 22 ± 1 |
| GS077§ | 38 ± 2 | 40 ± 1 | 38 ± 1 | 37 ± 2 |
| GS077 pBAD22‡ | 39 ± 1 | 41 ± 1 | 38 ± 1 | 38 ± 1 |
| GS077 pBADEcoxyR** | 37 ± 2 | 40 ± 1 | 19* ± 1 | 18* ± 1 |
| GS077 pBADLpoxyR** | 39 ± 1 | 39 ± 1 | 29* ± 2 | 29* ± 2 |
Triplicate experiments were performed under anaerobic conditions on Mueller-Hinton agar without antibiotics (†) or supplemented with 100 μg/ml ampicillin (‡), with 40 μg/ml kanamycin (§), or with 100 μg/ml ampicillin and 40 μg/ml kanamycin (**).
*, significance (P < 0.01) was determined in relation to strain GS077.
Interaction of OxyRLp with the promoter region of ahpC2.
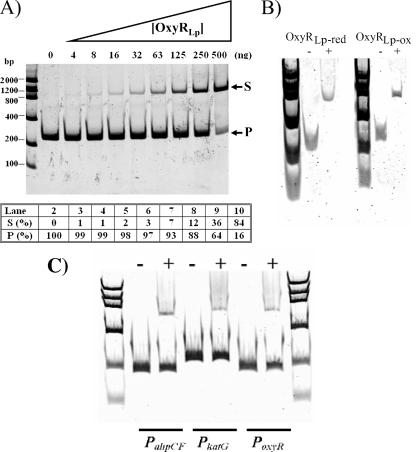
To both confirm ACDC results and extend functional studies, Ni2+-nitrilotriacetic acid-purified OxyRLp (overexpression from pETLpoxyR) was assessed by EMSA for DNA-binding specificity to the promoter regions (PahpC2, PahpC1, and PoxyR) of these genes. As seen in Fig. 2A, OxyRLp bound to a 226-bp DNA fragment containing PahpC2 (−199 to +27 relative to the ahpC2 coding sequence). This is evident in the sample containing 500 ng of His6-OxyRLp, where 84% of the DNA probe was in the bound state. No differences in binding were observed under oxidizing (no added reducing agent) or reducing conditions (200 mM DTT) (see Fig. 2B). Similar experiments were also carried out with PahpC1 and PoxyR; however, no DNA binding was observed (data not shown). EMSAs were also performed with amplicons containing promoter regions of known members of the E. coli OxyR regulon (ahpCF, katG, and oxyR) under oxidizing conditions. As seen in Fig. 2C, OxyRLp weakly bound these promoters and only at the highest concentrations of protein used (500 ng). The EMSA results indicate that the partial complementation of an E. coli oxyR mutant by oxyRLp might be due to a combination of OxyRLp overexpression and partial activation of OxyR-regulated promoters in GS077.
FIG. 2.
Interaction between OxyRLp and PahpC2. (A) Fifty ng of a 226-bp PahpC2 fragment spanning the region −199 to +27 (relative to the L. pneumophila ahpC2 initiation codon) was incubated in the presence of various amounts of His6-OxyRLp. Lanes: 1, low-mass ladder (Invitrogen); 2, DNA without protein; 3 to 10, DNA in the presence of 3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, or 500 ng of His6-OxyRLp, respectively. Arrows indicate bands representing the DNA probe (P) and mobility shift (S). Densitometry analysis reported as a percentage of DNA probe (P [%]) or shift (S [%]) relative to the total intensity of each lane. (B) Mobility shift assay performed in the presence of 25 ng PahpC2 with OxyRLp under reducing (200 mM DTT, OxyRLp-red) or oxidizing (no DTT, OxyRLp-ox) conditions. (C) Mobility shift assay performed in the presence of 50 ng each of E. coli PahpCF, PkatG, and PoxyR in the presence (+) or absence (−) of 500 ng His6-OxyRLp under nonreducing conditions.
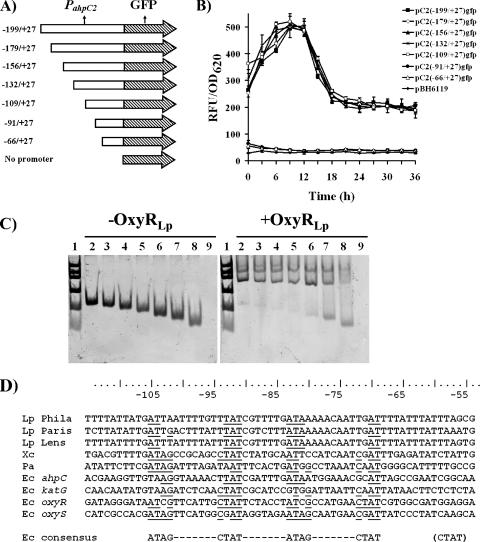
Mapping of the OxyR binding site within PahpC2.
To identify upstream DNA sequences necessary for ahpC2 expression, PCR-generated 5′-end deletion fragments of PahpC2 were cloned upstream of gfpmut3, encoding GFP. The promoter fragments shared a common 3′ end at +27 (relative to the ahpC2 initiation codon) and a variable 5′ end ranging from −199 to −66 (Fig. 3A). The ability of PahpC2 fragments to express GFP was assessed in the ahpC1 mutant (Fig. 3B), since ahpC2 levels are increased in this mutant (30). The 5′-end deletions of PahpC2 from base pairs −179 to −109 showed transcriptional activity similar to that of the previously studied full-length fragment (−199 to +27) (30). However, PahpC2 deletions of −91 or −66 no longer expressed GFP. It should be noted that similar results were obtained in the wild-type strain, albeit at a lower expression level (data not shown). In a parallel series of experiments, mobility shift assays were performed with various PahpC2 fragments (Fig. 3C). Deletion of nucleotides −199 to −109 had no effect on OxyRLp binding (Fig. 3C, lanes 2 to 5). However, further deletion of nucleotides −91 to −66 (which had ablated transcription) did not completely abolish DNA binding, suggesting that OxyRLp binding motif(s) remained. An alignment of DNA sequences for the −105 to −55 region of PahpC2 for the three genome sequences for L. pneumophila together with sequences from OxyR-regulated genes from E. coli and other species reveals some conservation with the E. coli OxyR consensus (Fig. 3D). Taken together, these findings suggest that the OxyRLp-protected region contains an OxyR motif that might include promoter sequences.
FIG. 3.
Promoter deletion analysis of the OxyRLp binding site within PahpC2. (A) Schematic representation of the 5′-end deletions of PahpC2 cloned upstream of promoter-less gfp. (B) Fluorescence detected from ahpC1::kan mutants harboring plasmids and PahpC2 promoter DNA-GFP constructs. (C) Mobility shift assay performed with the 5′-end deletions of PahpC2. Each lane contained 50 ng of the DNA fragments incubated with (right panel) or without (left panel) 500 ng of His6-OxyRLp. For both gels, lanes are as follows (symbols as shown in panel B): 1, low-mass ladder (Invitrogen); 2, PahpC2 fragment −199 to +27 (▪); 3, PahpC2 fragment −179 to +27 (⋄); 4, PahpC2 fragment −156 to +27 (▴); 5, PahpC2 fragment −132 to +27 (*); 6, PahpC2 fragment −109 to +27 (○); 7, PahpC2 fragment −91 to +27 (•); 8, PahpC2 fragment −66 to +27 (▵); and 9, no-DNA control (⧫). DNA fragments are annotated relative to the ahpC2 initiation codon. (D) DNA alignments of the ahpC2 upstream region (−115 to −55) from L. pneumophila Philadelphia-1, L. pneumophila Paris, and L. pneumophila Lens; ahpC from Xanthomonas campestris (Xc) and P. aeruginosa (Pa); and ahpC, katG, oxyR, and oxyS from E. coli. The consensus sequence from E. coli is provided, and the matching motifs are underlined.
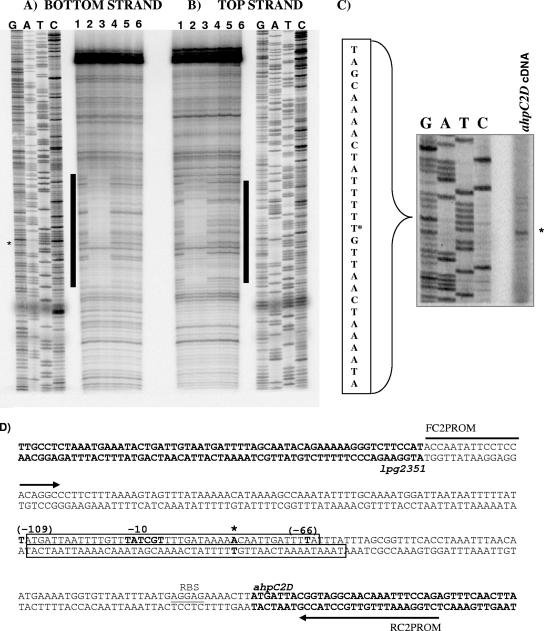
DNase I footprinting and primer extension.
To further delineate the specific DNA binding sequence of PahpC2 recognized by OxyRLp, the untreated and reduced (200 mM DTT) forms of OxyR (OxyRLp-ox and OxyRLp-red, respectively) were used in a footprinting assay. OxyRLp-red and OxyRLp-ox showed similar DNA footprints (Fig. 4 A and B) and confirmed that the protected region included the −91 and −66 regions. Primer extension analysis (Fig. 4C) revealed that the transcription start site as well as the −10 hexamer was located within the OxyRLp-protected region (Fig. 4D). The location of the promoter region and transcription start site within the OxyRLp-protected region is unusual, since in E. coli, reduced OxyR binds upstream of the promoter region of OxyR-regulated genes and represses transcription by overlapping the −35 hexamer (9, 31, 34, 36, 45, 46). In the case of PahpC2, the −35 hexamer is outside of the OxyRLp-protected region, suggesting a different regulatory mechanism from that reported for OxyREc (46, 47). However, if OxyRLp was not fully oxidized during the purification process, as is typical for other OxyR proteins (47, 48), then perhaps further oxidation with H2O2 might reveal more typical redox-mediated DNA-binding differences.
FIG. 4.
OxyRLp binding of PahpC2 and ahpC2 transcript start site. Shaded bars indicate OxyRLp DNA footprints for bottom strand (end labeled at BamHI) (A) and top strand (end labeled at EcoRI) (B). OxyRLp binding to PahpC2 occurred under the following conditions: lanes 1 and 6, no protein control reactions; lane 2, oxidizing conditions (no DTT); lane 3, 200 mM DTT; lanes 4, 1 mM H2O2; lane 5, treated with 1 mM H2O2 and then with 200 mM DTT to recover DNA-binding activity. In panels A and C, asterisk denotes ahpC2 transcript start site as indicated by alignment of primer extension product with the corresponding DNA sequence ladder. (D) Schematic of PahpC2. Arrows indicate FC2PROM and RC2PROM primers, open boxes denote OxyRLp sites as determined by DNase I protection assays shown in panels A and B, boldface indicates open reading frames of ahpC2 and lpg2351, and the ribosome-binding site (RBS) for ahpC2 is underlined. The transcription start site is indicated by an asterisk, and the −10 sequence is indicated in bold. The numbers in parentheses depict the numbering and sites of deletions shown in Fig. 3.
To test this possibility, we treated OxyRLp with a range of peroxide concentrations (10 μM to 50 mM) and determined that 1 mM completely abolished DNA binding (Fig. 4A, lane 4). While not shown, treatment of OxyRLp with 500 μM H2O2 reduced DNA binding by 50% but did not alter the protected area relative to untreated OxyRLp controls. To distinguish between reversible and irreversible oxidation of OxyRLp, we followed 1 mM H2O2 treatment with 200 mM DTT treatment to reduce the disulfide bonds, and as seen in Fig. 4A, lane 5, subsequent reduction could not restore DNA binding activity. These studies indicate that the OxyRLp footprint is unchanged by treatments with H2O2 at concentrations that should be sufficient to fully oxidize the protein.
OxyRLp represses ahpC2-gfp expression.
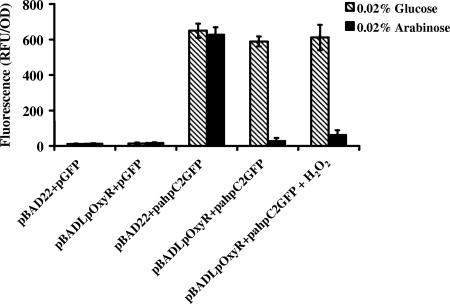
Since the OxyRLp footprint for ahpC2 appears to block the binding of the RNA polymerase complex, we sought to confirm that OxyRLp was indeed a repressor. We first introduced pahpC2gfp into E. coli GS077 (the oxyR mutant) and noted constitutive expression of GFP (Fig. 5). We then introduced pBADLpoxyR and pBAD22 vector controls into GS077 strains harboring pahpC2gfp and pgfp controls and induced each with either arabinose or glucose. As seen in Fig. 5, arabinose-dependent induction of OxyRLp from pBADLpoxyR repressed fluorescence from pahpC2gfp, consistent with our hypothesis that binding of OxyRLp to the promoter of ahpC2-gfp blocked transcription. Moreover, there was no change in ahpC2-gfp expression following challenge with 100 μM H2O2 (oxidative stress), further evidence that OxyRLp is not a redox-active regulator.
FIG. 5.
Regulation of ahpC2-gfp in E. coli GS077 by OxyRLp. Strains were grown in LB broth and subjected to repressing (glucose) or inducing (arabinose) conditions. Treatment with 100 μM H2O2 is indicated where appropriate. RFUs were normalized to 1.0 OD620 unit. Results are the means ± standard deviations of three independent experiments.
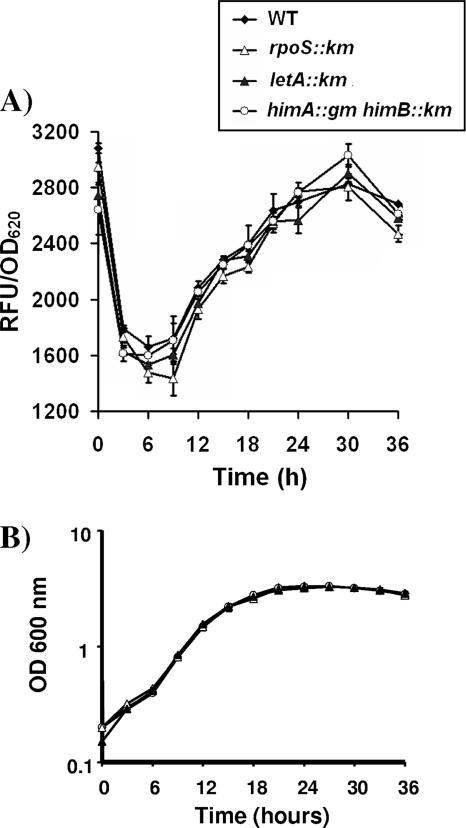
Cell cycle expression of oxyRLp.
To determine the temporal expression of oxyR during in vitro growth, a GFP reporter assay was performed. Figure 6 shows that the expression of oxyRLp is growth cycle-dependent, increasing during mid- and postexponential phases and decreasing slightly during late-stationary phase. Similar trends were observed in peroxide-sensitive ahpC1 and ahpC2D mutants. In an attempt to identify possible regulators of oxyR expression, the poxyRgfp reporter construct was transformed into mutant strains lacking known regulators of L. pneumophila virulence, rpoS, letA, and himAB (encoding integration host factor). No differences in oxyR expression were observed in any of the mutants analyzed (Fig. 6), indicating that the postexponential expression of OxyRLp is not activated by RpoS or the LetA/S regulatory cascade.
FIG. 6.
Cell cycle-dependent oxyR expression in L. pneumophila. (A) GFP reporter assays and (B) bacterial growth curves for PoxyR were performed in BYE broth, and fluorescence was determined for samples taken every 3 h, following the growth of the wild type (⧫) or the ahpC2D (•), rpoS (▵), letA (▴), and himAB (encoding integration host factor) (○) mutant strains. RFUs were normalized to 1.0 OD620 unit. Results are the means ± standard deviations of three independent experiments.
DISCUSSION
Our laboratory has characterized two alkyl hydroperoxide reductases (AhpC1 and AhpC2D) that are expressed at different stages of bacterial growth and provide essential peroxidatic functions in L. pneumophila. Loss of AhpC1 function results in increased expression of ahpC2D, suggesting a compensatory response to cellular oxidative stress (30). Using an ACDC assay, we identified OxyRLp as a regulatory factor controlling expression of the ahpC2D operon. OxyRLp bound to ahpC2D promoter sequences but did not bind to the promoter regions of ahpC1 or itself (autoregulation). Members of the OxyR family are well-characterized, redox-active transcriptional activators of genes associated with defense against oxidative stress. In contrast, none of the antioxidant defense genes (sodB, sodC, katA, katB, or ahpC1 and ahpC2D) of L. pneumophila appear to be activated in response to oxidative stress (4, 5, 30, 39, 43). Moreover, the expression of most of these genes, including oxyRLp, is aligned with growth stage, increasing dramatically around mid-exponential phase and peaking in early stationary phase, consistent with the view that oxidative stress is more acute for stationary-phase bacteria (4, 39). By demonstrating that OxyRLp repressed transcription from ahpC2D, we have partly resolved the basis for the reciprocal gene expression noted between ahpC1 and ahpC2D. Studies are in progress to identify regulatory mechanisms associated with postexponential activation of ahpC1 and oxyR.
OxyRLp appears to be functionally different from other OxyR proteins that have been shown to function as redox-active repressors (47, 48). Here we present several lines of evidence to suggest that OxyRLp may no longer function as a global regulator of antioxidant defenses: (i) oxyRLp expression is growth stage dependent, (ii) OxyRLp function is apparently essential, (iii) DNA-binding properties of OxyRLp are not redox dependent, (iv) OxyRLp contains key amino acid substitutions that likely ablate disulfide-bond formation or conformational changes that are required for activation, and (v) H2O2 treatment did not increase ahpC2-gfp levels in L. pneumophila or in an E. coli oxyR mutant (GS077) expressing oxyRLp. However, these findings do not exclude the possibility that OxyRLp retains some DNA sequence recognition for OxyR-regulated promoters of E. coli, as indicated by complementation and EMSA studies. Our studies indicate that OxyRLp may be locked in an activated DNA-binding conformation (29), independent of further activation by peroxide but sufficient to activate the antioxidant defense system of E. coli.
In E. coli, OxyR binds DNA as a dimer of dimers under both reducing and oxidizing conditions, yet only the latter promotes expression of its target genes (47). Treatment of E. coli OxyR with millimolar amounts of DTT was shown to promote the binding of OxyR to contact sites that blocked −35 sequences of OxyR-regulated promoters, whereas peroxide oxidation of OxyR led to a conformational change in the protein that contracted the DNA footprint and permitted strong induction of these promoters (44, 46, 47). In the case of L. pneumophila, treatment of OxyRLp with micromolar concentrations of H2O2 did not alter the footprint, which suggested either that OxyRLp was fully oxidized during protein purification or that DNA binding was independent of oxidation state. While millimolar concentrations of H2O2 indeed abolished DNA binding, the result is not considered physiologically relevant since biological activity was not recoverable by treatments with 200 mM DTT. In this regard, functional studies have shown that the redox-active thiols (C199 and C208) of OxyREc spontaneously oxidize to form disulfide bonds (redox potential of −185 mV) during protein purification (1, 50). In some organisms, such as Deinococcus radiodurans, oxidation of a single (peroxidatic) cysteine residue of OxyR is sufficient to promote activation (10). Thus, regardless of whether a single cysteine becomes oxidized or even a disulfide bond is formed, our studies suggest that the redox-sensing domain of OxyRLp no longer signals conformational changes that promote activation.
Members of the LysR family (which includes OxyR) use a ligand-binding conformational change mechanism to mediate DNA binding (11). Mutation-based studies have identified key amino acids required for the function of OxyR. For example, E. coli OxyR H125I and H218D mutations resulted in wild-type DNA binding but a loss of redox-mediated regulation (11). These amino acid substitutions occur naturally in the OxyRLp proteins from all four strains of L. pneumophila whose genomes have been sequenced (125 and 218) (Fig. 7). While the two redox-active cysteine residues involved in OxyR activation (Cys199 and Cys208) are conserved in OxyRLp, there is substantial amino acid sequence divergence in the activation domain that might affect redox activity. For example, a key arginine at position 220 in OxyREc (Fig. 7) that is predicted to interact with catalytic cysteines and peroxide (28) is changed to threonine in OxyRLp proteins. Interestingly, in the OxyR proteins of close relative C. burnetii and in Francisella tularensis, Arg220 is replaced by lysine, which might suggest a more traditional function in these organisms. Further comparisons revealed amino acid sequence variation within the helix-turn-helix region that might account for differences in the DNA-binding specificity noted between OxyRLp and OxyREc in complementation experiments and EMSAs. We suggest that the loss of the redox-sensing function of OxyRLp has occurred naturally through specific amino acid substitutions that have been demonstrated through empirical studies to alter disulfide bond formation and conformational changes required for activation.
FIG. 7.
Clustal W alignment of selected OxyR amino acid sequences. The alignments are of L. pneumophila Philadelphia-1 (LpnOxyR [OxyRLp]), Xanthomonas campestris (XanOxyR [OxyRXc]), E. coli (EcoliOxyR [OxyREc]), and C. burnetii (CbOxyR [OxyRCb]). The DNA binding helix-turn-helix (HTH) is shaded, and relevant amino acid positions are numbered and bolded.
In contrast to other bacteria, such as F. tularensis, C. burnetii, and Streptomyces species, in which oxyR is located upstream and divergently expressed from the ahpC2D system, the oxyRLp gene is located elsewhere in the chromosome. It is conceivable that the present oxyRLp was acquired laterally, perhaps concomitantly with the deletion or loss of an ancestral upstream ortholog, perhaps a necessary event associated with the evolution of a dimorphic lifestyle.
It seems unlikely that the only function for OxyRLp is to repress ahpC2D during the growth transition to stationary phase and into cyst morphogenesis. While downregulation of ahpC2D is consistent with metabolic changes and a shift to the NADPH-thioredoxin AhpC1 system that occur during the transition to stationary phase, ahpC2D deletion mutants are both viable and infectious (30). Moreover, high-level ahpC2D expression, noted with the ahpC1 deletion mutant, also suggested that the postexponential presence of AhpC2D was not toxic. Thus, OxyRLp must provide other essential regulatory functions to L. pneumophila, since this mutant could not be obtained by methods routinely used to generate mutants (30). We used the pattern search option of the L. pneumophila Paris and Lens genome website (http://genolist.pasteur.fr/LegioList/) to screen for additional genes, using motif patterns deduced from comparisons with either the E. coli or L. pneumophila OxyR binding motif. While these motifs are relatively degenerate, binding sites were observed upstream of approximately 50 genes, including the dps homolog, which has recently been found to prevent Fenton-mediated DNA damage by sequestering iron (37). Others included promoters of genes encoding efflux pumps (that exclude redox-cycling compounds and organic solvents); metal ion transporters (which regulate rates of Fenton chemistry); components of the respiratory chain, such as cytochrome oxidases, DNA repair, or modification enzymes; known virulence factors, such as the zinc metalloproteinase (shown to inhibit the oxidative burst); and substrates of the Dot/Icm type IVB secretion system (SidG, SdhB, and SdeA). Finally, OxyR binding motifs were also found upstream of genes encoding numerous transcriptional regulators, like Fur, FleQ, FleN, and other members of the LysR family, suggesting that there could be cross talk with other regulatory pathways. Interestingly, unlike in E. coli, fur is an essential gene in L. pneumophila (24). Further studies will be required to dissect additional regulatory functions associated with essentiality.
Our studies predict the participation of additional regulatory elements in controlling the expression of ahpC2D, since repression of ahpC2D by OxyRLp does not fully explain the upregulation of ahpC2D in an ahpC1 mutant or the postexponential activation of ahpC1 (30). In L. pneumophila, transition from exponential phase to stationary phase/cyst differentiation is controlled in part by postexponential regulators RpoS, LetA, and HimAB (2, 13, 20, 22, 32, 33, 52). However, the oxyRLp-gfp gene expression studies of rpoS, letA, and himAB mutants were indistinguishable from those of the wild type. While not directly examined, carbon storage regulator CsrA, an important repressor of stationary-phase genes during exponential phase (13, 33), may not be involved in regulating oxyRLp, since CsrA should have repressed postexponential expression of oxyRLp in letA and rpoS mutants. Future studies will employ the ACDC strategy to capture putative regulatory factors associated with the control of oxyRLp expression.
In summary, we have shown that OxyRLp represses ahpC2D expression and that oxyRLp is induced postexponentially. Our studies further establish that OxyRLp no longer functions as an oxidative response regulator in L. pneumophila, which is consistent with previous observations that L. pneumophila mounts no response to oxidative stress and that these bacteria are rather resistant to oxidative damage, even in phagocytic cells (30). We propose that OxyRLp has been adapted from an oxidative stress response regulator to a growth stage-specific regulator of genes mediating the transition from vegetative to resilient cyst-like transmissible forms.
Acknowledgments
We thank Gisela Storz and Michele Swanson for providing bacterial strains and plasmids.
This work was supported by CIHR grant MOP 14443 and NIH grant AI066058 to P.S.H. and a CIHR postdoctoral fellowship to A.K.C.B.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Aslund, F., M. Zheng, J. Beckwith, and G. Storz. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 966161-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, P., B. Byrne, Y. Chan, M. S. Swanson, and H. M. Steinman. 2003. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 714526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay, P., and H. M. Steinman. 2000. Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J. Bacteriol. 1826679-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandyopadhyay, P., and H. M. Steinman. 1998. Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J. Bacteriol. 1805369-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandyopadhyay, P., H. Xiao, H. A. Coleman, A. Price-Whelan, and H. M. Steinman. 2004. Icm/Dot-independent entry of Legionella pneumophila into amoeba and macrophage hosts. Infect. Immun. 724541-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 77-19. [DOI] [PubMed] [Google Scholar]
- 8.Bruggemann, H., A. Hagman, M. Jules, O. Sismeiro, M. A. Dillies, C. Gouyette, F. Kunst, M. Steinert, K. Heuner, J. Y. Coppee, and C. Buchrieser. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 81228-1240. [DOI] [PubMed] [Google Scholar]
- 9.Charoenlap, N., W. Eiamphungporn, N. Chauvatcharin, S. Utamapongchai, P. Vattanaviboon, and S. Mongkolsuk. 2005. OxyR mediated compensatory expression between ahpC and katA and the significance of ahpC in protection from hydrogen peroxide in Xanthomonas campestris. FEMS Microbiol. Lett. 24973-78. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., G. Xu, Y. Zhao, B. Tian, H. Lu, X. Yu, Z. Xu, N. Ying, S. Hu, and Y. Hua. 2008. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS ONE 13e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105103-113. [DOI] [PubMed] [Google Scholar]
- 12.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 623254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291353-360. [DOI] [PubMed] [Google Scholar]
- 14.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 2971189-1197. [DOI] [PubMed] [Google Scholar]
- 16.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garduño, R. A., E. Garduno, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 706273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halablab, M. A., M. Bazin, L. Richards, and J. Pacy. 1990. Ultra-structure and localisation of formazan formed by human neutrophils and amoebae phagocytosing virulent and avirulent Legionella pneumophila. FEMS Microbiol. Immunol. 2295-301. [DOI] [PubMed] [Google Scholar]
- 20.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 1814879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33721-731. [DOI] [PubMed] [Google Scholar]
- 22.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44107-118. [DOI] [PubMed] [Google Scholar]
- 23.Hassett, D. J., E. Alsabbagh, K. Parvatiyar, M. L. Howell, R. W. Wilmott, and U. A. Ochsner. 2000. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol. 1824557-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindre, T., H. Bruggemann, C. Buchrieser, and Y. Hechard. 2008. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 15430-41. [DOI] [PubMed] [Google Scholar]
- 26.Hishinuma, S., M. Yuki, M. Fujimura, and F. Fukumori. 2006. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 82115-2124. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs, R. F., R. M. Locksley, C. B. Wilson, J. E. Haas, and S. J. Klebanoff. 1984. Interaction of primate alveolar macrophages and Legionella pneumophila. J. Clin. Investig. 731515-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kona, J., and T. Brinck. 2006. A combined molecular dynamics simulation and quantum chemical study on the mechanism for activation of the OxyR transcription factor by hydrogen peroxide. Org. Biomol. Chem. 43468-3478. [DOI] [PubMed] [Google Scholar]
- 29.Kullik, I., M. B. Toledano, L. A. Tartaglia, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 1771275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBlanc, J., R. Davidson, and P. S. Hoffman. 2006. Compensatory functions of two alkyl hydroperoxide reductases in the oxidative defense system of Legionella pneumophila. J. Bacteriol. 1886235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loprasert, S., M. Fuangthong, W. Whangsuk, S. Atichartpongkul, and S. Mongkolsuk. 2000. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 371504-1514. [DOI] [PubMed] [Google Scholar]
- 32.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219241-248. [DOI] [PubMed] [Google Scholar]
- 33.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50445-461. [DOI] [PubMed] [Google Scholar]
- 34.Mongkolsuk, S., W. Whangsuk, P. Vattanaviboon, S. Loprasert, and M. Fuangthong. 2000. A Xanthomonas alkyl hydroperoxide reductase subunit C (ahpC) mutant showed an altered peroxide stress response and complex regulation of the compensatory response of peroxide detoxification enzymes. J. Bacteriol. 1826845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, C., S. Hendy, K. Reid, and J. Cavanagh. 2002. Acrylamide capture of DNA-bound complexes: electrophoretic purification of transcription factors. BioTechniques 32808-810, 812, 814-815. [DOI] [PubMed] [Google Scholar]
- 36.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 1824533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, M., S. T. Yun, S.-Y. Hwang, C.-I. Chun, and T. I. Ahn. 2006. The dps gene of symbiotic “Candidatus Legionella jeonii” in Amoeba proteus responds to hydrogen peroxide and phagocytosis. J. Bacteriol. 1887572-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rankin, S., Z. Li, and R. R. Isberg. 2002. Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect. Immun. 703637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadosky, A. B., J. W. Wilson, H. M. Steinman, and H. A. Shuman. 1994. The iron superoxide dismutase of Legionella pneumophila is essential for viability. J. Bacteriol. 1763790-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sisson, G., A. Goodwin, A. Raudonikiene, N. J. Birks, H. Han, A. Mukhopadhyay, D. E. Berg, and P. S. Hoffman. 2002. Enzymes associated with the reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob. Agents Chemother. 462116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer, B., S. Master, P. Sander, T. Zahrt, M. McFalone, J. Song, K. G. Papavinasasundaram, M. J. Colston, E. Boettger, and V. Deretic. 2001. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect. Immun. 695967-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St. John, G., and H. M. Steinman. 1996. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J. Bacteriol. 1781578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. The OxyR regulon. Antonie van Leeuwenhoek 58157-161. [DOI] [PubMed] [Google Scholar]
- 45.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248189-194. [DOI] [PubMed] [Google Scholar]
- 46.Tartaglia, L. A., G. Storz, and B. N. Ames. 1989. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210709-719. [DOI] [PubMed] [Google Scholar]
- 47.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78897-909. [DOI] [PubMed] [Google Scholar]
- 48.Tseng, H.-J., A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, G., P. Alamuri, and R. J. Maier. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 61847-860. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 2791718-1721. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 1834562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the dot/icm type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]