Abstract
Bacillus subtilis forms structured communities of biofilms encased in an exopolysaccharide matrix on solid surfaces and at the air-liquid interface. It is postulated that nonoptimal growth conditions induce this multicellular behavior. We showed that under laboratory conditions a strain deleted for sigB was unable to form a floating pellicle on the surface of a liquid medium. However, overexpression of yxaB, encoding a putative exopolysaccharide synthase, from a pSpac promoter in a sigB-deleted strain resulted in partial recovery of the wild-type phenotype, indicating the participation of the YxaB protein in this multicellular process. We present data concerning the regulation of transcription of genes yxaB and yxaA, encoding a putative glycerate kinase. Both genes are cotranscribed as a single transcription unit from a σA-dependent promoter during vegetative growth of a liquid bacterial culture. The promoter driving transcription of the yxaAB operon is regulated by AbrB. In addition, the second gene in the operon, yxaB, possesses its own promoter, which is recognized by RNA polymerase containing the σB subunit. This transcription start site is used under general stress conditions, resulting in increased expression.
Since bacteria in their natural setting face widely fluctuating conditions, including changes in pH, temperature, or limitation of nutrients, biofilms appear to be an excellent adaptive strategy to protect the inhabitants from diverse stress factors (11). Indeed, it is commonly known that the biofilm mode of growth is the major bacterial life-style in nature (9). In order to adapt to a wide diversity of stresses, Bacillus subtilis uses an alternative sigma factor, σB, to express a large set of genes in the σB regulon. The σB factor is estimated to control at least 5% of the coding capacity of the genome (36).
In B. subtilis, 17 sigma factors have been described, showing a wide range of recognition sequences. The major vegetative sigma factor is required for expression of so-called housekeeping genes, whereas exposure of bacteria to a wide variety of growth-limiting conditions requires the alternative sigma factor, σB (17, 19, 36). On the other hand, changes in gene expression that occur during the transition from a planktonic state to a biofilm mode of growth depend on several sigma factors associated mainly with sporulation (4, 15, 38, 39). Despite the fact that a large number of sporulation genes are induced in the wild-type biofilm, the forespore sigma factor σF and its counterpart σE, which is activated in the mother cell, seem not to play any role in biofilm formation (4, 15, 38). Surprisingly, the absence of σH, which is involved in the early stages of sporulation, nevertheless does affect this process. The most characteristic defect associated with the lack of σH is the absence of fruiting-body structures typical of mature biofilms of natural isolates of B. subtilis (4). Recent data showed that a strain with inactivated sigma factors σX, σM, and σW displayed a complete loss of the pellicle formation (26). According to Kobayashi (24) a mutation in the gene encoding σX is likely to affect cell envelope structure, which influences biofilm formation.
Biofilms are often formed under nonopitmal growth conditions, and this encouraged us to examine whether the general stress response factor σB plays an important role in this process. In this report we provide evidence that σB is crucial for biofilm formation by B. subtilis (strain 168), since a strain with an impaired general stress response was unable to form floating pellicles. Moreover, we showed that overexpression of yxaB, encoding a putative exopolysaccharide (EPS) synthase (28, 40), in a strain deleted for σB resulted in partial recovery of the wild-type phenotype, suggesting the participation of the YxaB protein in this multicellular phenomenon.
The results presented here indicated that yxaB possesses its own promoter which is recognized by RNA polymerase containing the σB subunit. This transcription start site is used in response to stress conditions (e.g., high salt or alcohol), resulting in increased expression of that gene. However, during vegetative growth, yxaB is expressed together with yxaA, encoding a putative glycerate kinase. Both genes are transcribed as a single transcription unit and are under the control of a σA-dependent promoter. The global regulator AbrB was shown previously to repress other genes participating in biofilm formation (15, 16), and we found that the yxaAB operon is negatively controlled by AbrB. We found no influence of other important proteins known to regulate EPS synthesis.
MATERIALS AND METHODS
Strain and plasmid construction.
The Bacillus subtilis strains used in this study are listed in Table 1. For the cloning of selected B. subtilis genome fragments, DNA from strain 168 was used as a template in PCRs. The reactions were performed with Walk polymerase (A&A Biotechnology) and appropriate primers (Table 2).
TABLE 1.
Bacterial strainsa
| Strain | Genotype or description | Source |
|---|---|---|
| M | trpC2 ΔsinI::kan | R. Kort |
| 168 | trpC2 | BGSC |
| 1A147 | trpC2 ccpA1 alsR1 ilvBD1 | BGSC |
| 1S5 | trpC2 spo0A3 | BGSC |
| BAL373 | trpC2 ΔabrB::cat pheA1 | Grossman Strain Collection |
| MM233 | trpC2 ΔsigB | S. Séror |
| MM1701 | trpC2 yxaB::pKN3 erm | 168←pKN3 |
| MM1704 | trpC2 sigH::kan yxaB::pKN3 erm | MM1701←MO1615 |
| MM1702 | trpC2 ΔsigB yxaB::pKN3 erm | MM233←pKN3 |
| MM1705 | trpC2 spo0A3 yxaB::pKN3 erm | 1S5←MM1701 |
| MM1707 | trpC2 ΔabrB::cat yxaB::pKN3 erm | MM1717←MM1701 |
| MM1709 | trpC2 amyE::pyxaAB-lacZ spc | 168←pKN1 |
| MM1710 | trpC2 amyE::pyxaB-lacZ spc | 168←pKN2 |
| MM1712 | trpC2 sigH::kan amyE::pyxaAB-lacZ spc | MM1709←MO1615 |
| MM1713 | trpC2 spo0A3 amyE::pyxaAB-lacZ erm | 1S5←MM1709 |
| MM1715 | trpC2 ΔabrB::cat amyE::pyxaAB-lacZ spc | MM1717←MM1709 |
| MM1717 | trpC2 ΔabrB::cat | 168←BAL373 |
| MM1718 | trpC2 ccpA1 alsR1 ilvBD1 amyE::pyxaAB-lacZ spc | 1A147←MM1709 |
| MM1719 | trpC2 ccpA1 alsR1 ilvBD1 amyE::pyxaAB-3-lacZ spc | 1A147←MM1803 |
| MM1721 | trpC2 amyE::pSpac-yxaB cat | 168←pKN6 |
| MM1722 | trpC2 ΔsigB amyE::pSpac-yxaB cat | MM233←pKN6 |
| MM1801 | trpC2 amyE::pyxaAB-1-lacZ spc | 168←pKN1-1 |
| MM1802 | trpC2 amyE::pyxaAB-2-lacZ spc | 168←pKN1-2 |
| MM1803 | trpC2 amyE::pyxaAB-3-lacZ spc | 168←pKN1-3 |
| MM1804 | trpC2 amyE::pyxaAB-4-lacZ spc | 168←pKN1-4 |
| MM1805 | trpC2 ΔabrB::cat amyE::pyxaAB-1-lacZ spc | MM1717←MM1801 |
| MM1806 | trpC2 ΔabrB::cat amyE::pyxaAB-2-lacZ spc | MM1717←MM1802 |
| MM1807 | trpC2 Δ abrB::cat amyE::pyxaAB-3-lacZ spc | MM1717←MM1803 |
| MM1808 | trpC2 Δ abrB::cat amyE::pyxaAB-4-lacZ spc | MM1717←MM1804 |
| MM1809 | trpC2 ΔsinI::kan amyE::pyxaAB-lacZ spc | ΔsinI::kan←MM1709 |
| MM1811 | trpC2 ΔyxaB::spc | 168←pKN7 |
| MM1821 | trpC2 ΔsinI::kan yxaB::pKN3 erm | ΔsinI::kan←MM1701 |
| MM1823 | trpC2 ccpA1 alsR1 ilvBD1 yxaB::pKN3 erm | 1A147←MM1701 |
| MO1615 | trpC2 sigH::kan | P. Stragier |
For detailed information on fragments and restriction sites used for cloning, see Table 2.
TABLE 2.
Primers used for cloning, RT-PCR, and RACE PCR
| Primera | Gene | Primer sequence (5′→3′)b | Orientation | Position in B. subtilis chromosome |
|---|---|---|---|---|
| BZ-up | GTC CAG GAT CCC CAT AAA ATG C | Forward | 4110249 | |
| BZ-dn | yxaB | GAA CTG GTG AAG CTT ATC ATT CGT | Reverse | 4110776 |
| B-up | yxaB | CTT CGT CAG TCA CTT TCT CC | Forward | 4110377 |
| B-dn | yxaB | CCT TCA ACT CCA ATG TGA TC | Reverse | 4110502 |
| A-up | yxaA | CTC TGT ATC TCG GAA GGT TG | Forward | 4111511 |
| A-dn | yxaA | CCG TTA ACA GGA CCA AAG | Reverse | 4111657 |
| yxaB-2 | yxaB | GAT AAA TTT CTC TTC CGC ATA CG | Forward | 4110840 |
| pAB-dn | yxaA | GAC GGA TCC ACA GCC AAA CG | Reverse | 4111368 |
| pAG-up | TTT TCT GTC GAC AGC ACC CGG | Forward | 4112132 | |
| pAG-dn | CAT AAG GAT CCG CGA TTT TCT TCT C | Reverse | 4112717 | |
| pAB-up | CAT TTA GTC GAC CTT CCG GCT C | Forward | 4110926 | |
| pAR-up | yxaA | CTG TGG GAT CCT GCT CTA TGA | Forward | 4112047 |
| pAR-dn | gntR | GTA TGC GAA TTC AAC TCC GGT | Reverse | 4112515 |
| YxaB-up | yxaC | GCT CGT CGA CTG GCT GTT TC | Forward | 4109906 |
| YxaB-dn | CGA AGT CGA CGT TTC TGA TAT AGC | Reverse | 4111119 | |
| pAG3-up | ATT GAA TTC ATG CAA ACA C | 4112375 | ||
| pAG2 | yxaA | TAA TCA GGA TCC GGA AAG | 4112139 | |
| pAG5-dn | CAC TTT GAA TTC TAA TTT TTA TTT | 4112368 | ||
| pAG2-up | TTT GAA TTC AAT TTT TAC C | 4112352 | ||
| pAG2-dn | yxaA | TAA TCA GGA TCC GGA AAG | 4112139 | |
| pAG4-up | yxaA | TAA TCA GGA TCC GGA AAG ACC | 4112139 | |
| pAG4-dn | CCG AAT TCC ATG GTA TGT ATT TC | 4112335 | ||
| delC-up | yxaC | CAT TAG AAT TCT TTT AAC AGT TGC C | Forward | 4109264 |
| delC-dn | yxaC | CCG GGG AGC TCT TTC TTT G | Reverse | 4109952 |
| delA-up | yxaA | GGC ATC TGC AGC GGC TAT ATC AG | Forward | 4111083 |
| delA-dn | yxaA | GAT GCA AAG CTT GTG ATT ACC GG | Reverse | 4111396 |
| spc-up | CTG GAT CCA AAA TTT AGA AG | |||
| spc-dn | GTG TGT CGA CCA TTT TTT C |
Primers AAP and AUAP were provided with the RACE system by Invitrogen.
Restriction sites are underlined.
Construction of transcriptional fusions with the reporter gene lacZ.
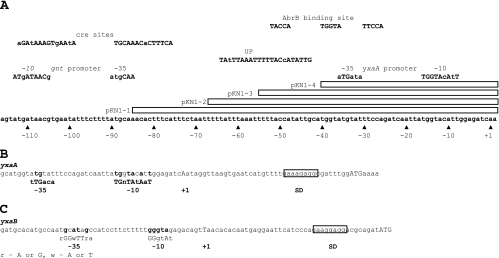
For the construction of transcriptional fusions of lacZ to the promoter regions of yxaA and yxaB, a fragment of B. subtilis DNA containing the putative promoter region was amplified by PCR and cloned into the integration vector pDG1728 (14). First, the fragment containing a putative promoter region of yxaA (pyxaAB) (569 bp), amplified using primers pAG-up and pAG-dn (Table 2), was inserted into the SalI/BamHI restriction sites of pDG1728. The resulting plasmid, pKN1, was linearized with XhoI and used to transform B. subtilis 168, with selection for spectinomycin resistance, generating strain MM1709. Additionally, four derivatives of the pKN1 plasmid, differing in the length of the upstream regulatory region, were constructed similarly (see Fig. 5A). The strains obtained after transformation of B. subtilis with those plasmids were designated MM1801, MM1802, MM1803, and MM1804. The inserted fragments contain the following regulatory sequences: the pyxaAB promoter with an upstream cre site (231 bp) in the case of pKN1-1, the pyxaAB promoter with the up element (UP) (212 bp) in the case of pKN1-2, the pyxaAB promoter with 16 nucleotides of the UP (199 bp) in the case of pKN1-3, and the core of the pyxaAB promoter (183 bp) in the case of pKN1-4. All listed fragments were generated by PCR with appropriate pairs of primers (Table 2): pAG3-up and pAG2 (pKN1-1), pAG2 and pAG5-dn (pKN1-2), pAG2-up and pAG2 (pKN1-3), and pAG4-up and pAG4-dn (pKN1-4). The PCR products obtained were then integrated into the EcoRI/BamHI restriction sites of pDG1728.
FIG. 5.
(A) Detailed structure of part of the region present in plasmid pKN1. All sites are listed above the sequence with appropriate descriptions. Upper- and lowercase letters in descriptions denote consensus and nonconsensus bases, respectively. Boxes show part of the region present in clones pKN1-1, pKN1-2, and pKN1-3, respectively. Numbering of motifs is according to the mapped transcription start site of the pyxaAB promoter. (B and C) Mapping of the transcription start sites of yxaA (B) and yxaB (C) with the 5′ RACE method. Conserved bases from the promoter region are in bold, and the +1 nucleotide is in uppercase. Ribosome binding sites are boxed, and the ATG start codon is indicated by uppercase letters. The most important bases in the consensus sequence are capitalized. R, A or G; W, A or T.
To obtain plasmid pKN2, a fragment containing the putative promoter region of yxaB (pyxaB), was amplified by PCR using the pAB-up and pAB-dn primers (Table 2) and chromosomal DNA. The amplified fragment was digested with appropriate enzymes and ligated into the pDG1728 vector (14). Next, DNA of pKN2, after linearization, was inserted into the amyE locus on the chromosome of B. subtilis 168, generating strain MM1710.
To monitor expression levels of yxaB, another transcriptional fusion, pKN3, was made. In this case, an internal fragment of the yxaB gene (508 bp), amplified with BZ-up and BZ-dn primers (Table 2) and then digested with HindIII and BamHI, was inserted into the pMutin4 integrative vector (42). The DNA of pKN3 was used to transform the wild-type B. subtilis (strain 168), with selection for erythromycin resistance, to generate strain MM1701.
Construction of plasmid pKN4 used in DNase I footprinting.
The pKN4 plasmid, used for footprinting experiments, was constructed by insertion of a 452-bp fragment containing the pyxaAB promoter into the EcoRI/BamHI restriction sites of the high-copy-number pUC19 vector (45). The cloned fragment was amplified with the pAR-up and pAR-dn primers (Table 2).
Construction of plasmids for overexpression and deletion of yxaB.
To overexpress yxaB, we amplified a 1,200-bp fragment with primers YxaB-up and YxaB-dn (Table 2) from the chromosome of 168. The product obtained was ligated into the SalI/SalI sites of the pDR66 vector (20), generating plasmid pKN6. This construct has yxaB under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pSpac promoter. The DNA of the pKN6 plasmid, after linearization with Eam1105I (Fermentas), was used for transformation of wild-type B. subtilis (strain 168), selecting for chloramphenicol resistance, to generate strain MM1720.
The pKN7 plasmid used for yxaB deletion was constructed as follows. The fragment containing the spectinomycin resistance gene was amplified from plasmid pDG1728 (14) by PCR with primers spc-up and spc-dn (Table 2). The PCR product (881 bp) was inserted into the BamHI/SalI restriction sites of pUC19 (45), generating pKN7A. Then, a distal part of yxaC (677 bp) was produced by PCR with the forward primer containing an EcoRI sequence at its 5′ end (delC-up) and a reverse primer containing a SacI sequence at its 5′ end (delC-dn). This PCR product was introduced in the pKN7A vector digested with SacI and EcoRI to generate pKN7B. As a last step in pKN7 construction, a 293-bp fragment containing the distal part of yxaA was amplified from the strain 168 chromosome with the forward primer containing a PstI sequence at its 5′ end (delA-up) and a reverse primer containing a HindIII sequence at its 5′ end (delA-dn). This PCR product was introduced in the pKN7B vector digested with PstI and HindIII to generate pKN7. The DNA of the pKN7 plasmid, after linearization with Eam1105I, was used for transformation of wild-type B. subtilis (strain 168), selecting for spectinomycin resistance, to generate strain MM1811.
β-Galactosidase measurements.
Cultures were grown at 37°C with shaking. Samples were taken from different phases of growth and stored at −20°C until enzyme assays were carried out. After thawing, bacterial pellets were suspended in buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) containing 1 mM dithiothreitol (DTT), and 1/100 volume of lysis solution (1 mg/ml DNase, 10 mg/ml lysozyme) was added. The mixture containing lysed cells was incubated for 20 min at 37°C and then centrifuged for 10 min at 10,000 × g and 4°C to remove the cell debris. The supernatant was used for measurement of protein concentration and β-galactosidase activity. Protein concentrations were measured using the Bradford reagent (Bio-Rad) as recommended by the manufacturer. Supernatants (200 μl) were mixed with 600 μl of buffer Z containing 1 mM DTT. Samples were placed in a 37°C water bath, and 200 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml) was added. After 60 min of incubation, the reaction was stopped by addition of 500 μl of 1 M Na2CO3, and the absorbance was measured at 420 nm. β-Galactosidase activity, in nmol of o-nitrophenol produced min−1 mg−1, was calculated using the following formula: (absorbance at 420 nm × 1.5)/(concentration of protein in mg/ml × volume of sample in ml × reaction time in min × 0.00486).
β-Galactosidase measurements with 4-methylumbelliferyl-β-d-galactoside were performed as follows. After thawing, bacterial pellets were suspended in reaction buffer (25 mM Tris [pH 7.5], 125 mM NaCl, 2 mM MgCl2, 12 mM 2-mercaptoethanol) with 1 mM DTT and 1/100 volume of lysis solution (1 mg/ml DNase, 10 mg/ml lysozyme). Samples (40 μl) were placed in an Eppendorf tube, and 160 μl reaction buffer with 0.3 mM 4-methylumbelliferyl-β-d-galactoside was added. After 30 min of incubation, the reaction was stopped by addition of 50 μl of 25% TCA, and the solution was cooled on ice and clarified by centrifugation. Next, 0.1 ml of supernatant was added to 1.9 ml glycine-carbonate reagent (133 mM glycine, 83 mM Na2CO3, pH 10.7). Fluorescence was measured in a DyNA Quant 200 fluorometer (Amersham). The concentration of 4-methylumbelliferone was determined according to a standard concentration curve of 0 to 400 nM 4-methylumbelliferone.
RNA isolation and 5′ rapid amplification of cDNA ends (RACE) PCR.
RNA was isolated from strain 168 cultivated in rich medium (nutrient broth; Difco) until stationary phase. The cells were harvested by centrifugation at 4°C (10 min, 5,000 × g). To lyse the cells, 1 ml of Trizol (Roche) and 0.2 ml of 0.1-mm zirconium/silica beads were added to the 2-ml tubes containing the pellet. The tubes were beat for 45 s at maximum speed in a Mini-Bead-Beater (Biospec Products). Total RNA was isolated by following the protocol of A&A Biotechnology (total RNA isolation kit).
RACE is a procedure for amplification of nucleic acid sequences from an mRNA template inserted between a defined internal site and an unknown 5′ end. For such an analysis, we used the RACE system produced by Invitrogen (version 2.0), and experiments were performed according to the manufacturer's manual. The specific primers for cDNA synthesis (RevT) and PCR amplification (cDAMP and AAP; 9d and AUAP) are listed in Table 2.
DNase I footprinting.
The 452-bp EcoRI/BamHI fragment of pKN4, which was end labeled using [α-32P]dATP (Amersham) and the Klenow enzyme, was used in the DNase I footprinting reactions. The protein binding buffer composition (1×) was 50 mM Tris (pH 8.0), 10 mM MgCl2, 100 mM KCl, 10 mM 2-mercaptoethanol, and 100 μg/ml bovine serum albumin. AbrB binding (10 min) and DNase I cleavage reactions (30 μg/ml DNase I for 10 s) were performed at room temperature (22°C) as previously described (41, 44). Maxam-Gilbert purine and pyrimidine reactions were performed on the labeled fragments (27) and used for sequence orientation.
Pellicle formation.
Pellicle formation experiments were performed in either MSgg or B-medium, which differ in the main source of carbon. In the case of B-medium (50 mM Tris-HCl [pH 7.5], 27 mM KCl, 7 mM sodium citrate, 8 mM MgSO2, 2 mM CaCl2, 1 μM FeSO4, 10 μM MnSO4, 0.6 mM KH2PO4, 0.5% glucose, 4.5 μM glutamic acid, 860 μM lysine, 780 μM tryptophan) (1), the wild-type and mutant strains were grown overnight at 37°C with shaking in medium containing antibiotics when required (first-day culture). The overnight culture was diluted to an optical density at 575 nm (OD575) of 0.2 and incubated for 24 h under stationary conditions at room temperature (second-day culture). Next, bacteria (100 μl of a second-day culture for each 2 ml of fresh medium) were transferred to either sterile glass beakers (for visual observation) or 24-well plates (for microtiter plate assay of pellicle formation) and incubated at 30°C without shaking for the appropriate period of time.
For MSgg medium, the following procedure was used. An overnight culture of bacteria was diluted in fresh LB medium. After reaching an OD575 of 1, bacteria were diluted 350-fold in minimal MSgg medium (5 mM potassium phosphate, 100 mM MOPS [morpholinepropanesulfonic acid] [pH 7], 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnSO4, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg ml−1 tryptophan, 50 μg ml−1 phenylalanine, and 50 μg ml−1 threonine) (4) supplemented with 200 μM NaCl. Cells were incubated without shaking in 24-well plates for 70 h at 37°C to estimate the relative amount of biofilm according to the microtiter plate assay described below.
Microtiter plate assay of pellicle formation.
At appropriate times the medium was carefully removed from the wells in which the pellicle was formed (see “Pellicle formation” above), and the wells were rinsed with water. The plate containing biofilm was allowed to dry at 37°C for 15 min. Next, methanol was added to fix the biofilm to the walls of the wells. After removing methanol and rinsing the wells with water, the plates were allowed to dry again at 37°C for 20 min, and then 1% crystal violet (CV) solution was added to stain the cells that had adhered to the wells. Excess CV was then removed, and the wells were rinsed with water. The CV that had bound the pellicle was then solubilized in an ethanol-acetone solution (4:1, vol/vol). Biofilm formation was quantified by measuring the OD500 for each well using a plate reader.
Confocal scanning laser microscopy.
For observation of B. subtilis biofilms by confocal scanning laser microscopy, pellicles were grown using a modification of the method described above (see “Pellicle formation”). The bacteria from a second-day culture growing on B-medium were transferred to sterile petri dishes with a glass slide at the bottom, immersed in 25 ml of medium. The slides had a cavity in the middle to prevent damage of the three-dimensional structure of the biofilm during observation. Biofilms were allowed to develop at the air-liquid interface at 30°C under stationary conditions for the appropriate period of time. Then, the medium was carefully removed from the plate, enabling the biofilm to settle on the microscope slide. Samples were observed using an Olympus BX51 confocal laser microscope after staining with a Live/Dead BacLight bacterial viability kit (Molecular Probes). Images were analyzed using the EZ2000 program, and individual scans through the Z section were obtained with this software.
RESULTS
General stress response factor σB plays a crucial role in pellicle formation.
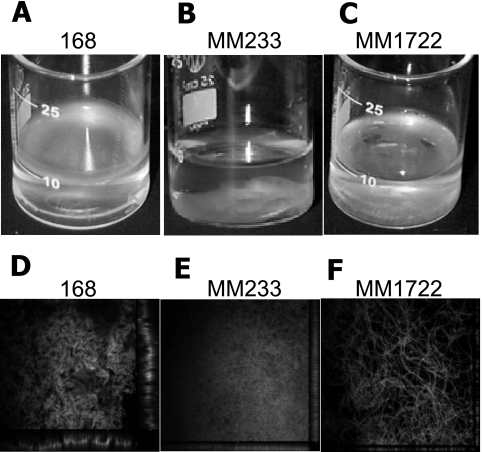
The synthesis of σB is activated by diverse environmental and energy stresses and governs the transcription of a large set of general stress response genes (for a review, see reference 16). There are reports showing a direct contribution of σB to the formation of adherent biofilms by Staphylococcus epidermidis and Staphylococcus aureus (22, 23, 37, 43). Given these results, it was of interest to determine whether a B. subtilis sigB mutant (MM233) was able to form a biofilm under laboratory conditions. As seen in Fig. 1B, MM233 was unable to form a pellicle at an air-liquid interface. The cells formed structures which looked like strands of cotton wool accumulating at the bottom of the cultivation beaker.
FIG. 1.
Biofilm formation. Strains 168, MM233 (ΔsigB), and MM1722 (ΔsigB amyE::pSpac-yxaB) were analyzed for biofilm formation on synthetic medium at 30°C. (A to C) Overview of the pellicles at 24 to 26 h after inoculation. (D to F) Structure of 24- to 26-h-old biofilm as seen by confocal laser scanning microscopy at a magnification of ×600. Bacteria were visualized using the Live/Dead BacLight kit (Molecular Probes). The IPTG concentration was 1 mM.
Overexpression of the putative EPS synthase yxaB partially suppresses the defect in formation of biofilm by a sigB mutant.
There are some data indicating that yxaB is activated under stress conditions as detected by microarray experiments (33, 35). However, the cellular role of yxaB and the mechanism of regulation remain unknown. The product of the yxaB gene shows similarity to enzymes involved in EPS production. The closest described homologs are EpsL from Streptococcus thermophilus and PssK from Rhizobium leguminosarum bv. trifolii (28, 40), which show 59% and 47% similarity, respectively. Moreover, we found two paralogs of YxaB among other proteins of B. subtilis: YveS (EpsI) and YvfF (EpsO), both of which are part of the eps operon, which is necessary for EPS production (4, 21).
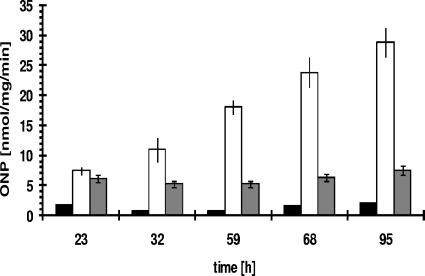
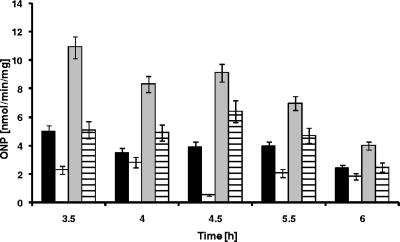
Since the product of yxaB has similarity to enzymes involved in EPS synthesis, it was possible that the inability of a sigB mutant (MM233) to form a robust pellicle was due to a lack of this enzyme. To test this, we first analyzed the transcription of a yxaB-lacZ fusion in the biofilm by measuring β-galactosidase activity in strain MM1701. The results showed that the transcription of yxaB increased significantly in biofilm cells, reaching the maximum level in biofilms grown for an extended time (Fig. 2). We deleted yxaB from the chromosome and searched for phenotypic effects associated with pellicle formation (data not shown). The yxaB-deleted strain (MM1811) also grew normally under all tested laboratory conditions both in liquid culture and on plates. To our surprise, this strain showed normal biofilm formation and structure in MSgg and B-medium (which differ in the main source of carbon), as estimated by CV staining and by confocal microcopy (data not shown). A possible explanation for the lack of a specific phenotypic effect observed for MM1811 may be the presence of the two paralogs of yxaB, epsI and epsO, in the large eps operon. These paralogs may be capable of taking over the function of yxaB.
FIG. 2.
Transcription of yxaA and yxaB during biofilm formation. Biofilm pellicles were collected at appropriate times, disrupted by vigorous shaking with glass beads, and analyzed for β-galactosidase activity as described in Materials and Methods. Black bars, 168; open bars, MM1701 (yxaB::pKN3); gray bars, MM1709 (amyE::pyxaAB-lacZ). ONP, o-nitrophenol. Error bars indicate standard deviations.
Our next step was to analyze the effect of overexpression of yxaB in a strain (MM233 ΔsigB) unable to induce the general stress response. To achieve this, we constructed a 168 derivative carrying the yxaB gene under the control of an IPTG-inducible promoter, pSpac (MM1722). The results obtained with this strain showed that IPTG (1 mM)-dependent transcription of yxaB in MM1722 partially restored the ability to form a floating pellicle on the surface of the medium (Fig. 1). Moreover, detailed analysis of the biofilm by confocal microscopy revealed that induction of expression of yxaB largely restored the ability of B. subtilis 168 cells to form long chains in the biofilm (Fig. 1F). While the sigB mutant strain (MM233) formed a nonstructured mass of cells, the overexpression of yxaB in this strain background largely restored the wild-type organization of the biofilm (Fig. 1D and E). Nevertheless, the pellicle formed by strain MM1722 was more fragile than that formed by strain 168 but still was able to remain on the liquid surface, in contrast to the case for the mutant strain deleted for sigB.
Genes yxaA and yxaB form an operon.
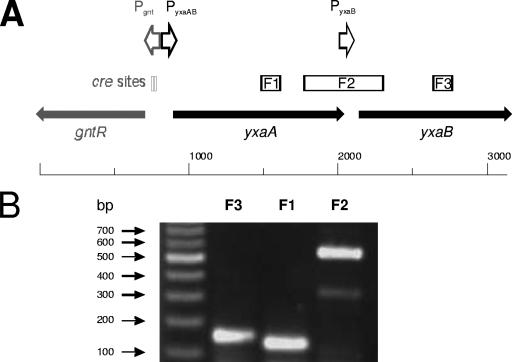
In order to obtain more information about the cellular function and regulation of yxaB transcription, we decided to determine whether yxaB forms an independent transcriptional unit or is cotranscribed as a member of a larger operon. We focused on the yxaA gene, which encodes a putative glycerate kinase and is the only gene in the neighborhood of yxaB that is transcribed in the same direction (counterclockwise) relative to other genes in that region of the B. subtilis chromosome. To determine whether yxaA and yxaB might be cotranscribed from a promoter localized upstream of yxaA, we designed three pairs of primers for reverse transcription-PCR (RT-PCR) analysis. The first pair was used to amplify a 125-bp fragment inside the yxaA gene (designated F1); the second, a 145-bp fragment from the internal part of yxaB (designated F3); and the third, a 545-bp fragment containing the distal part of yxaA together with the proximal part of yxaB (Fig. 3A). The results of RT-PCR analysis revealed amplification of cDNA in reactions with all pairs of primers (Fig. 3B). The RT-PCR product with primers for fragment F3 suggested that the transcript originating from the pyxaAB promoter extended into the yxaB gene. However, under stress conditions the pyxaB promoter becomes activated, resulting in increased transcription of the yxaB gene (33, 35). To confirm the results indicating that yxaA and yxaB are cotranscribed, we also performed a negative control experiment, running the RT-PCR in the absence of reverse transcriptase, and no product was obtained.
FIG. 3.
Analysis of cotranscription of yxaA and yxaB. (A) The identified promoter in the analyzed region is marked by arrows. Fragments amplified by RT-PCR analysis are marked by boxes. (B) RT-PCR amplification. MW, DNA molecular mass marker (Fermentas); F3, inside of yxaB; F1, inside of yxaA; F2, distal part of yxaA and proximal part of yxaB.
This operon organization of yxaA and yxaB appears to be specific to B. subtilis, as confirmed by analysis of available bacterial databases (http://www.ebi.ac.uk/Databases/genomes.html).
A σA-dependent promoter is localized upstream of the yxaA gene.
There are no available data describing the enzymatic activity or the transcriptional regulation of the yxaA gene product in B. subtilis. However, some information was acquired from sequence comparisons. The YxaA protein shows high similarity to the GarK and GlxK proteins from Escherichia coli (72% and 65% similarity, respectively), which were characterized as glycerate kinases (EC 2.7.1.31) (10, 31). Glycerate kinases are highly conserved enzymes playing a variety of roles in different organisms but with an unidentified role in many bacteria, including B. subtilis.
To determine the transcriptional start site for the yxaA gene, we constructed strain MM1709 (amyE::pyxaAB-lacZ). The β-galactosidase activity analysis for strain MM1709 showed that the cloned region indeed contains an active promoter (Fig. 4). For precise mapping of the transcription start site, we employed the 5′ RACE method. This approach resulted in identification of the +1 nucleotide (position 4112285 in the B. subtilis chromosome) of the yxaA transcript (Fig. 5B). Analysis of the sequence upstream of the +1 nucleotide allowed us to find a strong match (TGGTAcAtT), seven of nine bases, to a canonical extended −10 sequence (TGNTATAAT) and a partial match (aTGta) to a canonical −35 sequence (TTGACA) of a σA-dependent promoter. The −10 and −35 sequences were separated from each other by an appropriate spacing of 17 bp (18). Examination revealed the presence of an AT-rich UP immediately upstream of the pyxaAB promoter (at positions −43 to −64), identical to analogous motifs in E. coli, representing 20 out of 22 consensus bases (6). UP elements have been shown to stabilize binding of RNA polymerase to DNA through direct interaction with the C-terminal domain of the α subunit of this enzyme (6). The function of such a UP in B. subtilis was previously demonstrated for promoters of phage φ29 (29). Our finding that the promoter for yxaA is recognized by the housekeeping σA factor is consistent with a postulated role of YxaA as a glycerate kinase in central metabolism. The results based on monitoring β-galactosidase activity in strain MM1709 showed also that the transcription of yxaA remained unchanged during biofilm development, contrasting with the pattern of yxaB expression (Fig. 2).
FIG. 4.
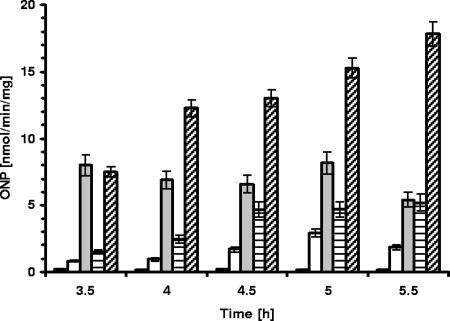
Transcriptional activity of the pyxaAB promoter and its derivatives in the presence and absence of AbrB in the bacterial cell. β-Galactosidase activity in strains carrying a lacZ transcriptional fusion to the pyxaAB promoter and its derivatives during planktonic growth in NB medium supplemented with 0.5% glucose with shaking at 37°C is shown. Black bars, 168; open bars, MM1709 (amyE::pyxaAB-lacZ); gray bars, MM1715 (ΔabrB::cat amyE::pyxaAB-lacZ); open bars with horizontal lines, MM1804 (amyE::pyxaAB-4-lacZ); open bars with diagonal lines, MM1808 (ΔabrB::cat amyE::pyxaAB-4-lacZ). The transition state of growth was reached between 4 h and 4.5 h. ONP, o-nitrophenol. Error bars indicate standard deviations.
yxaB possesses its own σB-dependent promoter.
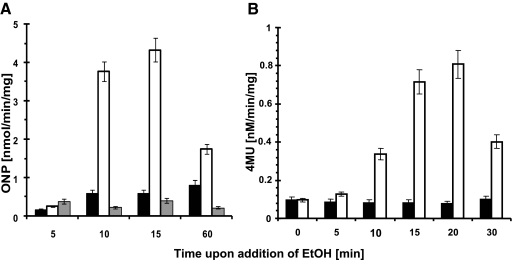
To confirm that the transcription of yxaB increases during ethanol stress, as postulated previously form microarray experiments by Petersohn et al. (33) and Price et al. (35), we constructed strain MM1701 (yxaB::pKN3), which allowed us to monitor the level of yxaB transcription. Indeed, we observed an increase in β-galactosidase activity after addition to the medium of either 4% ethanol (Fig. 6 A) or 4% NaCl (data not shown). However, taking into consideration the fact that the activity of the pyxaAB promoter was stress independent in our study (data not shown) and that we did not observe any ethanol induction of β-galactosidase activity in the strain deleted for sigB (MM1703) (Fig. 6A), we hypothesized that yxaB transcription under stress conditions may occur due to the presence of promoter in addition to that upstream of yxaA. To investigate this, we constructed a transcriptional fusion of the lacZ gene with the putative promoter region (pyxaB), generating strain MM1710 (amyE::pyxaB-lacZ). Analysis of β-galactosidase activity in this strain showed that this region contains a promoter which is activated under stress conditions (Fig. 6B). We next used the 5′ RACE technique to map the 5′ end of the putative transcript. The results indicated that the transcript start site (+1 nucleotide) is localized at position 4111046 in the chromosome. Analysis of the sequence upstream of the +1 nucleotide identified a strong match (GGGTAg) to an extended canonical −10 sequence (GGGTAT) and a partial match (tGcATaGc) to a canonical −35 sequence (rGGwTTrA) of a σB-dependent promoter (32). As shown in Fig. 5C, the −10 and −35 sequences were separated from each other by an appropriate spacing of 13 bp (19). Northern blotting analysis confirmed the presence of an approximately 1,100-bp band that corresponds exactly to the expected size of the yxaB transcript, indicating the presence of an active promoter region upstream of this gene (data not shown).
FIG. 6.
Analysis of pyxaB promoter activity during ethanol stress. (A) β-Galactosidase activity during ethanol (EtOH) stress in strain MM1701 (yxaB::pKN3). (B) β-Galactosidase activity during ethanol stress in strain MM1710 (amyE::pyxaB-lacZ). Black bars, β-galactosidase activity of the analyzed strains in the absence of ethanol; open bars β-galactosidase activity upon addition of ethanol (4%) to the medium (LB); gray bars, β-galactosidase activity upon addition of ethanol (4%) in the strain deleted for the sigB gene (strain MM1702). ONP, o-nitrophenol; 4MU, 4-methylumbelliferone. Error bars indicate standard deviations.
We also observed that yxaB transcription in a shaking liquid culture is maintained at a low level (data not shown). We interpret this as basal transcription from the σA-dependent pyxaAB promoter. The transcription of many general stress genes is known to depend upon σA-dependent promoters during nonstress conditions (1a, 33).
AbrB inhibits yxaAB transcription.
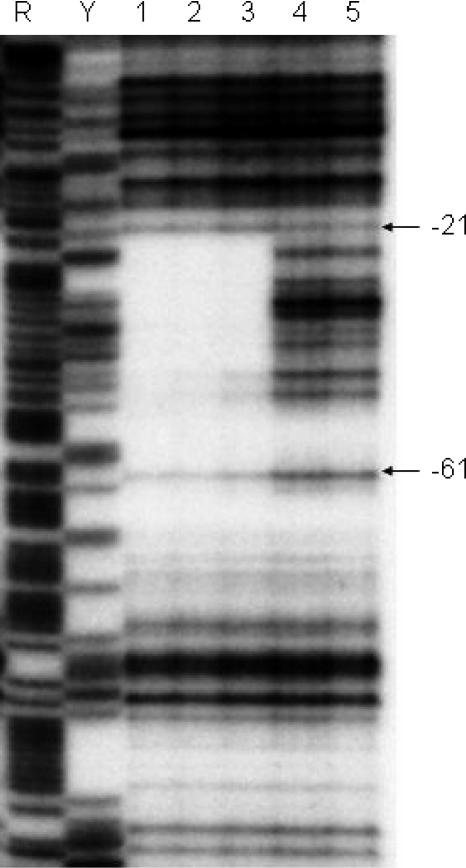
The finding that yxaB was involved in pellicle formation in the σB-deficient strain raised the question whether the yxaAB operon is under the control of the global transcriptional regulator AbrB, as in the case of many other biofilm associated genes (15, 16). As a preliminary test of this hypothesis, we performed β-galactosidase activity analysis of the pyxaAB promoter in the strain deleted for abrB (MM1715). The results obtained showed a threefold increase in β-galactosidase activity at the transition phase, which was reached at between 5 and 5.5 h of bacterial growth (Fig. 4). This indicated that AbrB is a negative regulator of yxaAB. The binding of AbrB in the proximity of the pyxaAB promoter was confirmed by footprint analysis (Fig. 7). The region protected by the AbrB protein extends from −21 to −68 relative to the transcription start site. The protected region contains also the postulated UP, together with the −35 region, of the pyxaAB promoter.
FIG. 7.
AbrB footprint on the yxaAB promoter. The results were obtained when the template strand was labeled at its 3′ end. Lanes: 1, 25 μM AbrB; 2, 8 μM AbrB; 3, 2.5 μM AbrB; 4 and 5: no AbrB. Maxam-Gilbert purine (R) and pyrimidine (Y) sequencing ladders are shown for reference.
We also decided to examine the effect of the absence of the UP region on yxaAB expression in the presence or absence of AbrB. In order to estimate the influence of UP on the transcription of yxaAB, we assayed derivatives of the pyxaAB promoter which contained different lengths of the upstream regulatory sequences: the promoter region with the UP (pKN1-2, strain MM1802), the promoter region with a fragment of UP (pKN1-3, strain MM1803), and only a core of the pyxaAB without UP (pKN1-4, strain MM1804). We introduced all these truncated promoter fusions into an abrB background, constructing strains MM1805 to MM1808. We found in fact that lacZ expression in the absence of functional AbrB was significantly increased, in particular as more upstream region was deleted, compared to that in strains carrying the same fusions in the 168 abrB+ background (MM1801 to MM1804) (Table 3 and Fig. 4). These data suggest that the presence of AbrB also facilitates the binding of other negative regulators to the UP region. Alternatively, we cannot exclude the possibility that AbrB itself controls the synthesis of such a negative regulator. This also could explain the lack of a positive effect of UP on transcription of the yxaAB operon under the tested conditions (Fig. 4). However, an equally plausible hypothesis is that the lack of a UP in the abrB+ background could increase transcription of yxaAB due to a reduction of the binding of AbrB to DNA, since the footprint analysis showed that AbrB binds to the DNA sequence overlapping the AT-rich UP element.
TABLE 3.
Transcriptional activities of derivatives of the pyxaAB promoter in different genetic backgroundsa
| Plasmid | Description | Mean β-galactosidase activity (nmol of produced ONP mg−1 min−1) ± SDb in:
|
|
|---|---|---|---|
| Wild type | ΔabrB mutant | ||
| pKN1 | yxaAB gene promoter with all surrounding sequences | 0.8 ± 0.3 | 3.1 ± 0.8 |
| pKN1-1 | yxaAB gene promoter with all surrounding sequences; gnt operon promoter inactivated by deletion of −10 region | 1.6 ± 0.3 | 5.1 ± 1.1 |
| pKN1-2 | yxaAB gene promoter with predicted UP | 4.4 ± 0.9 | 10.1 ± 1.8 |
| pKN1-3 | yxaAB gene promoter with shortened predicted UP | 6.1 ± 1.1 | 12.1 ± 1.7 |
| pKN1-4 | Core of yxaAB gene promoter | 8.2 ± 1.5 | 17.8 ± 3.2 |
For details concerning the pyxaAB derivatives, see Fig. 3 and the text.
Data were collected at the transition stage of growth from three independent experiments.
Searching for other regulators of the yxaAB operon.
We found that the transcription of the yxaAB operon increased when the medium was supplemented with 0.5% glucose. The gene ccpA is known to encode the catabolite control protein A, which plays a central role in sugar metabolism in B. subtilis (for a review, see reference 12). In fact, we found that transcriptional activity of the pyxaAB promoter was decreased twofold in the strain (MM1718) lacking ccpA when grown in the presence of glucose (Fig. 8). No effect was observed in the absence of glucose (data not shown). These data suggested that CcpA can act as an activator of yxaAB transcription. Since CcpA can function as a catabolite repressor or activator depending on relative position of cre sites (catabolite-responsive elements), it was of interest to determine if such a binding sequence is present in close proximity to the pyxaAB promoter (for a review, see reference 12). Indeed, two cre sites are located just upstream of yxaA at positions −76 to −89 and −99 to −112 in the gnt promoter (Fig. 5). Miwa and coworkers have shown that only the cre sequence located at position −76/−89 functions as an active regulatory site leading to repression of the gntR operon upon binding of CcpA (30). When we carried out a footprint experiment, we found that RNA polymerase protected the promoter region up to position −78 (data not shown); consequently, we cannot exclude a possible interaction between CcpA and RNA polymerase on the pyxaAB promoter.
FIG. 8.
Transcriptional activity of the pyxaAB promoter and its derivatives in the presence and absence of CcpA. β-Galactosidase activity in strains carrying a lacZ transcriptional fusion to the pyxaAB promoter and its derivatives was measured during planktonic growth in NB medium supplemented with 0.5% glucose with shaking at 37°C. Black bars, MM1709 (amyE::pyxaAB-lacZ); open bars, MM1718 (ccpA1 amyE::pyxaAB-lacZ); gray bars, MM1803 (amyE::pyxaAB-3-lacZ); bars with horizontal lines, MM1719 (ccpA1 amyE::pyxaAB-3-lacZ). ONP, o-nitrophenol. Error bars indicate standard deviations.
In order to confirm that the indicated effect of CcpA on yxaAB expression is directly dependent upon interaction of CcpA with RNA polymerase, we examined the activity of the promoter region lacking the cre site (pKN1-3 construct) in a strain deleted for ccpA and in the 168 background. Surprisingly, we noted a decrease in β-galactosidase activity (MM1719) compared to the ccpA+ background (MM1803) (Fig. 8). This effect was observed only in the presence of glucose (data not shown). From these data we cannot conclude what is the role of CcpA concerning transcription in the yxaAB operon. However, it is still plausible that the CcpA protein does play a direct role in the regulation of transcription of the yxaAB operon, but this will require further experiments.
We finally determined whether yxaAB expression was subject to control by other important regulators of the eps operon, including the σH, Spo0A, and SinI proteins (4, 15, 21). The results obtained with a strain deleted for sigH showed that expression of both yxaA and yxaB was independent of this regulator (data not shown). Surprisingly, we observed no effect of mutations in spo0A or sinI on expression of yxaAB in the tested conditions (data not shown). However, it should be emphasized that we performed these β-galactosidase measurements in a rich medium, which is not conducive for efficient sporulation, and the samples were collected mainly from the exponential and early stationary phases of growth. These conditions are associated with a low level of phosphorylated Spo0A in the cell. Consequently, the absence of an effect of a spo0A mutation on an AbrB-dependent promoter (such as yxaAB) may simply reflect the sampling conditions.
DISCUSSION
Biofilm communities are able to withstand harsh environmental conditions, as shown by the fact that the bacteria therein are often much more resistant to antimicrobial compounds or mechanical injury than planktonic cells (9) Formation of these communities is often perceived as an answer to stress conditions (39). In order to better understand this connection, we searched for an influence of the σB stress factor and yxaB, encoding a putative EPS synthase under σB control, on biofilm formation in B. subtilis.
Entry into the stationary phase of growth as well as diverse environmental stresses cause activation of σB. Nevertheless, the precise function and physiological roles of σB remain debatable (33). There are some reports showing a direct contribution of σB to the formation of adherent biofilms by human pathogens or to the virulence of Bacillus anthracis (13, 22, 37). We found that deletion of the σB gene has a severe effect on biofilm formation, leading to production of very fragile pieces of pellicle which accumulate at the bottom of the container (Fig. 1). These results indicate that σB makes a significant contribution to this multicellular process.
Among genes with unknown function belonging to the σB regulon (33, 35), we searched for possible genes connected to the synthesis of one of the major components of biofilm matrix, EPS (21). We focused on the yxaB gene, encoding a putative EPS synthase. This protein shares homology with EpsL from Streptococcus thermophilus and PssK from Rhizobium leguminosarum bv. trifolii (28, 40). Analysis of the region immediately upstream of yxaB revealed the consensus sequence of a σB-dependent promoter, whose presence has also been suggested by other authors (33). As expected for many general stress genes, whose basal expression depends upon σA under nonstress conditions (1a), we were able to map a σA regulatory sequence close to the 3′ end of yxaA, the gene upstream of yxaB. Transcription initiating at the σA-dependent promoter under nonstress conditions produced a bicistronic mRNA encoding both genes.
In contrast to yxaA, yxaB was highly induced during biofilm formation (Fig. 2). Nevertheless, a strain deleted for yxaB showed relatively normal biofilm formation and structure. One of the most interesting findings of this study was that expression of yxaB from the pSpac promoter in a strain deleted for sigB was able to partially restore formation of a floating biofilm pellicle (Fig. 1). Therefore, we conclude that YxaB is involved in formation and/or maintenance of the biofilm structure in B. subtilis; however, its function may be redundant since the cell possesses two paralogs. Another hypothesis is that YxaB may play a role in the stress resistance of the biofilm but is not required for biofilm formation itself. Further investigations are planned to test whether yxaB plays a role in biofilm-specific enhancement of resistance to antimicrobial agents and stresses.
The yxaA gene, which has not been reported to be stress inducible, encodes a putative glycerate kinase. This type of enzyme is widely present in all kingdoms of living organisms and plays an important function in cell metabolism. Correct functioning of such enzymes is necessary for the C2 respiratory cycle in Arabidopsis thaliana (2) and has a role in serine metabolism in Methylobacterium extorquens (8). In humans, a deficiency of glycerate kinase leads to d-glyceric aciduria, which is characterized by massive amounts of d-glyceric acid in urine (3).
It has been reported that a wide range of genes participating in different metabolic pathways, not directly associated with EPS synthesis, are specifically required for biofilm formation, biofilm maintenance, and/or the resistant properties of biofilm. For example, in B. subtilis, in addition to the epsA to -O and yqxM-sipW-tasA genes required for matrix formation (5), a number of genes such as ampS, gltAB, yoaW, and pgcA, which seemingly fulfill other crucial, often unknown, functions, somehow participate in biofilm development (7, 16, 25). We observed that transcription starting at pyxaAB is unchanged during biofilm formation, suggesting that the gene is not required for this process. However, a constant level of expression of the pgcA or gtaB gene, crucial for biofilm development, has been reported (25). Thus, we cannot rule out a possible role for yxaA in biofilm formation.
Detailed analysis of the pyxaAB promoter region using footprinting and genetic analysis revealed an AbrB binding region. AbrB has been shown to influence the transcription of genes encoding enzymes participating in EPS synthesis in addition to extracellular degradative processes, nitrogen and carbon utilization, amino acid metabolism, and antibiotic resistance under biofilm formation conditions (16, 34). Further investigation of transcription originating from the pyxaAB promoter showed that this is induced by the presence of 0.5% glucose in NB medium and that the maximal activity occurs at the transition into the stationary phase. The stimulatory effect of glucose on biofilm formation is in agreement with the observation that the absence of UDP-Glc in cells affects biofilm formation in B. subtilis. Therefore, it has been postulated that UDP-Glc might act as a metabolic signal regulating this multicellular process (25). The influence of glucose on biofilm formation is complex and depends on other conditions. Whereas Stanley and coworkers (39) showed that addition of glucose (1%) to rich media suppresses biofilm formation, Chagneau and Saier (7), using the same concentration of glucose, showed that it has also a stimulatory effect on this process when added to minimal medium (39).
Although we are not yet able to answer the question concerning the precise function of the yxaAB operon, our observations indicate that it plays a crucial role in the analyzed social behavior. Our future investigations will be directed toward elucidating the specific roles played by the two gene products during biofilm formation in attempts to further unravel the complexities of this multicellular phenomenon.
Acknowledgments
These studies were supported by grants from the Ministry of Science and Higher Education, project no. 2 P04A 039 30 and N301 002 32 (to M.O.), and by National Institutes of Health grant GM46700 (to M.A.S.).
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of stress-and starvation-induced dps/pexB-homologous gene is controlled by alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 1797251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bernhardt, J., U. A. Volker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress protein in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology 143999-1017. [DOI] [PubMed] [Google Scholar]
- 2.Boldt, R., C. Edner, U. Kolukisaoglu, M. Hagemann, W. Weckwerth, S. Wienkoop K. Morgenthal, and H. Bauwe. 2005. d-Glycerate 3-kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 172413-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonham, J. R., T. J. Stephenson, K. H. Carpenter, J. M. Rattenbury, C. H. Cromby, R. J. Pollitt, and D. Hull. 1990. d(+)-Glyceric aciduria: etiology and clinical consequences. Pediatr. Res. 2838-41. [DOI] [PubMed] [Google Scholar]
- 4.Branda, S. S., J. E. González-Pastor, S. E. Ben-Yehuda, S., R. Kolter, and R. Losick. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9811621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 591229-1238. [DOI] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition and transcription activation in prokaryotes. Cell 79743-746. [DOI] [PubMed] [Google Scholar]
- 7.Chagneau, C., and M. H. Saier, Jr. 2004. Biofilm-defective mutants of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 8177-188. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova, L., and M. E. Lidstrom. 1997. Identification and mutation of a gene required for glycerate kinase activity from a facultative methylotroph, Methylobacterium extorquens AM1. J. Bacteriol. 1794946-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49711-745. [DOI] [PubMed] [Google Scholar]
- 10.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantonin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 1817479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 1825036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 18057-61. [DOI] [PubMed] [Google Scholar]
- 15.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 421199-1209. [DOI] [PubMed] [Google Scholar]
- 16.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecker, M., J. Pané-Farré, and U. Völker. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61215-236. [DOI] [PubMed] [Google Scholar]
- 18.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequence: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 232351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmann, J. D., and C. J. Moran. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Shonenshein (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, DC.
- 20.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7283-294. [DOI] [PubMed] [Google Scholar]
- 21.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55739-749. [DOI] [PubMed] [Google Scholar]
- 22.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 1832624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knobloch, J. K., S. Jäger, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 723838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, K. 2007. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 1894920-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarevic, V., B. Soldo, N. Medico, H. Pooley, S. Bron, and D. Karamata. 2005. Bacillus subtilis α-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 7139-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascher, T., A. B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function σ factors. J. Bacteriol. 1896919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65499-560. [DOI] [PubMed] [Google Scholar]
- 28.Mazur, A., J. E. Król, and A. Skorupska. 2001. Isolation and sequencing of Rhizobium leguminosarum bv. trifolii PssN, PssO and PssP genes encoding the proteins involved in polymerization and translocation of exopolysaccharide. DNA Seq. 121-12. [DOI] [PubMed] [Google Scholar]
- 29.Meijer, W. J. J., and M. Salas. 2004. Relevance of UP elements for three strong Bacillus subtilis phage f29 promoters. Nucleic Acids Res. 321166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miwa, Y., K. Nagura, S. Eguchi, H. Fukuda, J. Deutscher, and Y. Fujita. 1997. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol. Microbiol. 231203-1213. [DOI] [PubMed] [Google Scholar]
- 31.Monterrubio, R., L. Baldoma, N. Obradors, J. Aguilar, and J. Badia. 2000. A common regulator for the operons encoding the enzymes involved in d-galactarate, d-glucarate, and d-glycerate utilization in Escherichia coli. J. Bacteriol. 1822672-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran, C. P., Jr. 1993. RNA polymerase and transcriptions factors, p. 653-682. In A. L. Sonenshien, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 33.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 1835617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, Z. E. V., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41757-774. [DOI] [PubMed] [Google Scholar]
- 36.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Shonenshein (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, DC.
- 37.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 1826824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren, D., L. A. Bedzyk, P. Setlow, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Bacillus subtilis surface biofilms with and without sporulation and the importance of yveR for biofilm maintenance. Biotechnol. Bioeng. 86344-364. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 1851951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stingele, F., J. R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 1781680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 81615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 43.Valle, J., M. Vergara-Irigaray, N. Merino, J. R. Penadés, and I. Lasa. 2007. σB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 1892886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, K., and M. A. Strauch. 2001. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 1834094-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]