Abstract
A novel strictly anaerobic bacterium designated strain SSD-17BT was isolated from the hypersaline brine-sediment interface of the Shaban Deep, Red Sea. Cells were pleomorphic but usually consisted of a central coccoid body with one or two “tentacle-like” protrusions. These protrusions actively alternated between a straight, relaxed form and a contracted, corkscrew-like one. A peptidoglycan layer was not detected by electron microscopy. The organism forms “fried-egg”-like colonies on MM-X medium. The organism is strictly anaerobic and halophilic and has an optimum temperature for growth of about 30 to 37°C and an optimum pH of about 7. Nitrate and nitrite are reduced; lactate is a fermentation product. The fatty acid profile is dominated by straight saturated and unsaturated chain compounds. Menaquinone 4 is the major respiratory quinone. Phylogenetic analysis demonstrated strain SSD-17BT represents a novel and distinct lineage within the radiation of the domain Bacteria. The branching position of strain SSD-17BT was equidistant to the taxa considered to be representative lineages of the phyla Firmicutes and Tenericutes (with its sole class Mollicutes). The phenotypic and phylogenetic data clearly show the distinctiveness of this unusual bacterium, and we therefore propose that strain SSD-17BT (= DSM 18853 = JCM 14575) represents a new genus and a new species, for which we recommend the name Haloplasma contractile gen. nov., sp. nov. We are also of the opinion that the organism represents a new order-level taxon, for which we propose the name Haloplasmatales.
The divergent movement of the African and Arabian tectonic plates, which is associated with deposition of new oceanic crust, led to the formation of several isolated topographical depressions within the Red Sea basin (4). Redissolution of buried Miocenic evaporites and/or hydrothermal phase separation, followed by upward migration to the seafloor and later accumulation in enclosed depressions, filled as many as 25 such deeps with highly saline water (13, 15). In addition to their common extreme salinity (up to 27% [wt/vol] NaCl), all of these brines are anoxic, are enriched in metals and hydrocarbons, and have temperatures higher than 21.7°C, which is the mean deep-seawater temperature of the Red Sea (13, 29).
The brine-filled deeps of the Red Sea represent a rare type of extreme environment and were considered to be sterile as recently as the late 1960s (34). Few studies have focused on the microbiology of these biotopes, resulting in only a few microbial isolates (1, 2, 10). Phylogenetic studies have, however, revealed an unexpectedly high microbial diversity (10, 11). Parallel studies, focusing on the microbiology of analogous deep-sea brines located in the eastern Mediterranean Sea further corroborate the extensive microbiological diversity in such biotopes (8).
Samples for geochemical and microbiological studies were retrieved from the northernmost brine-filled deeps of the Red Sea during the RV Meteor Cruise 52/3 in 2002. Isolation attempts with a sample of the previously unexplored brine-sediment interface of the Shaban Deep led to the recovery of strain SSD-17BT. This isolate displayed a highly unusual morphology with contractile “tentacle-like” structures and minibodies, and it represents a novel phylogenetic lineage. The phenotypic and phylogenetic data point to the distinctiveness of this organism, which we believe represents a novel taxon at the order level.
MATERIALS AND METHODS
Sampling, storage, and transport.
The Shaban Deep, one of the northernmost deep-sea anoxic brine pools of the Red Sea, was sampled during RV Meteor Cruise 52/3 in 2002. The sampling of undisturbed surface sediments and overlying bottom brine (i.e., the brine-sediment interface) of this deep was achieved with the deployment of a Multicorer unit (GeoB, University of Bremen, Bremen, Germany). The samples were maintained under anaerobic conditions at 19°C for 15 days before being transported, for about 24 h at room temperature, to the laboratory, where the samples were stored at 19°C.
Growth conditions and isolation.
Strain SSD17BT was routinely grown on anaerobic MM-X medium containing the following (per liter): NaCl, 57.5 g; KH2PO4, 0.5 g; yeast extract, 0.2 g; peptone, 0.2 g; starch, 5.0 g; and artificial seawater, 250 ml (17). It was enriched in anaerobic MM-X medium containing 12% (wt/vol) NaCl. Strictly anaerobic culture medium was prepared as described previously (3). The medium was reduced by adding 0.05% Na2S·9H2O, with resazurin (5 μg liter−1) added as the redox indicator. Prior to autoclaving, the medium was dispensed in 10-ml aliquots into 28-ml stoppered serum tubes, and the gas phase was exchanged with N2 (250 kPa). Solidified MM-X medium was made by the addition of 1.5% (wt/vol) agar. A pure culture was obtained by visual controlled separation of the desired cell type from freshly grown mixed cultures, by means of a laser microscope-assisted procedure (16).
Morphological studies.
Cultures were routinely observed using phase-contrast microscopy. For transmission electron microscopy cells were fixed, immediately after harvesting by centrifugation, with 2.5% glutardialdehyde in 75 mM sodium cacodylate, 2 mM MgCl2, and 1.0 M NaCl (pH 7.0) for 1 h at room temperature; rinsed several times in cacodylate buffer containing 1.0 M NaCl; and postfixed for 1 h with 1% osmium tetroxide in the same buffer at room temperature. Cells were then stained with uranyl acetate, dehydrated, embedded in low-viscosity resin, and poststained as described previously (35) Thin sections were observed with an EM 912 electron microscope (Zeiss, Oberkochen, Germany).
For scanning electron microscopy, bacteria were concentrated by filtration through 0.4-μm Nuclepore membrane filters, which were fixed with 2.5% (vol/vol) glutardialdehyde in 75 mM cacodylate buffer (pH 7.0) supplemented with 1.0 M NaCl. All other steps were performed as described previously (18). The specimens were examined with a Hitachi S-4100 field emission scanning electron microscope operated at 5 to 8 kV.
Physiological and biochemical studies.
All studies were performed at 30°C and pH 6.5 in medium containing 6% NaCl under strictly anaerobic conditions, unless otherwise stated. The presence of cytochrome oxidase and catalase was determined as described elsewhere (30). Growth at different temperatures, pH values, and NaCl concentrations was determined in MM-X liquid medium. Growth was assessed by measuring the turbidity at 600 nm or by counting the number of cells in a field with a Neubauer counting chamber (Weber, United Kingdom).
Hydrolysis of arginine and urea and the presence of phosphatase were assessed after growth on MM-X agar (19). Cholesterol was added to the medium at final concentrations of 1, 5, 10, and 20 μg ml−1 (28). Sensitivity to antimicrobial agents was tested by addition of 25 μg/ml of filter-sterilized ampicillin, bacitracin, cephalosporin C, chloramphenicol, and penicillin G to MM-X liquid medium. Metabolism of carbon sources was assessed using a Biolog GN2 microplate (Biolog) under anaerobic conditions according to the manufacturer's instructions but using the growth medium without organic compounds as the inoculating fluid. The reduction of nitrate was examined using 1.0, 0.1, and 0.01 g liter−1 of KNO3, and the nitrite produced quantified as described previously (30). The formation of d- and l-lactate and acetate was assessed in 10-day-old cultures using the Boehringer/Mannheim/R-Biopharm kits 11112821035 and 10148261035. The effect of the growth medium on the enzyme activities was controlled by adding the reaction mixtures to medium containing known amount of the substrates.
Chemotaxonomic characterization.
Cultures for fatty acid analysis were grown on MM-X liquid medium containing 6.0% (wt/vol) NaCl at 30°C for 72 h. Fatty acid methyl esters were obtained and separated, identified, and quantified with the standard MIS library generation software as described by the manufacturer (Microbial ID Inc.). The respiratory quinones were determined as described elsewhere (31). The polar lipid profile of isolate SSD-17BT was determined by the Identification Service of the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany.
Determination of G+C content and 16S rRNA gene sequence and phylogenetic analysis.
DNA for determination of G+C content was isolated as previously described (25). The G+C content of DNA was determined by high-pressure liquid chromatography as described elsewhere (24). Extraction of nucleic acids, PCRs, and sequencing of the 16S rRNA gene (with primers 9bF and 1406uR) were carried out as described previously (9, 10). The 16S rRNA gene sequences were aligned against representative reference sequences of members of the Bacteria using the ae2 editor (6). The method of Jukes and Cantor (20) was used to calculate evolutionary distances. Phylogenetic dendrograms and bootstrap analyses were generated using neighbor-joining, maximum-likelihood, and maximum-parsimony methods contained in the PHYLIP and ARB software packages (12).
Determination of physical and chemical parameters of the sample and site.
Salinity and pH values were promptly measured using a hand refractometer (Atago, Tokyo, Japan) and Neutralit pH strips (Merck Eurolab, Darmstadt, Germany), respectively.
Concentrations of several major inorganic constituents (e.g., Na, Mg, K, Ca, and B) and some trace elements (e.g., Li, Fe, Mn, and Ba) in the pore water were determined with a Unicam 701 ICP-AES spectrometer and a Unicam 939 AAS spectrometer, while chlorinity was determined titrimetrically. Mineralogical analysis of the sediment fraction was performed by semiquantitative X-ray diffraction, performed on a Philips X'Pert Pro multipurpose diffractometer equipped with a Cu tube (kα 1.541 radiation, 45 kV, 40 mA), a fixed divergence slit of 1/4° 2θ, a secondary monochromator, and the X'Celerator detector system. Mineral identification and semiquantification was performed using the X'Pert HighScore software and referring to the “matrix-flushing method” (5).
Nucleotide sequence accession number.
The GenBank accession numbers for the 16S rRNA gene sequence of strain SSD-17BT is EF999972.
RESULTS
Physical and chemical parameters.
Sample SD-17 was collected from the brine-sediment interface of the eastern basin of the Shaban Deep (station no. 133-1; 26°13.9′N, 35°21.3′E) at a depth of 1,447 m. The sample had a pH of 6.0, a salinity of 24.4%, and a temperature of 24.1°C. Pore water analysis and comparison with values from normal Red Sea water and the lower brine-seawater interface revealed significant enrichment in sodium and chloride within the brine body and an increase in the concentration of iron and manganese with depth (Table 1). Analysis of the sediment fraction revealed a predominance of carbonates, mostly calcite (trigonal-rhombohedral CaCO3), Mg-calcite (Mg0.1Ca0.9CO3), and aragonite (orthorhombic CaCO3), together with significant amounts of muscovite {KAl2[(OH)2/Si4O10]} and quartz (SiO2).
TABLE 1.
Concentrations of selected major constituents and some trace elements
| Element | Concn in:
|
||
|---|---|---|---|
| Red Sea watera | Shaban Deep brinea | Shaban Deep sediment pore water | |
| Na | 564 mM | 4,410 mM | 4,680 mM |
| Mg | 62.2 mM | 77.3 mM | 76.8 mM |
| K | 12.2 mM | 44.5 mM | 53.3 mM |
| Ca | 11.6 mM | 20.3 mM | 91.6 mM |
| B | 0.5 mM | 0.61 mM | 0.69 mM |
| Cl | 634.4 mM | 4,538 mM | 4,614 mM |
| Li | 33.1 μM | 122.4 μM | |
| Fe | <0.1 μM | 143.2 μM | 1,015 μM |
| Mn | <0.05 μM | 30.0 μM | 96.1 μM |
| Ba | <0.05 μM | 0.7 μM | |
Data are from reference 11.
Enrichment, isolation, and morphological characteristics of strain SSD-17BT.
Initial enrichment cultures were established in 10 ml of MM-X medium containing 12% (wt/vol) NaCl and inoculated with 0.2 ml of the original sample. Microbial growth was detected by the presence of turbidity and by phase-contrast microscopy after approximately 30 days of incubation at 22°C. After several transfers of the enrichment, a pure culture of strain SSD-17BT was obtained using the laser microscope-assisted “optical tweezers” method.
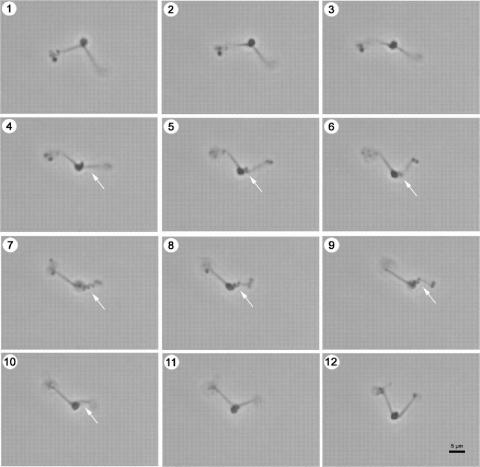
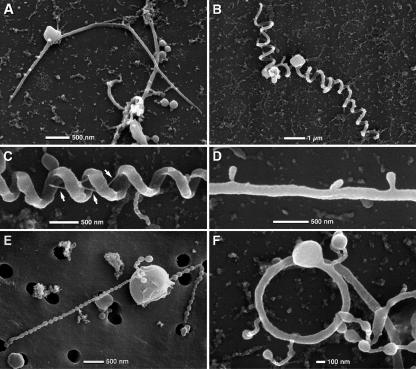
After 3 days of incubation, at 30°C on MM-X agar, very small colonies with a diameter of approximately 0.03 to 0.05 mm could be observed under a dissecting microscope. The colonies were gold-yellow and round, and most had a “fried-egg”-like appearance. Cells of isolate SSD-17BT were pleomorphic, but almost all consisted of a central coccoid body (normally between 0.5 and 1.8 μm in diameter) with one or two “tentacle-like” protrusions at all salinities examined between 1.5 and 18% (wt/vol). These “tentacle-like” protrusions actively alternated between a straight, relaxed form and a contracted corkscrew-like form with a periodicity of 5 to 10 seconds (Fig. 1 and 2A and B). Scanning electron microscopy corroborated the pleomorphism of isolate SSD-17BT (Fig. 2). Although two protrusions were most frequently observed, cells with only one or several protrusions were also observed. There was a wide range of structural variants of protrusions: straight or slightly curved with a length of up to 20 μm and diameters of 0.05 to 0.15 μm (Fig. 2A), helical (with a radius of a few micrometers) with one or two coils (Fig. 2F), helical with 5 to 10 coils (Fig. 2B and C), chains of connected microspheres with diameter of 0.15 μm (Fig. 2E), helical or straight protrusions with secondary protuberances ranging from bulges to extended polyp-like structures (Fig. 2D and F), or combinations of the above (Fig. 2F). Secondary protuberances frequently terminate in bulbous structures of various diameters (Fig. 2F). Helical protrusions occasionally exhibit thin filaments between coils (Fig. 2C). Taking sputter coating into consideration, the diameter of the filaments is in the range of 0.01 to 0.015 μm.
FIG. 1.
Series of consecutive light micrographs (phase contrast) of strain SSD-17BT, illustrating the contraction of the “tentacle-like” protrusions. The time span between the first and last micrographs was about 8 s.
FIG. 2.
Scanning electron micrographs showing examples of morphological variants of the central body and protrusions of strain SSD-17BT. (A) Central body with two protrusions (relaxed state). (B) Central body with two coiled protrusions (contracted stage). (C) Detail of coiled protrusions showing helical winding; occasionally thin filaments are visible (arrows). (D) Protrusions exhibiting lateral polyp-like protuberances. (E) Central body exhibiting protrusions with chains of microspheres. (F) Terminal bacterial body with a helical protrusion (one coil) and several extended polyp-like secondary protuberances, frequently with terminal bulbous structures of various diameters.
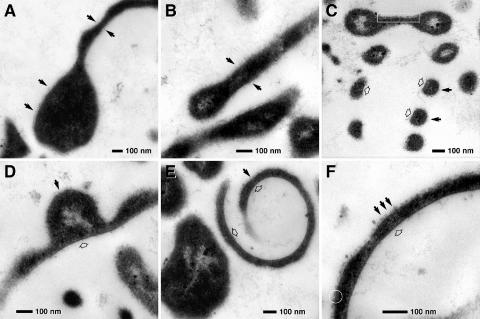
Transmission electron microscopy of ultrathin sections provided additional information on the structural composition of protrusions (Fig. 3A to F). In particular, secondary protuberances are integral elements of the protrusions (Fig. 3A, B, and D). Cross sections of the helical protrusions show a flat inner surface and rounded outer surface (Fig. 2C and 3C). Regardless of their shape, protrusions appear to be coated by a diffuse envelope (Fig. 3A to F). This diffuse coating is visible only on the outer side of the coils, while the inner side appears to be uncoated. The matrixes of both the bacterial bodies and their protrusions are very electron dense (osmiophilic). Due to the high contrast, the cytoplasmic membrane can only occasionally be seen in favorable sections (Fig. 3C and F). Fibrous electron-transparent areas indicating a nucleoid are found in the cell bodies (Fig. 3E), in the helical protrusions (Fig. 3C), and in the polyp-like structures and other secondary protuberances (Fig. 3B and D). These nucleoids indicate that the protrusions are extensions of the cytoplasm and allow the DNA to move through them. These observations suggest that viable cells can form from these secondary structures of the protrusions.
FIG. 3.
Transmission electron micrographs of ultrathin sections of morphological variants of protrusions from strain SSD-17BT. (A) Central body with one “tentacle-like” protrusion (relaxed state); the surface of the cell is coated by a diffuse electron-dense layer (arrows). (B) Detail of “tentacle-like” protrusions (relaxed state); the surface of the cell is coated at both sides by a diffuse electron-dense layer (arrows). (C) Longitudinal section of a helical protrusion (contracted stage); several windings are cross sectioned. (D to F) Cross sections of helical protrusions. The outer side of the rings (C), lateral protuberances (D), and helixes (E and F) are “rough” (black arrows) compared to the inner surface (open arrows). The cytoplasmic membrane can be recognized locally (C, framed area; F, circle).
Physiological and biochemical characteristics.
Strain SSD-17BT grew very poorly under all conditions examined, with a maximum turbidity of about 0.09 at 600 nm. Omission of yeast extract or peptone from the medium composition resulted in no growth, while omission of starch resulted in decreased growth yields. The strain grew at between 10 and 44°C, with optimal growth at between 30 and 37°C. The optimum pH for growth was about 7.0, with growth occurring between pH 6.0 and 8.0. The NaCl range for growth was determined to be between 1.5 and 18% (wt/vol), with optimal growth occurring at around 8% (wt/vol). The addition of cholesterol had no effect on growth. Strictly anaerobic conditions were necessary for growth, with no growth occurring under aerobic or microaerophilic conditions. Cells were filterable through 0.8-μm and 0.45-μm but not through 0.2-μm pore membrane filters. Strain SSD-17BT was resistant to penicillin, ampicillin, and cephalosporin but susceptible to bacitracin and chloramphenicol. Oxidase and catalase were negative. The organism reduced nitrate slowly to nitrite, but the levels of nitrite then decreased with prolonged incubation, indicating that the organism denitrified, although an increase in turbidity of the cultures during denitrification was not observed. The production of acetate was not detected, but l-lactate was produced, indicating that the organism is able to ferment some components of the medium.
Strain SSD-17BT was able to metabolize a very restricted range of carbon sources using Biolog GN2 microplates. Clear positive reactions were obtained only for l-arabinose and d-psicose, with positive weak reactions observed for α-ketobutyric acid, α-ketoglutaric acid, and α-ketovaleric acid. SSD-17BT was also unable to hydrolyze urea or arginine, and phosphatase activity was not detected.
Chemotaxonomic characteristics.
The fatty acid composition of strain SSD-17BT was dominated by straight saturated and unsaturated chain compounds, of which the major fatty acids were 16:0 (35.7%), 18:1 ω9c (23.9%), and 18:0 (18.1%). Other fatty acids detected included summed feature 5 (most likely 18:2 ω6,9c [12.8%]), 14:0 (4.6%), summed feature 3 (most likely 16:1 ω7c [2.3%]), 13:0 anteiso (1.1%), 19:1 iso (1.0%), and 20:0 (0.6%). The major polar lipids were phosphatidylglycerol and bisphosphatidylglycerol, but three minor glycolipids were also present. The major respiratory quinone was menaquinone 4 (MK-4).
16S rRNA gene sequence determination and phylogenetic analyses.
A nearly complete 16S rRNA gene sequence comprising 1,345 nucleotides was determined for strain SSD-17BT and compared to those of representatives of the major lineages within the Bacteria and the Archaea (Fig. 4). Phylogenetic analysis demonstrated strain SSD-17BT to represent a novel and distinct lineage within the radiation of the domain Bacteria. The branching position of strain SSD-17BT was equidistant to the taxa considered to be representative lineages of the phyla Firmicutes and Tenericutes (with the sole class Mollicutes). Pairwise similarity values to representatives of these lineages were in the range of 80.1 to 91.5%, with highest similarity to Gemella haemolysans (91.5%), which is a member of the phylum Firmicutes. Within this distinct lineage, strain SSD-17BT clustered with two environmental clone sequences from different sources, one from Lake Van, Turkey (91.0%) (accession no. AY642583) and the other from a mouse gastrointestinal tract (93.8%) (AJ400262). The DNA G+C content of strain SSD-17BT was found to be 33.6 mol%.
FIG. 4.
Phylogenetic tree based on a comparison of the 16S rRNA gene sequences of strains SSD-17BT and representatives of the main lineages within the domain Bacteria. Additional species of the phyla Firmicutes and Tenericutes were included to establish the relationship of this novel lineage to these previously described phyla. The tree was reconstructed from distance values using the neighbor-joining method. Scale bar, 5 inferred nucleotide substitutions per 100 nucleotides.
DISCUSSION
The brine-filled deeps of the Red Sea represent unusual environments which remain largely unexplored. During the course of a microbial diversity assessment study of the Shaban Deep, an organism designated SSD-17BT was recovered from the brine-sediment interface of this deep, which was striking in both its morphology and its dynamic contractility. Phylogenetic studies showed strain SSD-17BT to represent a new lineage between the phylum Firmicutes and the phylum Tenericutes (Mollicutes). The Firmicutes currently comprise the low-G+C gram-positive organisms such as the bacilli and clostridia. The Tenericutes lineage comprises wall-less species of genera such as Mycoplasma and Acholeplasma, as well as cell-walled lineages such as the misnamed taxa Clostridium ramosum, Clostridium innocuum, Eubacterium biforme, and the species of the genera Solobacterium, Coprobacillus, and Erysipelothrix. Even though the branching point of the lineage representing strain SSD-17BT is equidistant to the lineages of the Firmicutes and the Tenericutes (Mollicutes), the sequence similarities between the new organism and representatives of the Firmicutes are higher. This is in part due to the short branch lengths of genera such as Gemella, Globicatella, Aerococcus, and Bacillus, in contrast to the extended lineages of genera of the phylum Tenericutes (Mollicutes) such as Acholeplasma, Ureaplasma, and Coprobacillus. This new lineage, in addition to strain SSD-17BT, contains a number of environmental 16S rRNA gene clone sequences at the 91.0 to 94.0% similarity level. These include an environmental clone sequence retrieved during a study of the halophilic and alkalophilic microbial communities from Lake Van in Turkey (23). There is a significant ecological correlation between the two saline environments, but the other branch within this novel lineage comprises two highly similar environmental clone sequences retrieved from mammalian gastrointestinal tracts, which are not presently known to harbor halophilic taxa.
The species represented by strain SSD-17BT shares several morphological characteristics with the wall-less species of the class Mollicutes, including the absence of a cell wall, which was not observed by electron microscopy. Resistance to penicillin and cephalosporin also argues for the absence of a peptidoglycan layer, as in the wall-less mollicutes but unlike the members of the Firmicutes, namely, the species of the genus Gemella (26) and walled mollicutes where penicillin sensitivity has been examined (21). The ability to be filtered through 450-nm pore membrane filters, typical colonies with a “fried-egg”-like appearance, and the unusual cellular contractility and dynamic morphology of this isolate also indicates the lack of a rigid cell wall and evokes the flagellum-independent movements seen in the strains of the genus Spiroplasma (22, 32). However, the new organism possesses a central coccoid body which is not found in the species of the genus Spiroplasma. The chains of microspheres which appear to derive from the “tentacles” protruding from the central body of strain SSD-17BT might result from an atypical binary fission process, similar to that observed in isolates of the genus Mycoplasma (27). These characteristics of strain SSD-17BT, like for other wall-less organisms, are probably due to the lack of a rigid cell wall.
The fatty acid composition indicates that the organism from the Shaban Deep, like several wall-less members of the class Mollicutes, a few walled relatives (namely, Erysipelothrix and the most closely related members of the Firmicutes, such as Gemella), the lactic acid bacteria, and Aerococcus, possesses straight-chain saturated and unsaturated fatty acids (21, 33, 35). MK-4 has been rarely detected in bacteria but has been found in Mycoplasma arthriditis and Acholeplasma axanthum (14), although most authors have not found respiratory quinones in wall-less members of the class Mollicutes (26). On the other hand, trace amounts of MK-7 have been found in Erysipelothrix tonsilarum (33), although respiratory quinones have not been detected or sought in other walled mollicutes or the most closely related genera of the Firmicutes (7).
Despite many similarities to the wall-less members of the Mollicutes, the ability of isolate SSD-17BT to grow at between 1.5 and 18% NaCl clearly clashes with the characteristic osmotic fragility and the obligate association with plant or animal hosts for growth, which are hallmarks of members of the class Mollicutes (26, 27). The relatively simple medium for growth (which does not contain complex organic components such as serum), the extreme environment from which the organism was isolated, and the lack of host dependency of strain SSD-17BT, in addition to the phylogenetic position, strongly support the proposal of a novel genus and species which we name Haloplasma contractile gen. nov., sp. nov. Moreover, the distinct phylogenetic lineage of strain SSD-17BT argues for the proposal of a higher-level taxon to accommodate this bacterium, perhaps at the phylum level. However, only one strain of this lineage has been isolated, and the phylogenetic data available indicate that other organisms of the same lineage inhabit not only saline environments, such as deep sea brines and Lake Van (AY642583), but also mammalian intestines (AJ400262), clearly indicating that the organisms of this distinct lineage have characteristics which could be very different from those of strain SSD-17BT. The absence of other isolates precludes the description of a novel class or phylum at this point. Nevertheless, the organism from the Shaban Deep has distinct physiological and biochemical characteristics and phylogenetic position to justify the proposal of a new order. We are therefore of the opinion that strain SSD-17BT belongs to a novel family, for which we propose the name Haloplasmataceae fam. nov., within a novel order, for which we propose the name Haloplasmatales ord. nov.
Description of Haloplasmatales ord. nov. (Rainey, da Costa, Antunes, and Huber).
Haloplasmatales (Ha.lo.plas.ma.ta′les. N.L. n. Haloplasma, type genus of the order; suff. -ales, ending denoting an order; N.L. fem. pl. n. Haloplasmatales, the Haloplasma order). The description of this order is the same as that for the genus Haloplasma. The order contains the family Haloplasmataceae. The type genus of the order is Haloplasma.
Description of Haloplasmataceae fam. nov. (Rainey, da Costa, Antunes, and Huber).
Haloplasmataceae (Ha.lo.plas. ma.ta′ce.ae. N.L. n. Haloplasma, type genus of the family; suff. -aceae, ending to denote a family; N.L. fem. pl. n. Haloplasmataceae, the Haloplasma family). The description is the same as that for the genus Haloplasma. The family contains the type genus Haloplasma.
Description of Haloplasma gen. nov. (Antunes. Rainey, da Costa, and Huber).
Haloplasma gen. nov. (Ha.lo.plas′ma. Gr. n. hals, salt; Gr. neut. n. plasma, something formed or molded, a form; N.L. neut. n. Haloplasma, a salt-loving form). Gram-negative, non-spore-forming, pleomorphic cells sometimes with “tentacle-like” protrusions. Moderately halophilic. Mesophilic. Grows in medium with neutral pH. Strictly organotrophic. Strictly anaerobic. Denitrifying and fermentative. Oxidase and catalase negative. Cell wall is not detected. Straight chain fatty acids are present. The major polar lipids are phosphatidylglycerol and bisphosphatidylglycerol (DPG). MK-4 is the major respiratory quinone. The G+C content of the DNA of the type species is 33.6 mol%. The type species of this genus is Haloplasma contractile.
Description of Haloplasma contractile sp. nov. (Antunes. Rainey, da Costa, and Huber).
Haloplasma contractile sp. nov. (con.trac.ti′le. N.L. neut. adj. contractile, contractile). Haloplasma contractile possesses, in addition to the characteristics in the description of the genus, pleomorphic cells, usually consisting of a central coccoid body (0.5 to 1.8 μm) with one or two “tentacle-like” protrusions (up to 20 μm in length and 0.1 to 0.25 μm in diameter). Motility may occur by cellular contraction of the protrusions. Colonies in MM-X agar are very small (0.03 to 0.05 mm in diameter), gold-yellow, and round, often with a “fried-egg”-like appearance. The NaCl range for growth is 1.5 to 18% (wt/vol); optimum growth is at about 8% (wt/vol) NaCl. Growth occurs at between 10 and 44°C (optimal temperature, 30 to 37°C), and the pH range is between about 6.0 and 8.0 (optimal pH, 7.0). Cells are able to be filtered through 450-nm pore membrane filters. Resistant to penicillin G, ampicillin, and cephalosporin but susceptible to bacitracin and chloramphenicol. Nitrate and nitrite are reduced; lactate was detected from the fermentation of the medium components. Urea and arginine were not hydrolyzed. Phosphatase was not detected. l-Arabinose, d-psicose, α-ketobutyric acid, α-ketoglutaric acid, and α-ketovaleric acid were metabolized with the Biolog GN2. Peptone and yeast extract are required for growth. The predominant cellular fatty acids are 16:0, 18:0, and 18:1 ω9c. The G+C content of the DNA of the type species is 33.6 mol%. Isolated from the anoxic brine-sediment interface of the Shaban Deep, Red Sea. The type strain, SSD-17BT, has been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany, as strain DSM 18853 and in the Japan Collection of Microorganisms (JCM), Saitama, Japan, as strain JCM 14575.
Acknowledgments
We are grateful for the valuable help of the scientists and crew on board RV Meteor (M 52/3 cruise). We also thank G. Gmeinwieser (U. Regensburg), M. Bock (U. Regensburg), and S. Dobler (LMU Munich) for their excellent technical assistance. The X-ray diffraction analyses were performed by C. Vogt (Crystallography Group, Dept. of Geosciences, University of Bremen).
This work was supported by the Deutsche Forschungsgemeinschaft (DFG HU 711/2-1 and HU 711/2-2), Fundação para a Ciência e a Tecnologia (POCI/BIA-BDE/56014/2004), and by Governor's Biotechnology Initiative of the Louisiana Board of Regents (BOR 021—Moving an Established Marine Biotechnology Program to the Next Level: Natural Product Screening and Development). A. Antunes was supported by a postdoctoral scholarship from Fundação para a Ciência e a Tecnologia (SFRH/BPD/22576/2005).
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Antunes, A., W. Eder, P. Fareleira, H. Santos, and R. Huber. 2003. Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine-seawater interface of the Shaban Deep, Red Sea. Extremophiles 729-34. [DOI] [PubMed] [Google Scholar]
- 2.Antunes, A., M. Taborda, R. Huber, C. Moissl, M. F. Nobre, and M. S. da Costa. 2008. Halorhabdus tiamatea sp. nov., a non-pigmented, extremely halophilic archaeon from a deep-sea hypersaline anoxic basin of the Red Sea. Int. J. Syst. Evol. Microbiol. 58215-220. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonatti, E. 1985. Punctiform initiation of seafloor spreading in the Red Sea during transition from a continental to an oceanic rift. Nature 31633-37. [Google Scholar]
- 5.Chung, F. H. 1974. Quantitative interpretation of X-ray diffraction patterns. I. Matrix-flushing method of quantitative multicomponent analysis. J. Appl. Crystal. 7513-519. [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, M. D., and D. Jones. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45316-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffonchio, D., S. Borin, T. Brusa, L. Brusetti, P. W. J. J. van der Wielen, H. Bolhuis, M. M. Yakimov, G. D'Auria, L. Giuliano, D. Marty, C. Tamburini, T. J. McGenity, J. E. Hallsworth, A. M. Sass, K. N. Timmis, A. Tselepides, G. J. de Lange, A. Hübner, J. Thomson, S. P. Varnavas, F. Gasparoni, H. W. Gerber, E. Malinverno, C. Corselli, and Biodeep Scientific Party. 2006. Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature 404203-207. [DOI] [PubMed] [Google Scholar]
- 9.Dyall-Smith, M. 2001. The halohandbook: protocols for halobacterial genetics, ed. 4.5. University of Melbourne, Melbourne, Australia.
- 10.Eder, W., L. L. Jahnke, M. Schmidt, and R. Huber. 2001. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 673077-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eder, W., M. Schmidt, M. Koch, D. Garbe-Schönberg, and R. Huber. 2002. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine-seawater interface of the Shaban Deep, Red Sea. Environ. Microbiol. 4758-763. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 13.Holländer, R., G. Wolf, and W. Manheim. 1977. Lipoquinones of some bacteria and mycoplasmas, with considerations on their functional significance. Antonie van Leeuwenhoek 43177-185. [DOI] [PubMed] [Google Scholar]
- 14.Hovland, M., T. Kuznetsova, H. Rueslåtten, B. Kvamme, H. K. Johnsen, G. E. Fladmark, and A. Hebach. 2006. Sub-surface precipitation of salts in supercritical seawater. Basin Res. 18221-230. [Google Scholar]
- 15.Huber, R., H. Huber, and K. O. Stetter. 2000. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev. 24615-623. [DOI] [PubMed] [Google Scholar]
- 16.Huber, R., C. R. Woese, T. A. Langworthy, J. K. Kristjansson, and K. O. Stetter. 1990. Fervidobacterium islandicum sp. nov., a new extremely thermophilic eubacterium belonging to the “Thermotogales.” Arch. Microbiol. 154105-111. [Google Scholar]
- 17.Huber, R., E. Wolfgang, S. Heldwein, G. Wanner, H. Huber, R. Rachel, and K. O. Stetter. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl. Environ. Microbiol. 643576-3583, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson, J. A., H. W. Morgan, and R. M. Daniel. 1986. A numerical classification of some Thermus isolates. J. Gen. Microbiol. 132531-540. [Google Scholar]
- 19.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 20.Kandler, O., and N. Weiss. 1986. Regular, nonporing, Gram-positive rods, p. 1208-1260. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 21.Kürner, J., A. S. Frangakis, and W. Baumeister. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Nature 307436-438. [DOI] [PubMed] [Google Scholar]
- 22.López-García, P., J. Kazmierczak, K. Benzerara, S. Kempe, F. Guyot, and D. Moreira. 2005. Bacterial diversity and carbonate precipitation in the giant microbialites from the highly alkaline Lake Van, Turkey. Extremophiles 9263-274. [DOI] [PubMed] [Google Scholar]
- 23.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G-C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39159-167. [Google Scholar]
- 24.Nielsen, P., D. Fritze, and F. G. Priest. 1995. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 1411745-1761. [Google Scholar]
- 25.Razin, S. 2000. The genus Mycoplasma and related genera (class Mollicutes), release 3.3. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, NY. http://link.springer-ny.com/link/service/books/10125.
- 26.Razin, S., and E. A. Freundt. 1989. The mycoplasmas, p. 740-789. In M. P. Bryant, J. G. Holt, N. Pfennig, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 27.Razin, S., and J. G. Tully. 1970. Cholesterol requirement of mycoplasmas. J. Bacteriol. 102306-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, M., R. Botz, E. Faber, M. Schmitt, J. Poggenburg, D. Garbe-Schönberg, and P. Stoffers. 2003. High-resolution methane profiles across anoxic brine-seawater boundaries in the Atlantis-II, Discovery, and Kebrit deeps (Red Sea). Chem. Geol. 200359-375. [Google Scholar]
- 29.Smibert, R. M., and N. R. Krieg. 1981. General characterization, p. 409-443. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC.
- 30.Tindall, B. J. 1989. Fully saturated menaquinones in the archaebacterium Pyrobaculum islandicum. FEMS Microbiol. Lett. 60251-254. [Google Scholar]
- 31.Trachtenberg, S., R. Gilad, and N. Geffen. 2003. The bacterial linear motor of Spiroplasma melliferum BC3: from single molecules to swimming cells. Mol. Microbiol. 47671-697. [DOI] [PubMed] [Google Scholar]
- 32.Verbarg, S., H. Rheims, S. Emus, A. Frühling, R. M. Kroppenstedt, E. Stackebrandt, and P. Schumann. 2004. Erysipelothrix inopinata sp. nov., isolated in the course of sterile filtration of vegetable peptone broth, and description of Erysipelotrichaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54221-225. [DOI] [PubMed] [Google Scholar]
- 33.Watson, S. W., and J. B. Waterbury. 1969. The sterile hot brines of the Red Sea, p. 272-281. In E. T. Degens, and D. A. Ross (ed.), Hot brines and recent heavy metal deposits in the Red Sea. Springer-Verlag, New York, NY.
- 34.Witte, A., G. Wanner, U. Bläsi, G. Halfmann, M. Szostak, and W. Lubitz. 1990. Endogenous transmembrane tunnel formation mediated by φX174 lysis protein E. J. Bacteriol. 1724109-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worliczek, H. L., P. Kämpfer, R. Rosengarten, B. J. Tindall, and H. J. Busse. 2007. Polar lipid and fatty acid profiles—re-vitalizing old approaches as a modern tool for the classification of mycoplasmas? Syst. Appl. Microbiol. 30355-370. [DOI] [PubMed] [Google Scholar]