Abstract
Fluorescence microscopic observation of individual T4 DNA molecules revealed that the MukBEF complex (bacterial condensin) and its subunit, the MukB (a member of the SMC [structural maintenance of chromosomes] superfamily) homodimer, of Escherichia coli markedly shrunk large DNA molecules in the presence of hydrolyzable ATP. In contrast, in the presence of ADP or ATP-γS, the conformation of DNA was almost not changed. This suggests that the ATPase activity of subunit MukB is essential for shrinking large DNA molecules. Stretching experiments on the shrunken DNA molecules in the presence of ATP and MukBEF indicated a cross-bridging interaction between DNA molecules.
The conformational transitions of DNA molecules have been intensively studied to understand the role of DNA compaction in various biological processes (2, 6, 23). Long DNA molecules are particularly interesting because their compaction is accompanied by an extraordinary increase in DNA molecular density on the order of 10,000-fold, and the resulting condensates can be organized into characteristic structures such as the toroid and rod. In vivo, DNA compaction is achieved by complexation with cationic proteins, while in vitro it can be realized by the addition of a variety of condensation agents, such as multivalent cations, cationic surfactants, neutral hydrophobic polymers, etc. (24).
With regard to DNA compaction per se, regulation of the manner of DNA condensation—for example, the segregation between the tightly packed and swollen parts along a single genomic DNA—is considered to play an essential role in the selective activation/inactivation of desired genes.
The MukB protein of Escherichia coli is known to be a structural and functional analogue of the members of the ubiquitous SMC (structural maintenance of chromosomes) family of proteins, which are responsible for maintaining the higher-order structure of DNA in chromosomes (8, 12, 17, 19). MukB is the first bacterial SMC-like protein identified by Niki and coworkers in 1991 (13). It, along with associated proteins, forms a coiled-coiled dimer, as shown in Fig. 1A. It has been shown that MukB indeed binds and condenses DNA but is not stimulated by ATP in these reactions (12, 14). In the MukB protein, the N-terminal globular domain has a nucleotide binding consensus sequence (14, 21, 22), and the C-terminal domain participates in DNA binding activity. It has also been shown that MukB binds to DNA even in the absence of ATP (14). While the physical origin of the driving force of the MukB protein function is unclear, a large number of reports have described the ATP-dependent activities of SMC proteins (3-5), including ATP hydrolysis, as causing a supercoiling reaction (7).
FIG. 1.

Properties of MukB and MukBEF. (A) Schematic representation of the structure of MukB and MukBEF. (B) Electrophoresis of the purified MukBEF complex in a sodium dodecyl sulfate-polyacrylamide gel. (C) ATPase activity of purified MukB and MukBEF. ss, single stranded; ds, double stranded.
The MukBEF complex is constructed through the binding of both MukF (51 kDa) and MukE (27 kDa) proteins to the globular domains of MukB (170 kDa) (22). MukE, MukF, and the MukEF complex do not demonstrate DNA binding activity (22), but the MukBEF complex, thus formed, participates in the organization of sister chromosomes and in partitioning into both daughter cells in Escherichia coli (10, 20-22). While the MukB homodimer has a long, rod-hinge-rod, V-shaped structure with small globular domains at both ends, the MukBEF complex has a similar structure but larger globular domains than those of the MukB homodimer (10). A mutant with a deletion of the mukB gene exhibits hypersensitivity to novobiocin, which inhibits DNA gyrase, suggesting that MukB relates to superhelicity or the three-dimensional structure of chromosomal DNA (1, 18, 19). Understanding how MukBEF operates may answer decades-old questions on how chromosomes are organized on a global scale. A major point of difficulty is that MukBEF, like other condensins, seems to be optimized to act on the chromosome-size DNA. Traditional biochemical assays, however, have utilized much shorter DNA, and ATPase-dependent compaction of DNA has not been found so far. Therefore, it is important to develop new in vitro techniques for chromosome-size, large DNA to understand how MukBEF organizes chromosomal DNA.
In the present study, we performed single-molecule observations on the conformational behavior of Yoyo-labeled T4 phage DNA (ca. 166,000 base pairs; 57 μm) in the presence of MukB protein and the MukBEF protein complex with or without ATP by fluorescence microscopy. It has been found that both MukB and MukBEF promote marked shrinkage of large DNA depending on the presence of ATP. However, no apparent shrinkage was observed in the presence of ADP or ATP-γS, which are not hydrolyzed by ATPase. This indicates that the shrinkage of large DNA depends on the ATPase function of MukB.
MATERIALS AND METHODS
Materials.
Bacteriophage T4 DNA (ca. 166,000 base pairs; Nippon Gene Co., Ltd., Japan) and the fluorescent dye Yoyo (Molecular Probes, Inc., OR) were used as received from the manufacturer.
Purification of the MukB protein.
MukB protein was purified as described previously (22).
Purification of the MukBEF complex.
The MukBEF complex was purified as follows. Strain YM1078, which harbors plasmid pMY177 carrying the mukBEF operon, was grown in L medium containing ampicillin (50 μg/ml) and treated with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h at 37°C to overproduce the MukBEF complex. Cells were collected by centrifugation and washed with 25 mM HEPES-KOH (pH 7.6). Cells suspended in buffer Y (see Table 1) (14) were treated with 10 mg/ml of lysozyme for 1 h on ice, and the cells were lysed by repeated freeze-thaw cycles. The cell lysate was centrifuged at 157,000 × g for 40 min at 4°C. The supernatant (fraction I) was collected, treated with polyethyleneimine, and centrifuged. The recovered supernatant (fraction II) was precipitated with ammonium sulfate (50% saturation) and centrifuged. The pellet was suspended in buffer Y overnight at 4°C. The sample was centrifuged, and the supernatant (fraction IV) was recovered. Fraction IV was purified sequentially by Sephacryl S500HR column chromatography, Fractogel DEAE column chromatography, HiTrap desalting column chromatography, Fractogel DEAE column chromatography (for a second time), and sucrose gradient sedimentation (10 to 40% sucrose gradient). The purified sample was dialyzed against buffer Y and concentrated with a Centricon 30 device.
TABLE 1.
Buffer compositions
| Buffer | Composition |
|---|---|
| A (−ATP) | 20 mM Tris-HCl (pH 7.5), 7.5 mM KCl, 3.5 mM MgCl2 |
| B (+ATP) | 20 mM Tris-HCl (pH 7.5), 7.5 mM KCl, 2.5 mM MgCl2, 1 mM Mg-ATP |
| C (+ATP-γS) | 20 mM Tris-HCl (pH 7.5), 7.5 mM KCl, 3.5 mM MgCl2, 1 mM Li4-ATP-γS |
| D (+ADP) | 20 mM Tris-HCl (pH 7.5), 7.5 mM KCl, 3.5 mM MgCl2, 1 mM ADP |
| Y | 25 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, 2 mM dithiothreitol, 25 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol |
ATPase assay.
An ATPase assay was performed as described by Hirano and Hirano (3) with minor modifications. In brief, MukB or MukBEF (420 nM) was mixed in buffer B (see Table 1) and [α-32P]ATP (100 nCi). The total volume of the reaction mixtures was 10 μl. After incubation at 37°C for 60 min, sodium dodecyl sulfate was added to stop the reaction. One microliter of the sample was spotted on a polyethyleneimine-cellulose thin-layer chromatography plate (Merck KGaA, Darmstadt, Germany). The hydrolyzed product, [α-32P]ADP, was analyzed by autoradiography. When necessary, 31.2 μM nucleotides of double-stranded or single-stranded T4 DNA was added to the reaction mixtures.
Sample preparation.
For the fluorescence microscopic observations, a solution containing 0.02 μM Yoyo (a DNA fluorescent dye) and 0.1 μM nucleotides of phage T4 DNA was used in four different buffer solutions, as listed in Table 1. After 30 min, 0.5 ml of a solution of 11.3 μg/ml MukBEF or 6.0 μg/ml MukB (diluted 100-fold from the original solution and incubated at 37°C for 5 min) was added to the solution, and the mixture was gently stirred and incubated at 37°C for 60 min before the observations.
Fluorescence microscopy of DNA in solution.
Yoyo-labeled T4 DNA molecules in the solutions were observed with a fluorescence microscope, Axiovert 135 TV (Carl Zeiss, Germany), equipped with a 100×, oil-immersed lens at room temperature. Fluorescent images of Yoyo-labeled T4 DNA were recorded using an electron bombardment charge-coupled-device camera and an Argus 10 image processor (Hamamatsu Photonics, Japan). The apparent long-axis length of the DNA, which is defined as the longest distance in the outline of the fluorescent image, was evaluated by measuring about 100 DNA molecules. Fluorescence microscopy observations were used to confirm that no breakage of DNA molecules occurred during sample preparation, i.e., that no short DNA fragments were detected together with the main population of DNA molecules.
Fluorescence microscopy of DNA molecules fixed onto microscope glass slides.
Yoyo-labeled T4 DNA molecules were fixed onto microscope glass slides as follows. Microscope glass slides were pretreated with a poly-l-lysine solution (0.1% vol/vol), washed repeatedly with distilled water, and dried in room air. Five microliters of the sample was placed on a cationically modified glass slide (30 by 40 mm) to fix the DNA molecules and was covered with a slip glass (22 by 22 mm) by sliding it on gently.
RESULTS
ATPase activity of purified MukB and MukBEF.
Electrophoresis of the purified MukBEF complex is shown in Fig. 1B. Purified MukB was previously described (22). The purified MukB and MukBEF samples had >95% purity. These samples' molecular architectures were previously analyzed by electron microscopy (10). We analyzed the ATPase activity of these samples under the buffer condition that was used in the study of the ATPase activity of SMC protein in Bacillus subtilis (3). This reaction condition resulted in a much higher ATPase activity of MukB than in previous reports (9, 15). As shown in Fig. 1C, considerable ATPase activity was detected in both samples. The ATPase activity of the MukBEF complex was approximately twofold higher than that of MukB. No remarkable difference was observed in the presence or absence of double-stranded or single-stranded DNA.
Shrinkage of T4 DNA molecules by MukB and MukBEF in solution.
To monitor the conformational state of long T4 DNA molecules in a bulk solution upon the addition of proteins, we performed single-molecule observations of Yoyo-labeled T4 DNA molecules (0.1 μM in bp) at different concentrations of proteins with a fluorescence microscope connected to a video camera. Four buffers of the following different compositions were used for the observations: 20 mM Tris-HCl (pH 7.5), 7.5 mM KCl, 2.5 or 3.5 mM MgCl2 (for buffer B or buffers A, C, and D, respectively), with the addition of Mg-ATP (buffer B), ATP-γS (buffer C), or ADP (buffer D) (Table 1).
In the buffer solutions, single DNA molecules moved freely with a characteristic long-axis length of about 4 μm (data not shown). With the addition of MukBEF proteins, the conformational behavior of DNA depended on the buffer used for the experiment. Figure 2 shows time-dependent fluorescent images of a single DNA molecule interacting with MukBEF in buffer A and buffer B, or in other words, in the presence and in the absence of ATP, respectively. In both cases, DNA preserved the conformation of coils characterized by a large repulsion of molecular segments. The addition of MukBEF to the DNA in buffer A (without ATP) (Fig. 2A) had no effect on the DNA conformation or DNA long-axis length. In contrast, fluorescence microscopic observations in the ATP-containing buffer B (Fig. 2B) showed a marked decrease in the apparent size of the DNA coils. A comparison of Fig. 2A and Fig. 2B reveals that the time-averaged DNA long-axis length decreases when buffer with ATP is used. This shrinking of DNA is also accompanied by the generation of a local bright spot(s) on the fluorescent image in the fluctuating DNA coils, suggesting partial compaction of DNA molecules. In the case of MukB protein being added to the DNA in the two buffers, no significant changes in DNA long-axis length were observed in either buffer A or buffer B solutions (data not shown).
FIG. 2.
Fluorescence microscopy images and the corresponding quasi-three-dimensional profiles of single T4 DNA molecules. Brownian motion after complexation with the MukBEF protein (2.26 μg/ml) in buffer A (A) and in buffer B (B).
Figure 3 shows histograms of the distributions of DNA based on long-axis length after the addition of MukB (Fig. 3A) and of MukBEF (Fig. 3B) in both the ATP-free buffer A and the ATP-containing buffer B. As shown in Fig. 2, DNA interaction with MukBEF leads to a significant shrinkage of DNA coils only in buffer containing ATP. Figure 3 indicates that the degree of DNA coil shrinkage induced by ATP is about 1 μm, which corresponds to the approximately 25% decrease in the DNA long-axis length. In contrast, the same concentration of MukB protein added to DNA induced less-profound changes in DNA long-axis length even when the ATP-containing buffer B was used (Fig. 3A). Therefore, the most pronounced shrinking effect on the T4 DNA conformation is generated by the addition of the MukBEF complex in the presence of ATP.
FIG. 3.
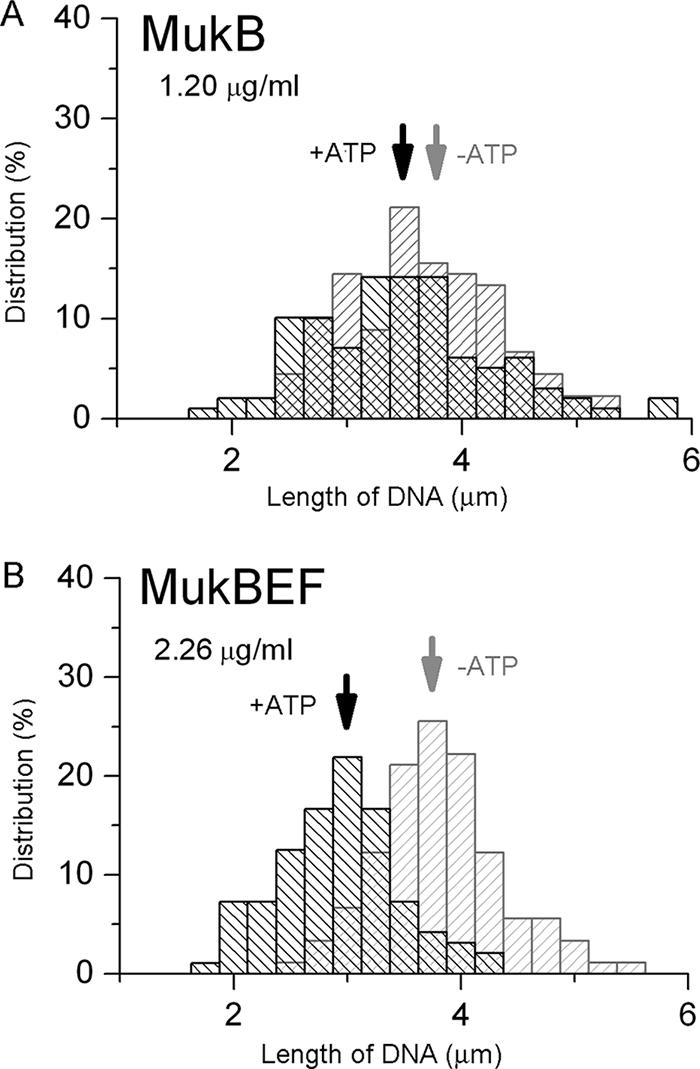
Distributions of T4 DNA based on long-axis length with the MukB homodimer (A) and the MukBEF complex (B) in ATP-free and ATP-containing solutions. Arrows indicate the mean values of the corresponding distributions based on DNA long-axis length.
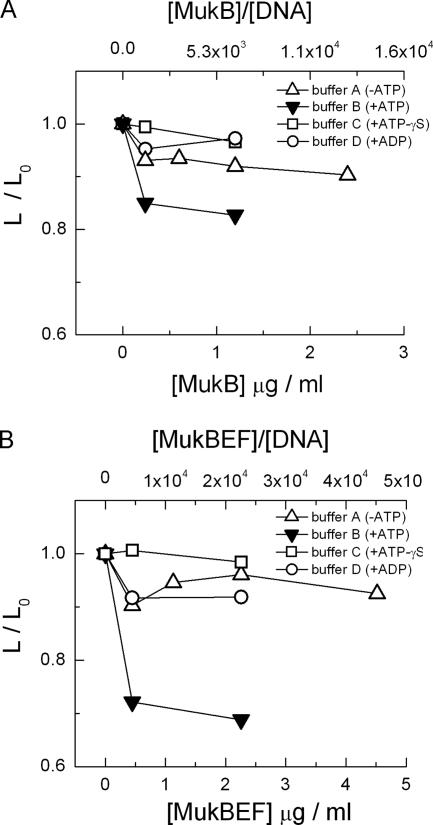
The calculated average DNA long-axis lengths at different concentrations of proteins are summarized in Fig. 4A and B. Although the conformational state of DNA did not change, the average size of DNA coils slightly decreased upon the addition of both types of protein in buffer A (without ATP). In contrast, the addition of the proteins to buffer B, containing ATP (Fig. 4A and B), led to a significant decrease in the size of T4 DNA. By comparing the activities of MukB and MukBEF, it becomes clear that MukBEF is more effective than MukB in providing a higher degree of DNA shrinkage. These results of single-molecule-DNA observations in buffer A and buffer B demonstrated that the MukB and MukBEF proteins induced strong DNA shrinkage and the formation of compact DNA regions on DNA coils, which are observed as bright spots.
FIG. 4.
Changes in DNA conformation (as a ratio of DNA long-axis length [L] at different concentrations of proteins to the original long-axis length [L0]) with increases in MukB (A) and MukBEF (B) depending on the concentration of proteins, where the ratios of protein to DNA molecules are also given. The L0 values are 3.9, 4.0, 3.6, and 3.8 μm for buffers A, B, C, and D, respectively.
Next, we performed additional experiments with buffer C, which contained ATP-γS (a nonhydrolyzable analogue of ATP) instead of ATP. The addition of the MukB and MukBEF proteins did not produce a strong decrease in the size of the DNA coils in the presence of ATP-γS (Fig. 4A and B). Finally, we performed experiments with buffer D, which contains ADP (a hydrolyzed form of ATP) instead of ATP. The results of these experiments are shown in Fig. 4A and B. In the case of the ADP-containing buffer D, strong DNA shrinkage by MukB and MukBEF did not occur, and only a very slight decrease in DNA size was seen, similar to the results with buffer A.
As indicated in Fig. 4A and B, it was clear that the two proteins have similar shrinkage effects on DNA. An effective decrease in the length of T4 DNA coils was observed only with buffer B solution, containing ATP; the length of T4 DNA coils decreased by 20% and 30% with the addition of MukB and MukBEF, respectively. Buffer A and buffer D showed almost equal, slight decreases in the size of the DNA coils with the addition of the two proteins. Buffer C, which contained ATP-γS, was the least effective buffer for DNA shrinkage.
Compaction of T4 DNA molecules by MukB and MukBEF in stretching experiments.
To gain deeper insight into the intramolecular structures along the T4 DNA chain, we performed a stretching experiment with Yoyo-labeled T4 DNA molecules on a glass surface by adapting a shear flow technique described in Materials and Methods which has been reported in detail previously (11). The adapted flow caused the extension of the T4 DNA molecules to a length of approximately 57 μm in the absence of protein (data not shown). Typical fluorescent images of stretched T4 DNAs after interaction with the MukBEF complex in buffer B are shown in Fig. 5. We frequently observed structures that seemed to be two DNA molecules forming a cross or a branch in the presence of ATP with a frequency of approximately 45% (52/115 structures) of crossed and branched molecules (Fig. 5A). In contrast, after interaction with MukBEF in the absence of ATP, crossed or branched molecules constituted only a minor fraction, with a frequency of approximately 4% (5/136 structures). A control experiment without protein but in the presence of ATP revealed no formation of the above-mentioned crossed or branched structures. Other extended structures observed were thicker, fiber-like, condensed DNA structures and globular structures on an extended thin DNA filament, typical examples of which are illustrated in Fig. 5B to D. “Crosses,” “thick fibers,” or globular parts were not observed in the absence of protein.
FIG. 5.

Typical examples of fluorescent images of T4 DNA after interaction with 2.26 μg/ml MukBEF in buffer B and stretching of a droplet of sample solution onto a cationically modified glass surface.
DISCUSSION
We found that MukB and MukBEF induce conformational changes in large DNA molecules of the T4 bacteriophage by decreasing the length of the DNA coils. At neutral pHs, the MukB and MukBEF proteins are negatively charged (e.g., MukB has a charge of −51 at pH 7) (19). Thus, DNA compaction cannot be explained by an overall electrostatic charge neutralization effect, but rather, it involves the interaction of DNA with positively charged patches on the proteins (19). Our observations demonstrate that hydrolyzable ATP plays a critical role in DNA compaction by the MukB and MukBEF proteins; i.e., effective DNA compaction was observed only in buffer B, which contained ATP. The compaction of DNA was not observed in the presence of ATP-γS, which is not hydrolyzed by ATPase. This indicates that the ATPase activity of MukB is important for the compaction of large T4 DNA. On the other hand, Petrushenko et al. (15) reported that MukB introduces right-handed knots into circular plasmid DNA in the presence of type 2 topoisomerase, indicating that the protein promotes intramolecular DNA compaction. Furthermore, the formation of knots in the plasmid DNA by MukB did not require ATP. This discrepancy between their results and our present results is probably due to the different functions of MukB in these assays. Our results may suggest a new ATP-dependent function of MukB and MukBEF in the condensation of large DNA molecules. The present results are consistent with the previous observations on the DNA binding activity of the Xenopus laevis condensin I complex reported by Strick et al. (16): the condensin can bind to nanomanipulated DNA in the absence of ATP, but it compacts the DNA only in the presence of hydrolyzable ATP.
Our observations confirm that the use of the MukBEF complex results in a significant enhancement of DNA compaction activity in comparison to that resulting from the use of the MukB protein. The linear size of DNA coils decreased by about 20% with MukB and by about 30% with MukBEF in the presence of ATP, which corresponds to approximately 50% and 70% increases, respectively, in average DNA molecular density in a coil state. Therefore, DNA compaction by MukBEF itself is not sufficient to account for the dramatic compaction, by several orders of magnitude, of bacterial chromosomes in cells. The amplification of DNA compaction found in our experiments might have been caused by further protein-dependent interactions between regions of the chromosomes.
It should be emphasized that the MukB homodimer itself induces partial DNA compaction in the presence of ATP. The ATPase activity of MukBEF was twofold higher than that of MukB in buffer B (Fig. 4), and thus, this difference in DNA compaction may correspond to a difference in the ATPase activity.
The samples of MukB and MukBEF used here had >95% purity. Acyl carrier protein was previously purified together with MukB (14, 15). However, acyl carrier protein does not have ATPase activity. Therefore, the ATPase activity observed in the MukB and MukBEF samples (Fig. 1C) might be due to the MukB subunit itself in the samples. The ATPase activity under the reaction conditions was much higher than that under previously described reaction conditions (14, 15).
As shown in Fig. 4B, the ATP-dependent compaction of T4 DNA by MukBEF was nearly saturated by 0.45 μg/ml of MukBEF. This corresponds to 2,200 molecules of MukBEF added for each T4 DNA molecule (57 μm), i.e., one MukBEF molecule per 25 nm (length) of DNA (75 bp). Similarly, the ATP-dependent compaction of T4 DNA by the MukB homodimer was also saturated by 0.25 μg/ml of MukB, which corresponds to about 2,500 molecules of MukB homodimer for each T4 DNA molecule (57 μm), i.e., one MukB molecule per 22 nm (length) of double-stranded DNA (66 bp).
The length of a stretched MukB homodimer is approximately 100 nm (10). Both globular domains at both ends of MukB and MukBEF have DNA binding activity (22). MukBEF and MukB can bind to small DNA molecules in the absence of ATP (14, 15, 22). ATP is not required for the knot formation of circular small DNA molecules (15). Furthermore, electron microscopy and gel filtration chromatography revealed previously that the purified MukBEF complex frequently forms large multimers of a chain or “rosette” form by binding a globular domain to a globular domain of another molecule in the absence of DNA and ATP (10). Thus, ATP is not required for intermolecular binding of MukBEF to form chain-like MukBEF multimers. Although the role of ATPase of MukBEF or MukB remains a riddle, we propose here a possible hypothesis based on the results described above. The ATPase activity may accelerate the intramolecular binding and release of both globular domains of MukBEF (Fig. 6A). The globular domains of MukBEF may intramolecularly associate with each other in the presence of ATP, resulting in close linking of two DNA regions that are more than 100 nm away from each other, i.e., the intramolecular compaction or intermolecular bridging of two DNA molecules (Fig. 6B and C). The ATP-dependent intramolecular compaction of DNA can be detected by using large DNA molecules such as T4 DNA but not, presumably, by using small DNA segments. Thus, the techniques used in the present work are important for understanding how bacterial chromosomal DNA is condensed by MukBEF in vivo.
FIG. 6.

Hypothesis on the role of ATPase activity in MukBEF complex formation. (A) ATPase activity may accelerate intramolecular binding and release of both globular domains of MukBEF. (B) ATP-dependent intramolecular compaction of two DNA molecules. (C) ATP-dependent intermolecular bridging of two DNA molecules. (D) Formation of thick, fiber-like structures of small regions on long T4 DNA. (E) Formation of globular structures of small regions on long T4 DNA.
Stretching experiments revealed that MukBEF induced the formation of “cross” and “branch” structures, thick fiber-like structures, and globular structures of T4 DNA molecules in the presence or absence of ATP (Fig. 5). These thick fiber-like structures and globular structures might correspond to the bright fluorescent spots in DNA coils that were observed in the presence of both MukBEF and ATP in bulk solution experiments (Fig. 2B). The thick, fiber-like structures and the globular structures in small regions on T4 DNA were presumably formed through the arrangement of MukBEF molecules as depicted in Fig. 6D and E. These structures are the structural basis of the formation of a “rosette” structure on MukBEF multimers as observed by electron microscopy (10). The intermolecular bridging of two DNA molecules in the presence of MukBEF was frequently observed in the stretching experiments but only in the presence of ATP (Fig. 5A to E), which indicates the importance of ATP, not only for DNA shrinking activity of MukBEF but also for realization of the cross-linking mechanism of the DNA-protein interaction. The formation of the cross regions by MukBEF might correspond to the development of complexes in which MukBEF binds two DNA molecules together through the formation of an ATP-dependent, intermolecular bridging ring that might trap the DNA molecules inside (Fig. 6C).
The MukBEF complex forms foci in ordered positions in cells (14a), and it participates in the organization of sister chromosomes. The present results in vitro correspond to the formation of foci by the MukBEF complex and the chromosomal organization in vivo. Our models in Fig. 6 show the possible mechanisms of the focus formation and the organization of chromosomal DNA in vivo.
Acknowledgments
S.A. is supported by a fellowship and grant from the Japan Society for the Promotion of Science for Young Scientists. S.H. is financially supported by the Center of Excellence of Kyoto University and by grants from the Ministry of Education, Science, Sports, Culture and Technology of Japan.
The experiments using radioisotopes were performed at the Radioisotope Research Center of Kyoto University.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Adachi, S., and S. Hiraga. 2003. Mutants suppressing novobiocin hypersensitivity of a mukB null mutation. J. Bacteriol. 1853690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloomfield, V. A. 1996. DNA condensation. Curr. Opin. Struct. Biol. 6334-341. [DOI] [PubMed] [Google Scholar]
- 3.Hirano, M., and T. Hirano. 1998. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 177139-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirano, M., and T. Hirano. 2004. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 232664-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopfner, K.-P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101789-800. [DOI] [PubMed] [Google Scholar]
- 6.Hud, N. V., and I. D. Vilfan. 2005. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu. Rev. Biophys. Biomol. Struct. 34295-318. [DOI] [PubMed] [Google Scholar]
- 7.Kimura, K., and T. Hirano. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90625-634. [DOI] [PubMed] [Google Scholar]
- 8.Koshland, D., and A. Strunnikov. 1996. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 12305-333. [DOI] [PubMed] [Google Scholar]
- 9.Lockhart, A., and J. Kendrick-Jones. 1998. Interaction of the N-terminal domain of MukB with the bacterial tubulin homologue FtsZ. FEBS Lett. 430278-282. [DOI] [PubMed] [Google Scholar]
- 10.Matoba, K., M. Yamazoe, K. Mayanagi, K. Morikawa, and S. Hiraga. 2005. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem. Biophys. Res. Commun. 333694-702. [DOI] [PubMed] [Google Scholar]
- 11.Matsuzawa, Y., and K. Yoshikawa. 1994. Change of the higher order structure in a giant DNA induced by 4′, 6-diamidino-2-phenylindole as a minor groove binder and ethidium bromide as an intercalator. Nucleosides Nucleotides 131415-1423. [Google Scholar]
- 12.Melby, T. E., C. N. Ciampaglio, G. Briscore, and H. P. Erickson. 1998. The symmetrical structure maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell. Biol. 1421595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niki, H., A. Jaffé, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niki, H., R. Imamura, M. Kitaoka, K. Yamanaka, T. Ogura, and S. Hiraga. 1992. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 115101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Ohsumi, K., M. Yamazoe, and S. Hiraga. 2001. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40835-845. [DOI] [PubMed] [Google Scholar]
- 15.Petrushenko, Z. M., C.-H. Lai, R. Rai, and V. V. Rybenkov. 2006. DNA reshaping by MukB: right-hand knotting, left-hand supercoiling. J. Biol. Chem. 2814606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strick, T. R., T. Kawaguchi, and T. Hirano. 2004. Real-time detection of single-molecule DNA compaction by condensin I. Curr. Biol. 14874-880. [DOI] [PubMed] [Google Scholar]
- 17.van den Ent, F., A. Lockhart, J. Kendrick-Jones, and J. Löwe. 1999. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Structure 71181-1187. [DOI] [PubMed] [Google Scholar]
- 18.Weitao, T., K. Nordstrom, and S. Dasgupta. 1999. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 34157-168. [DOI] [PubMed] [Google Scholar]
- 19.Weitao, T., K. Nordstrom, and S. Dasgupta. 2000. Escherichia coli cell cycle control genes affect chromosome superhelicity. EMBO Rep. 1494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka, K., T. Mitani, J. Feng, T. Ogura, H. Niki, and S. Hiraga. 1994. Two mutant alleles of mukB, a gene essential for chromosome partition in Escherichia coli. FEMS Microbiol. Lett. 12327-32. [DOI] [PubMed] [Google Scholar]
- 21.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250241-251. [DOI] [PubMed] [Google Scholar]
- 22.Yamazoe, M., T. Onogi, Y. Sunako, H. Niki, K. Yamanaka, T. Ichimura, and S. Hiraga. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 185873-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshikawa, K., and Y. Yoshikawa. 2002. Compaction and condensation of DNA, p. 137-163. In R. I. Mahato and S. W. Kim (ed.), Pharmaceutical perspectives of nucleic acid-based therapeutics. Taylor and Francis Inc., New York, NY.
- 24.Zinchenko, A. A., D. Baigl, and K. Yoshikawa. 2007. Nanostructures and organization of compacted single chains of polyelectrolytes, p. 402-442. In H. S. Nalwa (ed.), Polymeric nanostructures and their applications. American Scientific Publishers, Stevenson Ranch, CA.