Abstract
The phototrophic consortium “Chlorochromatium aggregatum” currently represents the most highly developed interspecific association of bacteria and consists of green sulfur bacteria, so-called epibionts, surrounding a central, motile, chemotrophic bacterium. In order to identify subcellular structures characteristic of this symbiosis, consortia were studied by a combination of high-resolution analytical scanning electron microscopy, transmission electron microscopy, and three-dimensional reconstruction and image analyses. Epibionts are interconnected and to a lesser extent are also connected with the central bacterium, by electron-dense, hair-like filaments. In addition, numerous periplasmic tubules extend from the outer membrane of the central bacterium and are in direct contact with the outer membrane of the epibionts. In each epibiont cell, the attachment site to the central bacterium is characterized by the absence of chlorosomes and an additional 17-nm-thick layer (epibiont contact layer [ECL]) attached to the inner side of the cytoplasmic membrane. The ECL is only occasionally observed in pure cultures of the epibiont, where it occurs in about 10 to 20% of the free-living cells. A striking feature of the central bacterium is the presence of one or two hexagonally packed flat crystals (central bacterium crystal [CBC]) per cell. The CBC reaches 1 μm in length, is 35 nm thick, and consists of bilayers of subunits with a spacing of 9 nm. A detailed model for consortia is presented, summarizing our conclusions regarding (i) cohesion of the cells, (ii) common periplasmic space between the central bacterium and the epibiont, (iii) ECL as a symbiosis-specific structure, and (iv) formation of the interior paracrystalline structures, central bacterium membrane layer, and CBC.
During the course of evolution, prokaryotes have entered into numerous symbiotic relationships. Until now, mostly symbioses between bacteria and eukaryotes have been investigated (see reference 23 for a review). These studies have revealed different ways in which prokaryotic cells have morphologically adapted to symbiosis with eukaryotes. Morphological changes have been especially well documented for cyanobacteria. Cyanobacteria of the genus Nostoc occur intracellularly in the fungus Geosiphon and the angiosperm Gunnera or extracellularly in bryophytes, in the water fern Azolla, and in cycads. In the symbiotic state, the bacteria are characterized by an increase in heterocyst frequency, an increased cell size and more-rounded cell shape, the appearance of shorter filaments or even single cells, a reduction in the thickness of the sheath and cell wall, and in some cases, altered thylakoid arrangements (29). During root nodule formation in clover, Rhizobium leguminosarum cells differentiate to bacteroids by enlargement and distortion, loss of the capability of cell division, and formation of polyhydroxybutyrate granules (18). In Azoarcus sp. strain BH72, the betaproteobacterial endophyte of kallar grass, low oxygen concentrations induce the formation of diazosomes, which are stacks of intracytoplasmic membranes in which the iron protein of nitrogenase is highly enriched (30). Morphological changes in the parabasalid flagellate Mixotricha paradoxa are induced due to the adhesion of numerous bacteria. Bacteroides spp. induce a striated calyx-like junction structure beneath the adhesion zone linked to the cortical microfibrillar network. Adhesion of long rod-shaped bacteria induce a dense plate under the adhesion site inside the bacterium and inside the flagellate. Slender spirochetes induce knobs on the cell surface (2). Verrucomicrobia-related ectosymbionts of Euplotidium have unique morphological features among prokaryotes. During stage II, a basket consisting of bundles of tubules surrounds an extrusive apparatus formed by unidentified specialized proteins. Multiplication of epixenosomes and also the differentiation from stage I to stage II are correlated with the cell host cycle (25).
However, various types of highly structured associations, so-called consortia, exist between different prokaryotes (19, 23). Typically, these consortia consist of two different types of prokaryotes which maintain a permanent cell-to-cell contact. In contrast to the interactions of prokaryotes with eukaryotes, no specific morphological adaptations have been reported for symbiotic prokaryotic consortia (14).
Phototrophic consortia consist of a heterotrophic, motile, colorless, rod-shaped central bacterium which is surrounded by up to 69 photoautotrophic green sulfur bacterial cells, the epibionts (6, 20, 23). Phototrophic consortia occur in numerous stratified lakes worldwide (11, 24), where they can amount to up to two-thirds of the total bacterial biomass in the chemocline (8). Based on 16S rRNA gene sequence analysis of natural populations of green sulfur bacteria, the epibionts do not occur as free-living cells (11), suggesting that they are specifically adapted to life in the association. The central bacterium belongs to the Comamonadaceae within the Betaproteobacteria (15, 27). As indicated by microautoradiography with labeled carbon substrates, the central bacterium is capable of assimilating 2-oxoglutarate (10).
Several independent experimental findings indicate that a rapid signal transfer occurs between the epibionts and the central bacterium in phototrophic consortia. First, cell division of both the central bacterium and the epibionts is highly coordinated (24). Second, intact consortia exhibit a scotophobic response and accumulate at wavelengths of light which correspond to the absorption maxima of the bacteriochlorophylls present in the immotile green sulfur bacterial epibionts (5), while only the central bacterium is flagellated and confers motility to the consortium (9). Third, the incorporation of 2-oxoglutarate by the central bacterium occurs only in the presence of light and sulfide used by the epibiont (10).
In the present study, detailed ultrastructural investigations combining high-resolution analytical scanning electron microscopy (SEM), transmission electron microscopy (TEM), and three-dimensional (3D) reconstruction and image analyses were conducted with the aim of elucidating the structural basis for the close cell-cell interaction in phototrophic consortia. By comparing intact consortia with epibiont cells in the recently established pure cultures (34), symbiosis-specific subcellular structures could be identified, serving as the basis for an ultrastructural model of phototrophic consortia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
“Chlorobium chlorochromatii”—an isolate from “Chlorochromatium aggregatum” from Lake Dagow (34)—was grown in standard SL10 medium for green sulfur bacteria (22) supplemented with 3 mM acetate and adjusted to pH 7.2. Cultures were incubated at 25°C with continuous illumination by a tungsten lamp (Osram; 60 W) at a light intensity of 50 μmol quanta·m−2·s−1 as determined with a Li Cor LI-189 quantum meter equipped with an LI-200 SA pyranometer sensor (Li Cor, Lincoln, NE). The consortium “Chlorochromatium aggregatum” was grown in K4 medium (15) in 10-liter glass vessels at 15°C under continuous illumination of 20 μmol quanta·m−2·s−1. Under these conditions, “Chlorochromatium aggregatum” forms an almost pure biofilm on the inner surface of the vessel (27). The biofilm was scraped off the glass wall and used in the subsequent ultrastructural investigations.
Cell fractionation and isolation of chlorosomes.
Cells were harvested by centrifugation at 10,000 × g for 30 min and then resuspended in 10 mM Tris buffer, pH 7.5, and broken by three passages through a French press cell at 110 MPa. Unbroken cells and cell debris were removed from the homogenate by centrifugation at 2,000 × g for 5 min; the supernatant was centrifuged again at 3,000 × g for 5 min, and the pellet was finally resuspended in 10 mM Tris. The resulting supernatants and resuspended pellets were used for ultrastructural analysis.
Electron microscopy.
For chemical fixation, immediately after collection, cells were fixed with 2.5% glutardialdehyde in fixative buffer (75 mM sodium cacodylate, 2 mM MgCl2, pH 7.0) for 1 h at room temperature. Afterwards, samples were rinsed several times in fixative buffer and postfixed at room temperature for 1 h with 1% osmium tetroxide in fixative buffer. After two washing steps in distilled water, the cells were stained en bloc for 30 min with 1% uranyl acetate in 20% acetone. Dehydration was performed with a graded acetone series. Samples were then infiltrated and embedded in Spurr's low-viscosity resin.
For high-pressure freezing, cellulose capillary tubes were filled by capillary forces with concentrated cell suspensions and the cells were immobilized by high-pressure freezing (Leica EMPACT2) as described previously (28, 31). Freeze substitution was performed in ethanol with 0.25% glutaraldehyde, 1% formaldehyde, and 0.5% uranyl acetate, including 5% water (3, 35).
After embedding the samples in Epon, ultrathin sections were cut with a diamond knife and mounted onto uncoated copper grids. The sections were poststained with aqueous lead citrate (100 mM, pH 13.0).
For negative staining, a drop of the sample of an appropriate dilution was placed on a 400 mesh carbon-coated copper grid, which had been freshly rendered hydrophilic, by glow discharge. After incubation for 2 min, the drop was quickly removed with a Pasteur pipette, and the grid was air dried and then stained with 2% uranyl acetate and 0.01% glucose.
TEM micrographs were taken with an EM 912 electron microscope (Zeiss, Oberkochen, Germany) equipped with an integrated OMEGA energy filter operated at 80 kV in the zero loss mode. Unless otherwise indicated, micrographs were recorded from conventionally fixed samples. Fast Fourier transformation (FFT) of digital images (1,024 by 1,024 pixels, taken with a charge-coupled-device camera; Proscan GmbH, Germering, Germany) was performed employing the analySIS 3.0 software. Autocorrelation of image details was performed with DigitalMicrograph 3.4 (Gatan, Pleasanton, CA) as follows. After Fourier transformation of the real image, the resulting image was multiplied by its complex conjugate and the inverse Fourier transform was calculated. Finally, the resulting image was normalized to a maximum value of 1. Cross-correlation was performed using DigitalMicrograph (Gatan) with real data in the following way: Fourier transformation was applied to each of the two source images; the Fourier transform of the first image was multiplied by the complex conjugate of the Fourier transform of the resulting image; and the inverse Fourier transform of the resulting image was calculated. Electron energy loss-specific imaging of the phosphorus distribution was performed with the three-windows method (analySIS 3.0 software; ΔE = 110.0 eV, 121.0 eV, and 153.0 eV).
For SEM, drops of the sample were placed onto a glass slide, covered with a coverslip, and rapidly frozen with liquid nitrogen. The coverslip was removed with a razor blade, and the glass slide was immediately fixed with 2.5% glutaraldehyde in 75 mM cacodylate buffer (pH 7.0), postfixed with 1% osmium tetroxide in fixative buffer, dehydrated in a graded series of acetone solutions, and critical-point dried after transfer to liquid CO2. Specimens were mounted on stubs, coated with 3-nm platinum using a magnetron sputter coater, and examined with a Hitachi S-4100 field emission SEM operated at 5 kV.
Calculation of surface and volume.
The surfaces and volumes of “C. aggregatum,” the epibionts (“Chlorobium chlorochromatii” cells), and the chlorosomes were calculated assuming a smooth surface with the formula for a cylinder and two hemispheres. To calculate the values of the central bacterium, a smooth surface was assumed and the formulas for ellipsoids were used.
RESULTS
The dimensions of “Chlorochromatium aggregatum” were determined by SEM of the cryopreparations of consortia on glass slides (Fig. 1A). Individual consortia were 5.2 ± 1.0 μm long and 2.5 ± 0.2 μm wide. On average, each consortium harbored 16 ± 3 epibionts (Table 1).
FIG. 1.
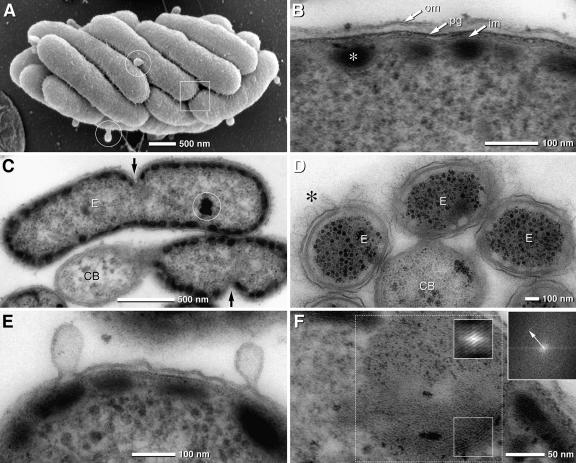
SEM and TEM micrographs of “Chlorochromatium aggregatum” and the epibionts, “Chlorobium chlorochromatii” cells, in the symbiotic state and in pure culture. (A) SEM of “Chlorochromatium aggregatum” showing the epibionts tightly packed; the cell surface appears rough and exhibits numerous thin filaments interconnecting neighboring cells (frame). Epibionts exhibit numerous bulb-shaped protrusions (circles). (B) TEM detail of the epibiont cell with typical gram-negative envelope (om, outer membrane; pg, peptidoglycan; im, inner membrane) and chlorosomes attached to the cytoplasmic membrane (asterisk). (C) TEM of epibionts (E)—attached to a central bacterium (CB)—revealing asymmetric cell division (arrows). (D) TEM of consortia after cryofixation by high-pressure freezing showing with high contrast the hair-like filaments (asterisk) and the undulating outer membrane (the chlorosomes appear electron translucent due to extraction during freeze substitution) of the epibionts (E, epibiont; CB, central bacterium). (E) TEM of epibiont bulb-shaped protrusions which are formed from the outer membrane by local enlargement of the periplasmic space. (F) Lipid body-like globule of pure culture of “Chlorobium chlorochromatii” exhibiting a myelin-like structure which is enhanced after autocorrelation (small insets) with a spacing of 3.5 nm, determined by FFT (upper right inset; the arrow indicates the first-order reflex) of the dotted area.
TABLE 1.
Dimensions for “Chlorochromatium aggregatum,” pure-culture “Chlorobium chlorochromatii,” and their respective chlorosomes
| Object | Dimensions
|
||||
|---|---|---|---|---|---|
| Length (μm) | Width (μm) | Surface (μm2) | Volume (μm3) | Cross section (μm2) | |
| “Chlorochromatium aggregatum” | 5.2 ± 1.0 | 2.5 ± 0.2 | 40.8 | 21.4 | |
| Epibiont (in symbiotic state) | 2.05 ± 0.5 | 0.7 ± 0.1 | 4.80 | 0.79 | |
| “Chlorobium chlorochromatii” | 1.2 ± 0.6 | 0.6 ± 0.05 | 2.30 | 0.29 | |
| Central bacterium | 2.9 ± 0.6 | 0.63 ± 0.06 | 5.71 | 0.60 | |
| Chlorosomes of “Chlorochromatium aggregatum” | 0.139 ± 0.041 | 0.053 ± 0.010 | 0.00027 | 0.023 | |
| Chlorosomes of “Chlorobium chlorochromatii” | 0.133 ± 0.040 | 0.060 ± 0.011 | 0.00032 | 0.025 | |
Ultrastructure of the epibiont cells.
In the symbiotic state, the rod-shaped epibionts are 2.05 ± 0.5 μm long and 0.7 ± 0.1 μm wide (Fig. 1A; Table 1). Their cell volume amounted to 0.79 μm3 and their surface to 4.8 μm2. Division stages were regularly observed. Interestingly, cell division of the epibionts proceeded in an asymmetric fashion: cell wall formation and cell division are initiated at the exterior (relative to the consortium), causing the daughter cells to bend at the constriction (Fig. 1C). Ultrathin sections revealed a typical gram-negative architecture for the 25-nm-thick bacterial envelope, which consisted of a 6- to 7-nm-thick outer membrane, an electron-dense peptidoglycan layer measuring 3 to 4 nm, and a cytoplasmic membrane with a thickness of 6 to 7 nm (Fig. 1B). Tangential sections demonstrated that the inner surface of the cytoplasmic membrane of epibiont cells is covered by chlorosomes which do not exhibit a particular orientation (see Fig. S1A in the supplemental material). In order to determine their dimensions in a reliable fashion, chlorosomes were isolated from “C. aggregatum” cultures and negatively stained (see Fig. S1B in the supplemental material), which yielded mean values for chlorosome length and width of 139 ± 41 nm and 53 ± 10 nm, respectively. These data are comparable with those of chlorosomes from free-living epibiont cells (133 ± 40 nm by 60 ± 11 nm; Table 1) isolated in the same manner.
At low magnification, epibionts in intact consortia exhibited a rough surface structure and bulb-shaped protrusions (Fig. 1A). Analysis of thin sections indicated that the outer membrane frequently undulates with distances from 10 to 35 nm to the peptidoglycan layer, resulting in local changes of the periplasmic space (Fig. 1E). Within intact phototrophic “C. aggregatum” consortia, neighboring epibionts are interconnected by thin filaments. To exclude artificial changes of the periplasmic space during conventional fixation, consortia were cryofixed by high-pressure freezing and freeze substituted. In these specimens, 150-nm-long and approximately 2.5- to 3-nm-wide electron-dense, hair-like filaments could be discerned which covered the entire surface (Fig. 1D). Furthermore, the variability of the periplasmic space was even more pronounced (Fig. 1D). The hair-like filaments also interconnect epibiont cells with the central bacterium (Fig. 1D). A characteristic feature of the epibionts was 50- to 200-nm large, bulb-shaped, bleb-like protuberances which occurred at a frequency of up to 20 per epibiont cell (Fig. 1A and E). These protuberances were also observed in pure cultures, where the abundance of these protrusions was negatively correlated with growth phase. Exponentially growing cells were almost free of protuberances, whereas during the transition from late exponential to stationary phase, the number of protuberances on cells increased. TEM of ultrathin sections demonstrated that these protuberances are contiguous with the outer membrane and represent a rather localized enlargement of the periplasmic space (Fig. 1E).
The cytoplasm of symbiotic as well as free-living epibionts contained one to three globules of a rather low electron density which resembled plant and fungal lipid bodies and were typically attached or positioned close to the cytoplasmic membrane (see Fig. S1C in the supplemental material). The diameters of these structures varied between 100 and 250 nm. Multiple parallel membrane-like layers were detected in the periphery of the globules, frequently exhibiting a myelin-like pattern with a periodicity of 3.5 nm as determined by FFT (Fig. 1F). In addition, osmiophilic globules with diameters of up to 170 nm were frequently observed in the cytoplasm of epibionts (Fig. 1C and Fig. 2B and D). Energy filtering (electron energy loss-specific imaging) of ultrathin sections (20 to 30 nm) of cells fixed only with glutaraldehyde revealed a high content of phosphorus within these globules, which can therefore be classified as polyphosphate.
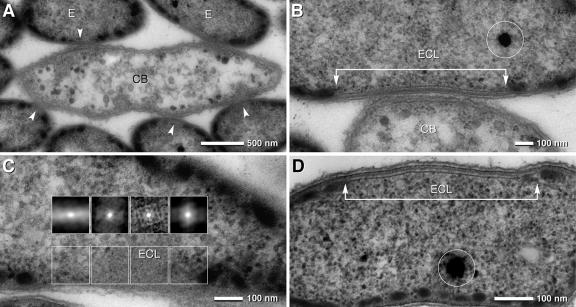
FIG. 2.
TEM micrographs of consortia and epibionts in the symbiotic state and in pure culture. (A) TEM of a longitudinal ultrathin section of a consortium with a central bacterium (CB) with the shape of an elongated ellipse (with the beginnings of cell division) and several attached epibionts (E); at the site of attachment, the cytoplasmic membrane of the epibionts is free of chlorosomes (arrowheads). (B) Area of attachment of the epibiont cell and the central bacterium (ECL, epibiont contact layer) which is characterized by a laminar layer (arrow bar) and the absence of chlorosomes; the osmiophilic globule (circle) represents polyphosphate. (C) Autocorrelation of an oblique section of an ECL revealing a pattern of regularly arranged particles (insets); at transition zones from the ECL to the cytoplasm, the pattern disappears (upper insets, outer left and right). (D) TEM of an ECL of “Chlorobium chlorochromatii” in pure culture; although the ECLs are present only in approximately 20% of the cells, their architecture is identical to that formed in symbiosis; the osmiophilic globule (circle) represents polyphosphate.
Differences between symbiotic and free-living epibiont cells.
Chlorosomes showed similar dimensions in symbiotic and free-living epibiont cells. Tangential sections revealed, however, that chlorosomes covered the inner face of the cytoplasmic membrane area at a density of 78 chlorosomes per μm2 (corresponding to 53% of the area; see Fig. S1A in the supplemental material), whereas the density was reduced in free-living epibiont cells to a value of 53 chlorosomes per μm2 of cytoplasmic membrane (38% of the area). When also accounting for the larger cell volume of epibionts in the symbiotic state (Table 1), the absolute number of chlorosomes per cell was 374 in the symbionts but only 121 in the free-living cells. Due to its limited area, the chlorosome-free contact site in symbiotic cells (see below) did not affect this estimate significantly.
The most conspicuous morphological feature of symbiotic epibiont cells is the absence of chlorosomes at the contact site with the central bacterium (Fig. 2A and B). Higher magnification revealed that this epibiont contact layer (ECL) is 17 nm thick and consists of two parallel 4.5-nm-wide electron-dense layers which are separated by an 8-nm-wide less-electron-dense zone (Fig. 2B). Reconstruction of serial sections demonstrated that each symbiotic epibiont cell contains one ECL and that the ECL has an ellipsoid shape about 100 nm wide and up to 800 nm long (see Fig. S2A in the supplemental material). Thus, the ECL occupies 1 to 2% of the total area of the cytoplasmic membrane. During cell division, the ECL is distributed to both daughter cells (see Fig. S2B in the supplemental material). Autocorrelation of the ECL in tangential sections revealed a pattern of regularly arranged particles which disappeared in the transition zones from the ECL to the cytoplasm (Fig. 2C). Electron microscopy of consortia after high-pressure freezing and freeze substitution confirmed the presence of the ECL in the epibionts. The ECL also occurred in free-living epibionts from pure cultures (Fig. 2D) but was detected only in 10 to 20% of the cells.
Ultrastructure of the central bacterium.
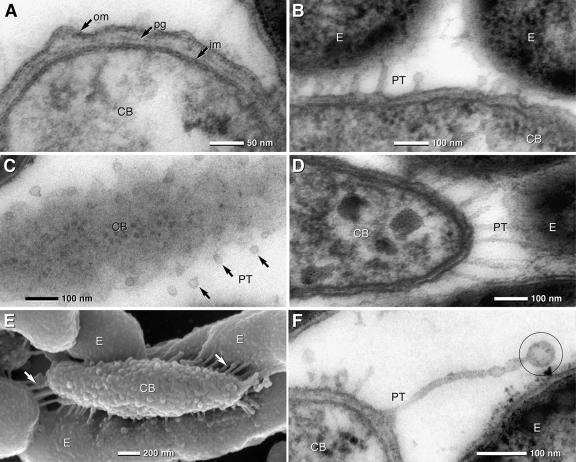
The shape of the central bacterium of “Chlorochromatium aggregatum” is a prolate ellipsoid with a length of 2.9 ± 0.6 μm and a width of 0.63 ± 0.06 μm (Fig. 3E; Table 1). The cell volume amounted to 0.60 μm3 and the surface to 5.71 μm2. SEM showed a rough surface on the central bacterium (Fig. 3E), which according to results from the ultrathin sections can be attributed to undulation of the outer membrane (Fig. 3A). High-pressure freezing and freeze substitution confirm the undulations of outer membranes. The cell envelope was 25 to 30 nm thick and had a typical gram-negative architecture consisting of a 7-nm-thick outer membrane, an electron-dense peptidoglycan layer of 3-nm thickness, and a 7-nm-thick cytoplasmic membrane (Fig. 3A).
FIG. 3.
TEM of ultrathin sections and SEM micrographs of the central bacterium (CB) and epibionts (E) of “C. aggregatum.” (A) TEM detail of the central bacterium cell with a typical gram-negative envelope (om, outer membrane; pg, peptidoglycan; im, inner membrane). (B and C) TEM of PT formed by the outer membrane of the central bacterium which are in contact with the outer membrane of the epibionts (B). In tangential sections of the central bacterium, PT are visible as small circular membrane cross sections with diameters in the range of 25 nm (C, arrows). (D and E) The lengths of the PT depend on the distance between the central bacterium and the epibionts. PT are rather straight and appear under tension when in contact with the epibionts, which is best seen in 150-nm sections (D) and SEM micrographs of a partially desegregated consortium (E). The central bacterium exhibits a knobby surface, with numerous PT (arrows) attached to the epibionts (E). (F) Unattached PT show terminal vesiculation (circle).
Two types of protrusions are typical for the cell envelope of the central bacteria: (i) small papillae, approximately 10 to 20 nm in length and regularly occurring at a distance of about 25 nm from each other, formed by protuberances of the outer membrane and local changes of the periplasmic space (Fig. 3A), and (ii) numerous periplasmic tubules (PT) formed by the outer membrane which are in linear contact with the epibionts (Fig. 3B and D to E; also see Fig. S3A to C in the supplemental material). Although PT are best observed at the poles of the central bacterium where the distance to the epibionts reaches a maximum of approximately 200 nm (Fig. 3D), they are distributed over the entire cell surface, with distances in the range of 50 to 100 nm (Fig. 3B to E). Tangential sections show that the diameters of PT cross sections are rather uniform; the mean diameter of the PT as calculated from their cross sections was 25 nm (Fig. 3C; also see Fig. S3B in the supplemental material). Subtracting the thickness of the outer membrane (7 nm) results in 11 nm for the inner tubular diameter. Occasionally, small membrane vesicles were observed at the end of the PT (Fig. 3F).
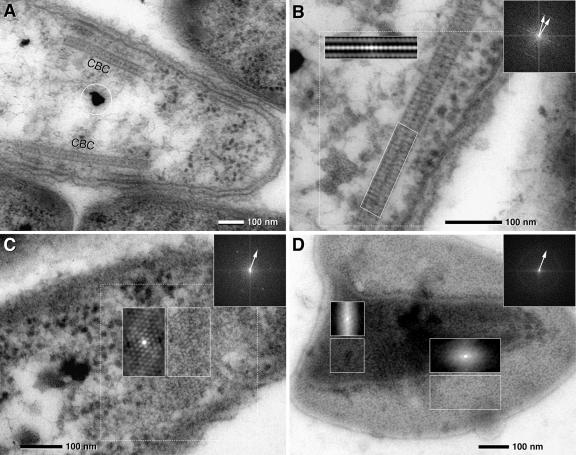
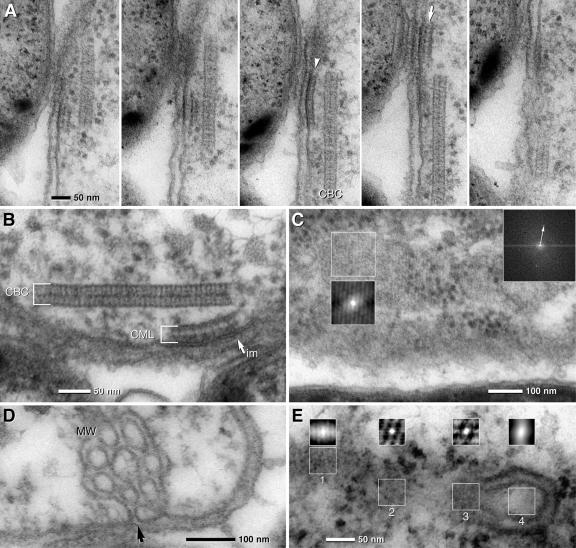
The most prominent ultrastructures within cells of the central bacterium were 35-nm-thick and up to 1-μm-long zipper-like crystalline structures. These structures either were located parallel to the cytoplasmic membrane (Fig. 4A) or occurred freely oriented within the cytoplasm. Results from serial sections of the entire “C. aggregatum” consortia showed that each central bacterium contained on average 1.5 central bacterium crystals (CBCs), with numbers ranging between 1 and 3 per cell. At higher magnification, CBCs were observed to consist of two parallel layers of subunits. Subunits were oriented in either an orthogonal or slightly oblique manner with regard to the axis of the CBC (Fig. 4B; also see Fig. S4A and B in the supplemental material). Employing FFT, the diameter of the subunits was determined to be 9 nm (Fig. 4B and C). Within the CBC, the relative orientation of the subunits changed by up to 20° over 100 nm (Fig. 4B). Tangential sections of the CBCs were obtained by serial sectioning and revealed a hexagonal pattern of subunits with a typical spacing of 9 nm (Fig. 4C; also see Fig. S4B in the supplemental material). This architecture of the CBCs was confirmed using consortia treated by high-pressure freezing and freeze substitution. Although the contrast of the CBCs in these latter preparations was very low, subsequent autocorrelation of CBCs yielded a pattern similar to conventionally fixed specimens (see Fig. S4C and D in the supplemental material).
FIG. 4.
TEM micrographs of ultrathin sections of the central bacterium of “C. aggregatum” revealing paracrystalline structures (CBC, central bacterium crystal) in situ and after isolation and negative staining. FFTs of dotted areas are presented as insets (upper right; arrows indicate first-order reflexes); autocorrelation is presented in the same size as the analyzed area. (A) Longitudinal section of a central bacterium with two cross-sectioned CBCs located parallel to the cytoplasmic membrane (circle, osmiophilic granules). (B) Detail of a CBC showing a paracrystalline zipper-like organization in the cross sections; two less-electron-dense layers are bordered and separated by electron-dense layers. Autocorrelation and FFT of the CBC reveal a 9-nm periodicity with the orientation of the subunits either vertical or oblique to the axis (two 9-nm FFT reflexes). (C) FFT and autocorrelation of a tangential section of the CBC shows a hexagonal pattern of subunits which are 9 nm in diameter. (D) Negative staining of the isolated CBC. Isolated CBCs are frequently observed as double or multiple layers. FFT of the whole image shows a Debye-Scherrer ring corresponding to a distance of 9 nm, indicating a lower degree of subunit orientation of the CBC than that found in situ; autocorrelation shows local patterns of subunit orientation similar to that of CBC in situ in a tangential ultrathin section (for comparison, see panel C).
In TEM, isolated CBCs are recognized after negative staining by their characteristic size and shape and a paracrystalline arrangement of subunits (Fig. 4D). The sheets are typically folded, forming locally double or multiple layers. This “overlay” impedes analysis of the subunit pattern by formation of a superimposed pattern. Only part of the CBC is accessible to FFT, resulting in a diffuse Debye-Scherrer ring corresponding to a distance of 9 nm of spacing (Fig. 4D). At favorable regions of the CBC, autocorrelation exhibits an arrangement of subunits similar to the CBC in situ in tangential ultrathin sections (compare Fig. 4D with Fig. S4B in the supplemental material).
Frequently underlying a CBC, additional membranous structures were observed at the inner face and parallel to the cytoplasmic membrane (Fig. 5A and B). These additional central bacterium membrane layers (CMLs) are 30 to 35 nm thick and 50 to 150 nm long, and at higher magnification they resembled the CBC at least in some of the preparations. Due to the small size, FFT analysis of the CML was only occasionally possible at favorable positions in tangential sections, but it revealed a hexagonal pattern which was 9 nm in diameter (Fig. 5C). Autocorrelation of oblique or tangential sections revealed locally restricted regular patterns (Fig. 5C; also see Fig. S5A in the supplemental material) with the same periodicity as detected in the CBC as shown by cross-correlation (see Fig. S5A in the supplemental material). The presence of the CML was confirmed by high-pressure freezing and freeze substitution (see Fig. S4C in the supplemental material).
FIG. 5.
TEM micrographs of ultrathin sections of the central bacterium of “C. aggregatum” revealing the CML and MW. (A) Serial sections show that the CML is only visible in two to three consecutive sections, proving that it is locally restricted. The CMLs are found with different thicknesses, suggesting the composition of a monolayer (arrowhead) or bilayer (arrow). (B) The electron density of the outer leaflet of the cytoplasmic membrane (im) is higher at the site of attachment of the CML (arrow). (C) FFT and autocorrelation of a tangential section of the CML shows a hexagonal pattern of subunits which are 9 nm in diameter (the arrow indicates the first-order reflex). (D) Details of the central bacterium showing MW which are formed by invaginations of the cytoplasmic membrane (arrow). (E) Autocorrelation of different areas of a section through a CBC in contact with a MW: an oblique section of the CBC is characterized by striations (1) and tangential sections with hexagonal patterns (2, 3); no significant pattern is visible at the area of the MW (4).
Typical for all central bacteria is the presence of two to three complex membranous whirls (MW) that are compact aggregates of tubules in continuity with the cytoplasmic membrane (Fig. 5D). 3D reconstruction from serial sections revealed that the size of the MW is variable in the range of 150 to 400 nm (see Fig. S5B in the supplemental material). Reconstruction from serial sections revealed that there is a significant contact or even continuity between the MW and the CBC (Fig. S5C in the supplemental material). Autocorrelation of transition areas (between the MW and the CBC) shows the regular arrangement of subunits, as typical for the CBC (compare Fig. 5E with Fig. S4B and D in supplemental material).
Osmiophilic globules, of various diameters (up to 100 nm), are frequently observed in the cytoplasm of central bacteria (Fig. 4A). 3D reconstruction from serial sections revealed that a central bacterium has zero to three osmiophilic globules.
In order to determine whether the ultrastructural features detected in laboratory-grown “Chlorochromatium aggregatum” also occur under natural conditions, natural populations of the phototrophic consortia “Pelochromatium roseum” and “Chlorochromatium aggregatum” were obtained from Lake Dagow (27) and their ultrastructures were investigated by electron microcopy. This revealed that all consortia in the samples shared the structural features described in the preceding sections for laboratory cultures of “Chlorochromatium aggregatum.”
DISCUSSION
The fact that “Chlorobium chlorochromatii” grows in pure culture demonstrates that this epibiont is not obligatorily symbiotic. Despite this capability, neither “Chlorobium chlorochromatii” nor any other epibiont of the known 19 types of phototrophic consortia has ever been detected in a free-living state in natural bacterial communities (11), indicating that epibionts of phototrophic consortia are specifically adapted to life in the symbiotic state. Thus far it was not known whether this adaptation to a symbiosis in phototrophic consortia involved specific morphological characteristics of the cells, like adhesion structures or specific organelles. In the course of the present study, different ultrastructures of the epibionts and the central bacterium were discovered (Fig. 6).
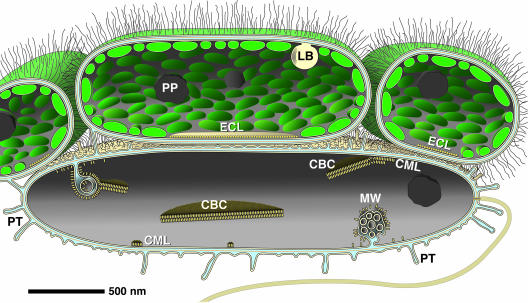
FIG. 6.
Schematic representation according to 3D reconstruction data for “Chlorochromatium aggregatum” summarizes the results of this study. Epibionts are connected by long, hair-like polysaccharide chains. The epibionts harbor lipid bodies (LB) with a myelin-like pattern which are attached to the cytoplasmic membrane and polyphosphate globules (PP). The attachment site of the epibiont is characterized by the absence of chlorosomes and a single contact layer (ECL, epibiont contact layer). Striking features of the flagellated central bacterium are (i) periplasmic tubules (PT), which can be in direct contact with the epibionts, postulated to form a common periplasmic space; (ii) complex invaginations (MW, membranous whirls) of the cytoplasmic membrane; (iii) subunits arranged in small monolayers or bilayers directly associated with the cytoplasmic membrane (CML, central bacterium membrane layer); and (iv) paracrystalline structures (CBC, central bacterium crystals) which are formed on the inner side of the cytoplasmic membrane (or membranous invaginations) by the accumulation of subunits. Globular structures occur in the cytoplasm of the central bacterium.
Symbiosis-specific ultrastructure of the epibiont in “Chlorochromatium aggregatum.”
SEM and TEM micrographs revealed up to 150-nm-long hair-like filaments which covered the surface of the epibionts and formed an interconnecting network between the cells of “C. aggregatum.” These filaments occur exclusively on epibionts, suggesting that the interconnected epibionts form an elastic cage which encloses the central bacterium. This conclusion is also supported by the observation that the epibionts stay associated with the central bacterium when consortia are subjected to shearing forces (by squeezing consortia on a glass slide with a coverslip, or by vacuum filtration of consortia through membrane filters) (23).
Recent disaggregation experiments with the consortium “C. aggregatum” (K. Vogl, M. Dreβen, R. Wenter, M. Schlickenrieder, M. Plöscher, L. Eichacker, and J. Overmann, submitted for publication) demonstrated that cell-cell aggregation is not susceptible to proteolysis with proteinase K, pepsin, trypsin, or chymotrypsin or to treatment with lysozyme, hyaluronidase, or β-glucuronidase, indicating that simple extracellular capsule polysaccharides or easily degradable proteins are not involved in the cell-cell binding. Extracellular appendages resembling the filaments seen on the epibiont surface have been identified for other bacteria as capsular polysaccharides (13) and may therefore also be involved in the adhesion in phototrophic consortia.
Underlying a typical outer membrane, peptidoglycan layer, and cytoplasmic membrane at the contact site, the ECL clearly differs in ultrastructure from typical biomembranes. Autocorrelation of cross sections did not reveal a characteristic pattern, whereas oblique sections showed that the ECL is composed of regularly arranged elements, possibly globular proteins. Within “C. aggregatum,” the ECL was invariably observed in all epibiont cells, indicating that the ECL is an ultrastructural complex which is essential for symbiosis. In contrast, ECL-like structures were observed only in a small fraction of the cells from pure cultures, suggesting that biosynthesis of this ultrastructure in epibionts is subject to regulation and induced in the symbiotic state.
Interior substructures of the central bacterium in “Chlorochromatium aggregatum.”
The CBC structures were found in various numbers and orientations. Since they were primarily cytoplasmic structures, their direct involvement in cell-cell adhesion is unlikely. Interestingly, the CBC structurally resembles the chemotaxis receptor Tsr of Escherichia coli (16, 36, 37, 38). In fact, overproduction of Tsr in E. coli results in internal membrane networks composed of stacks and tubular structures (16) which resemble the MW discovered in the central bacterium of “C. aggregatum.”
When comparing CBCs in situ with isolated CBCs, there are discrepancies in their appearances. Isolated CBCs are not flat bilayers or fragments thereof but rather are folded multilayers (Fig. 4D). Although a regular pattern of subunits is assumed at first glance, autocorrelation shows that only distinct areas show a pattern in the CBC (Fig. 4D). This explains why FFT analysis often fails or results in Debye-Scherrer rings corresponding to a distance of 9 nm (Fig. 4D) instead of hexagonally arranged first-order peaks (Fig. 4C): the areas of regular arrangement of subunits are too small.
In addition to the CBC, the CML, a layer parallel to the plasma side of the cytoplasmic membrane, is characteristic of the central bacterium. The different morphologies of the CML appear to be a transition from single- to double-laminated structures which resemble typical CBCs. Accordingly, CMLs may represent early stages of the CBC. So far, however, possible connections between the CML and the CBC cannot be investigated due to resolution limits of ultrathin sections.
Implications of ultrastructural features.
Although knowledge of the physiology of the central betaproteobacterium is still rather limited, experimental data indicate that it incorporates external 2-oxoglutarate (10). Theoretically, the tight arrangement of cells in phototrophic consortia as observed in TEM and SEM micrographs could lead to a diffusion limitation and hence a physiological isolation of the central bacterium. However, TEM analyses, including the cryofixed and cryosubstituted preparations, revealed that considerable intercellular space exists between the cells. Based on volumetric calculations, the central bacterium only occupies 25% of the volume available. Together with the gaps left by the epibiont cells forming the cortex of the consortia, this extracellular space likely prevents diffusion limitation of “C. aggregatum.”
A conspicuous feature of the intercellular space between the epibionts and the central bacterium is the occurrence of PT which extend from the outer membrane of the central bacterium toward the cell surface of the epibionts. Together with the numerous papillae, these outer membrane structures result in a surface area enlargement of approximately 300% compared to a smooth membrane, as calculated from TEM tangential sections and SEM micrographs. Based on the close association between the PT and the outer membrane of the epibionts, the two partner bacteria may actually share a common periplasmic space (Fig. 6).
The numbers of chlorosomes on the inner face of the cytoplasmic membrane were comparable for “Chlorobium chlorochromatii” (53 chlorosomes/μm2) and Chlorobaculum tepidum (51 chlorosomes/μm2, calculated based on the method in reference 4) when grown under saturating light intensities. Symbiotic epibiont cells grown at limiting light intensities reached a density of 78 chlorosomes/μm2. Based on these values, the epibiont does not represent one of the extremely low-light-adapted members of the green sulfur bacteria (7, 17, 21). This is consistent with the observation that growth of pure epibiont cultures becomes light saturated only at >10 μmol·m−2·s−1 (34), in contrast to low-light-adapted relatives whose growth becomes light saturated above 1 μmol·m−2·s−1 (17, 21). Although phototrophic consortia inhabit low-light environments (24), their limited adaptation to the in situ light intensities does not necessarily represent a disadvantage. Green sulfur bacteria are known to excrete considerable amounts of photosynthetically fixed organic carbon, which has recently also been confirmed for the epibiont (26), and it has been suggested that the epibiont supplies the central bacterium with these organic carbon excretion products (10). Although the rate of anoxygenic photosynthesis per cell decreases under light-limiting conditions, the surrounding epibionts together may still be able to maintain a considerable rate of carbon supply for the single central bacterium in phototrophic consortia. The disadvantage of carbon assimilation under limited light conditions is compensated by the acquired motility, enabled by the flagellated central bacterium, which ensures a habitat with favorable conditions, i.e., presence of sufficient sulfides, light, and absence of oxygen.
The CBC detected in the current study represents one of the most striking ultrastructural elements of phototrophic consortia. An exhaustive search of the literature revealed that there is a variety of striking internal structures in several other, nonrelated bacteria. Microtubule-like structures have been documented for numerous gram-negative and gram-positive bacteria (1), but they clearly differ in fine structure from the CBC detected in phototrophic consortia. For the sulfur-oxidizing epsilonproteobacterium “Arcobacter sulfidicus,” discrete 27- to 33-nm-thick structures underlying the cytoplasmic membrane at the cell pole have been described. Although they look similar to the CBC at lower magnification, they consist—in contrast to phototrophic consortia—of four to five membrane lamellae with a thickness of 7.5 nm each (33). Electron-dense, membrane-like elements have been observed in the thermophilic Persephonella marina, a member of the Aquificales (12). Some of these structures exhibited a bilayered substructure, thereby resembling the CBC in phototrophic consortia. Most notably, however, ultrastructural elements which exhibit an obvious similarity to the CBC have recently been documented for multicellular magnetotactic bacteria (32). These “striated structures” are identical to the CBC with respect to dimensions and shape. Although not explicitly described, in TEM micrographs published by Silva et al. (32), a structure which resembles the CML can also be recognized. The occurrence of structures similar to the CBC and CML in nonrelated bacteria forming multicellular associations suggests that these structures are involved in prokaryotic cell-cell interactions.
Our study has identified several ultrastructural elements of epibionts as well as of central bacteria. Some of these structures appear to be highly specific for phototrophic consortia and hence are likely to be involved in either the cell-cell aggregation or the physiological interaction in this most highly developed symbiosis between prokaryotes.
Supplementary Material
Acknowledgments
We thank Reinhard Rachel and Andreas Klingel for high-pressure freezing and freeze substitution of consortia and for valuable discussions. Silvia Dobler is gratefully acknowledged for skillful technical assistance, Elizabeth Schroeder-Reiter for carefully reading the manuscript, and H.-P. Grossart and Kristina Pfannes for providing samples from Lake Dagow.
This work was supported in part by grants of the Deutsche Forschungsgemeinschaft to J. Overmann (no. Ov 20/10-1 and 10-2).
Footnotes
Published ahead of print on 14 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bermudes, D., G. Hinkle, and L. Margulis. 1994. Do prokaryotes contain microtubules? Microbiol. Rev. 58387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugerolle, G. 2004. Devescovinid features, a remarkable surface cytoskeleton, and epibiotic bacteria revisited in Mixotricha paradoxa, a parabasalid flagellate. Protoplasma 22449-59. [DOI] [PubMed] [Google Scholar]
- 3.Burghardt, T., D. J. Näther, B. Junglas, H. Huber, and R. Rachel. 2007. The dominating outer membrane protein of the hyperthermophilic archaeum Ignicoccus hospitalis: a novel pore-forming complex. Mol. Microbiol. 63166-176. [DOI] [PubMed] [Google Scholar]
- 4.Frigaard, N.-U., A. G. M. Chew, H. Li, J. A. Maresca, and D. A. Bryant. 2003. Chlorobium tepidum: insights into the structure, physiology, and metabolism of a green sulfur bacterium derived from the complete genome sequence. Photosynth. Res. 7893-117. [DOI] [PubMed] [Google Scholar]
- 5.Fröstl, J., and J. Overmann. 1998. Physiology and tactic response of “Chlorochromatium aggregatum.” Arch. Microbiol. 169129-135. [DOI] [PubMed] [Google Scholar]
- 6.Fröstl, J., and J. Overmann. 2000. Phylogenetic affiliation of the bacteria that constitute phototrophic consortia. Arch. Microbiol. 17450-58. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrmann, S., J. Overmann, N. Pfennig, and U. Fischer. 1993. Influence of vitamin B12 and light on the formation of chlorosomes in green- and brown-colored Chlorobium species. Arch. Microbiol. 160193-198. [Google Scholar]
- 8.Gasol, J. M., K. Jürgens, R. Massana, J. I. Calderón-Paz, and C. Pedrós-Alió. 1995. Mass development of Daphnia pulex in a sulphide-rich pond (Lake Cisó). Arch. Hydrobiol. 132279-296. [Google Scholar]
- 9.Glaeser, J., and J. Overmann. 2003. Characterization and in situ carbon metabolism of phototrophic consortia. Appl. Environ. Microbiol. 693739-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaeser, J., and J. Overmann. 2003. The significance of organic carbon compounds for in situ metabolism and chemotaxis of phototrophic consortia. Environ. Microbiol. 51053-1063. [DOI] [PubMed] [Google Scholar]
- 11.Glaeser, J., and J. Overmann. 2004. Biogeography, evolution, and diversity of epibionts in phototrophic consortia. Appl. Environ. Microbiol. 704821-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Götz, D., A. Banta, T. J. Beveridge, A. I. Rushdi, B. R. T. Simoneit, and A.-L. Reysenbach. 2002. Persephonella marina gen. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 521349-1359. [DOI] [PubMed] [Google Scholar]
- 13.Graham, L. L., and T. J. Beveridge. 1990. Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J. Bacteriol. 1722141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 41763-67. [DOI] [PubMed] [Google Scholar]
- 15.Kanzler, B. E. M., K. R. Pfannes, K. Vogl, and J. Overmann. 2005. Molecular characterization of the nonphotosynthetic partner bacterium in the consortium “Chlorochromatium aggregatum.” Appl. Environ. Microbiol. 717434-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefman, J., P. Zhang, T. Hirai, R. M. Weis, J. Juliani, D. Bliss, M. Kessel, E. Bos, P. J. Peters, and S. Subramaniam. 2004. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 1865052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manske, A. K., J. Glaeser, M. M. M. Kuypers, and J. Overmann. 2005. Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at 100 m depth in the Black Sea. Appl. Environ. Microbiol. 718049-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2641-646. [DOI] [PubMed] [Google Scholar]
- 19.Overmann, J. 2001. Green sulfur bacteria, p. 601-605. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, NY.
- 20.Overmann, J. 2001. Phototrophic consortia. A tight cooperation between non-related eubacteria, p. 239-255. In J. Seckbach (ed.), Symbiosis. mechanisms and model systems. Kluwer, Dordrecht, The Netherlands.
- 21.Overmann, J., H. Cypionka, and N. Pfennig. 1992. An extremely low-light-adapted phototrophic sulfur bacterium from the Black Sea. Limnol. Oceanogr. 37150-155. [Google Scholar]
- 22.Overmann, J., and N. Pfennig. 1989. Pelodictyon phaeoclathratiforme sp. nov., a new brown-colored member of the Chlorobiaceae forming net-like colonies. Arch. Microbiol. 152401-406. [Google Scholar]
- 23.Overmann, J., and K. Schubert. 2002. Phototrophic consortia: model systems for symbiotic interrelations between prokaryotes. Arch. Microbiol. 177201-208. [DOI] [PubMed] [Google Scholar]
- 24.Overmann, J., C. Tuschak, J. Fröstl, and H. Sass. 1998. The ecological niche of the consortium “Pelochromatium roseum.” Arch. Microbiol. 169120-128. [DOI] [PubMed] [Google Scholar]
- 25.Petroni, G., S. Spring, K.-H. Schleifer, F. Verni, and G. Rosati. 2000. Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc. Natl. Acad. Sci. USA 971813-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfannes, K. 2007. Characterization of the symbiotic bacterial partners in phototrophic consortia. Ph.D. dissertation. Universität München, Munich, Germany.
- 27.Pfannes, K. R., K. Vogl, and J. Overmann. 2007. Heterotrophic symbionts of phototrophic consortia: members of a novel diverse cluster of Betaproteobacteria characterised by a tandem rrn operon structure. Environ. Microbiol. 92782-2794. [DOI] [PubMed] [Google Scholar]
- 28.Rachel, R., I. Wyschkony, S. Riehl, and H. Huber. 2002. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 19-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai, A. N., E. Söderbäck, and B. Bergman. 2000. Cyanobacterium-plant symbiosis. New Phytol. 147449-481. [DOI] [PubMed] [Google Scholar]
- 30.Reinhold-Hurek, B., and T. Hurek. 1998. Life in grasses: diazotrophic endophytes. Trends Microbiol. 6139-144. [DOI] [PubMed] [Google Scholar]
- 31.Rieger, G., K. Müller, R. Hermann, K. O. Stetter, and R. Rachel. 1997. Cultivation of hyperthermophilic archaea in capillary tubes resulting in improved preservation of fine structures. Arch. Microbiol. 168373-379. [Google Scholar]
- 32.Silva, K. T., F. Abreu, F. P. Almeida, C. Neumann Keim, M. Farina, and U. Lins. 2007. Flagellar apparatus of south-seeking many-celled magnetotactic prokaryotes. Microsc. Res. Tech. 7010-17. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, C. D., and C. O. Wirsen. 1997. Microbiology and ecology of filamentous sulfur formation. Science 2771483-1485. [Google Scholar]
- 34.Vogl, K., J. Glaeser, K. R. Pfannes, G. Wanner, and J. Overmann. 2006. Chlorobium chlorochromatii sp. nov., a symbiotic green sulfur bacterium isolated from the phototrophic consortium “Chlorochromatium aggregatum.” Arch. Microbiol. 185363-372. [DOI] [PubMed] [Google Scholar]
- 35.Walther, P., and A. Ziegler. 2002. Freeze-substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 2083-10. [DOI] [PubMed] [Google Scholar]
- 36.Weis, R. M., T. Hirai, A. Chalah, M. Kessel, P. J. Peters, and S. Subramaniam. 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 1853636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, P., E. Bos, J. Heymann, H. Gnaegi, M. Kessel, P. J. Peters, and S. Subramaniam. 2004. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J. Microsc. 21676-83. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, P., C. M. Khursigara, L. M. Hartnell, and S. Subramaniam. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 1043777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.