Abstract
Agrobacterium tumefaciens strain C58 can transform plant cells to produce and secrete the sugar-phosphate conjugate opines agrocinopines A and B. The bacterium then moves in response to the opines and utilizes them as exclusive sources of carbon, energy, and phosphate via the functions encoded by the acc operon. These privileged opine-involved activities contribute to the formation of agrobacterial niches in the environment. We found that the expression of the acc operon is induced by agrocinopines and also by limitation of phosphate. The main promoter is present in front of the first gene, accR, which codes for a repressor. This operon structure enables efficient repression when opine levels are low. The promoter contains two putative operators, one overlapping the −10 sequence and the other in the further upstream from it; two partly overlapped putative pho boxes between the two operators; and two consecutive transcription start sites. DNA fragments containing either of the operators bound purified repressor AccR in the absence of agrocinopines but not in the presence of the opines, demonstrating the on-off switch of the promoter. Induction of the acc operon can occur under low-phosphate conditions in the absence of agrocinopines and further increases when the opines also are present. Such opine-phosphate dual regulatory system of the operon may ensure maximum utilization of agrocinopines when available and thereby increase the chances of agrobacterial survival in the highly competitive environment with limited general food sources.
During agrobacterial infection of susceptible plants, a copy of the T region in a tumor-inducing plasmid (Ti plasmid), called the T-DNA, is transferred from the bacterium to the plant where it becomes integrated into the chromosome (10-12, 25). Expression of the genes on the integrated T-DNA results in the tumorous phenotype and also the production of unusual low-molecular-weight carbon compounds that belong to a large group of plant tumor-specific metabolites collectively called opines (15, 20, 58, 66).
Crown gall tumors induced by the classic nopaline-type Agrobacterium tumefaciens strain C58 synthesize and secrete two families of tumor metabolites: agrocinopines A and B and nopaline (20, 43). The genes responsible for the biosynthesis of agrocinopines and of nopaline are carried on the T region of pTiC58, harbored by the strain C58 (7, 14, 29). Strain C58 can catabolize nopaline and agrocinopines by using functions encoded by two different sets of genes located on the nontransferred region of the Ti plasmid (6, 27, 34, 76). These loci, called acc (agrocinopine catabolism) and noc (nopaline catabolism), code for the transport and catabolism of, as well as chemotaxis to, their cognate opines (34, 35, 51, 76).
The process by which A. tumefaciens transforms plant cells to produce opines has been termed genetic colonization of the plant (60). The genetic colonization theory leads to the opine concept, proposed by Tempé et al. (68), which states that the opines are the chemical mediators of the interaction between Agrobacterium spp. and its plant host. There are nonagrobacterial opine utilizers, including various pseudomonads and coryneforms, that can grow on common opines, such as octopine, indoleacetic acid, and succinamopine (49, 57, 69). However, these opines still comprise a small portion in the bacterial community in the rhizosphere and, further, the ability to utilize certain opines, including mannopine, is uncommon (49). Therefore, these compounds may serve as specific sources of nutrition to opine-utilizing agrobacteria, promoting their growth and dissemination.
Our previous studies revealed that acc is composed of eight genes: accR and accABCDEFG (34). accR codes for a repressor that regulates this locus, as well as the conjugative transfer of pTiC58 in response to the presence of agrocinopines A and B (3, 27, 34). The first five structural genes, accABCDE, code for an ABC-type transport system that takes up agrocinopines as well as a unique antibiotic called agrocin 84 (27, 34). Agrocin 84 is produced by A. radiobacter strain K84, which is antagonistic to A. tumefaciens strain C58 competing for agrocinopines that are produced by plants transformed by A. tumefaciens strain C58 (19, 36, 47, 48, 56, 64, 67). The last two genes, accF and accG, code for agrocinopine phosphodiesterase and arabinose phosphate phosphatase, respectively (34, 73). They are required to initiate catabolism of the opines in splitting them into an arabinose 2-phosphate and a fructose or a sucrose, depending on the type of the agrocinopine, and in further dephosphorylating the arabinose 2-phosphate. Interestingly, the product of accF also is required to activate agrocin 84, which most likely is not toxic to C58 in its native form (34). Strains with mutations in accF take up agrocin 84 but remain resistant to the antibiotic.
acc is expressed at a relatively high basal level in the absence of agrocinopines (28). In the presence of the opines, or when accR is mutated, the expression of acc is increased severalfold (3, 28). Analysis of lacZ fusions identified a second transcription unit located just 5′ to accR (34). This locus, which is oriented in the direction opposite to that of acc, contains the traR gene (55). traR codes for a transcriptional activator required for the expression of the tra and trb genes responsible for conjugative transfer of the Ti plasmid (54, 55). Expression of this set of genes also is regulated by AccR (50, 51, 55).
DNA sequence analyses revealed cis elements that might be involved in regulating acc (34). These include two related inverted repeat (IR) sequences located in the regions upstream of accR and accF. There also is a sequence similar to these IR sequences located upstream of the IR present in the 5′-untranslated region of accR. The intergenic region between accR and accA contains an unrelated IR (3). Finally, two overlapping putative pho boxes, related to those in Escherichia coli and to that associated with virG in the octopine type plasmid pTiA6 harbored by another A. tumefaciens strain (41, 42, 72), also were identified in the 5′-untranslated region of accR by Kim et al. and later by Yuan et al. (34, 75). This observation suggested that expression of acc is regulated by availability of phosphate, as well as by the opines (34).
In this report, we present genetic data showing that (i) the acc operon indeed is regulated by both agrocinopines and phosphate starvation, (ii) the main promoter is in the region upstream of accR, (iii) both of the IRs in the promoter are required for the full repression of the operon, and (iv) induction under low-phosphate conditions occurs in the absence of agrocinopines and further increases when the opines are also present. We also present other data showing that (i) two transcription start sites are present in the main promoter; (ii) two IRs in this promoter most likely are functional operators that bind repressor AccR in the absence of agrocinopines, but not when agrocinopines are present; and (iii) intact opines are the true inducers and that agrocin 84, which is another substrate of the acc operon, and l-arabinose and sucrose, the constituent sugars of agrocinopines, neither serve as inducers nor affect derepression of the operon by the opines.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used in the present study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype, phenotype, or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 59 |
| S17-1 | Pro− Res− Mod+recA; integrated RP4-Tcr::Mu-Kan::Tn7, Mob+; Smr | 63 |
| 2174(pPH1JI) | met pro Gmr Smr | 5 |
| Sm10::λpir | Derivative of C600 with Rp4-2(Tcr::Mu); Kmr, Thi, Thr, Leu, suIII, RecA | 32 |
| BL21(DE3)(pLysS) | F−ompThsdSB(rB− mB−) gal dcm (DE3) pLysS; Cmr | Novagen, Inc. |
| A. tumefaciens | ||
| C58 | Agrs Noc+ Acp+ (pTiC58) | Our collection |
| NT1 | pTiC58-cured C58 | 71 |
| NT1(pTiC58ΔaccR) | Agrs Noc+ Acp+ | 3 |
| NT1(pAgK84b-A1) | Agrr Agr production; Kmr | 22 |
| C58K476 | pTiC58 (accA::Tn3HoHo1 476); Cbr | 34 |
| C58KF | pTiC58 (accF::lacZY) | 34 |
| C58K11 | pTiC58 (accG::Tn3HoHo1 11); Cbr | 34 |
| C58KFΔR | pTiC58ΔaccR (accF::lacZY) | This study |
| C58K11ΔR | pTiC58ΔaccR (accG::Tn3HoHo1 11); Cbr | This study |
| T37 | Agrs Noc+ Acp+ (pTiT37) | Our collection |
| Plasmids | ||
| pLAFR6 | pLAFR1 with double terminators flanking polylinker; Tcr | 31 |
| pB153 | pLAFR6::part of BamHI fragment containing Tn3HoHo1 153; Tcr | This study |
| pBH153 | pLAFR6::HindIII fragment 31 containing Tn3HoHo1 153; Tcr | This study |
| pB476 | pLAFR6::part of BamHI fragment containing Tn3HoHo1 476; Tcr | This study |
| pBH476 | pLAFR6::HindIII fragment 31 containing Tn3HoHo1 153; Tcr | This study |
| pBHS476 | SalI deletion derivative of pBH476; Tcr | This study |
| pLFZY | pLAFR6:PstI-HindIII fragment containing lacZY; Tcr | This study |
| pTHB476 | pTHB112::Tn3HoHo1 476; Tcr Cbr | 28 |
| pHK11 | pTHB112::Tn3HoHo1 11; Tcr Cbr | 34 |
| pHK115 | pTHB112::Tn3HoHo1 115; Tcr Cbr | 34 |
| pHK153 | pTHB112::Tn3HoHo1 153; Tcr Cbr | 34 |
| pHKF | pTHB112(accF::lacZY); Tcr | 34 |
| pTHH206 | pSa4 containing HindIII partial of pTiC58; Cmr Kmr | 28 |
| pTHH206F | pTHH206 (accF::lacZY); Cmr Kmr | This study |
| pKFG | pKNG101::HindIII 11ΔSmaI::lacZ; Smr | 34 |
| pRK415 | Inc P1α cloning vector; Tcr Kmr | 33 |
| pRKW26 | pRK415K::EcoRI 26 from pTiC58; Kmr, accR | 3 |
| pET29b | ATG translational fusion vector for adding C-terminal His tag; Kmr | Novagen, Inc. |
| pEXPR | pET29b::accR; Kmr, His-tagged accR | This study |
| pTESTR | Cointegrate of pRK415 and pEXPR; Tcr Kmr, His-tagged accR | This study |
Abbreviations: Ampr, ampicillin resistance; Cbr, carbenicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance; Agrs, agrocin 84 sensitivity; Agrr, agrocin 84 resistance; Acp+, agrocinopine utilization; Noc+, nopaline utilization.
Media.
Strains of A. tumefaciens were grown in Luria-Bertani (LB) medium (Gibco-BRL, Gaithersburg, MD), nutrient agar (Difco Laboratories, Detroit, MI), or AB minimal (ABM) medium (9). Unless otherwise specified, mannitol at a final concentration of 0.25% was used as a carbon source in ABM medium. For studies of cells grown in low phosphate, ABM medium containing 0.1 mM phosphate and supplemented with 40 mM 4-morpholine-ethanesulfonic acid (pH 7.0) was used (72). E. coli strains were cultured in LB medium supplemented, when necessary, with antibiotics at the following concentrations: tetracycline, 10 μg/ml; ampicillin, 100 μg/ml; and kanamycin, 50 μg/ml. For A. tumefaciens, antibiotics were used at the following concentrations: kanamycin, 50 or 100 μg/ml; tetracycline, 2 μg/ml; and carbenicillin, 100 or 200 μg/ml.
Preparation of agrocinopines.
A mixture of agrocinopines A and B was purified from tomato crown galls induced by A. tumefaciens strain T37 as described by Ryder et al. (58). The agrocinopine opines were quantified by the phloroglucinol method (16) with arabinose (Sigma Chemical Co., St. Louis, MO) as the standard.
Genetic manipulations.
E. coli strains were transformed by using the CaCl2 procedure of Sambrook et al. (59). A. tumefaciens strains were electroporated according to the procedure of Cangelosi et al. (8). Recombinant plasmids based on IncP1 vectors were mobilized into A. tumefaciens by a biparental method using E. coli strain S17-1 (21). Tn3HoHo1 insertion mutant 11 of pTHB112 (for the map, see Fig. 1A) was marker exchanged into pTiC58ΔaccR as described previously (24), and the lacZY fusion to accF on pKFG (34) was marker exchanged into pTiC58ΔaccR and pTHH206 as described previously (34). The marker exchange mutations were confirmed by restriction endonuclease analysis of purified plasmid DNAs.
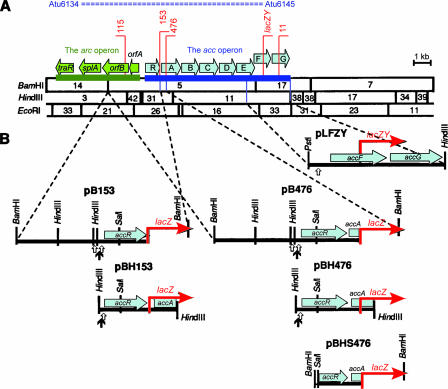
FIG. 1.
Reporter fusions and fragments of acc. (A) Physicogenetic map of pTHB112 which contains acc. Genes in acc, as well as those in the neighboring arc operon, are shown as open arrows above the restriction map. Matching locus tag numbers to the genes in the sequenced pTiC58, spanning from Atu6134 to Atu6145, are shown (73). Vertical lines denote the locations of Tn3HoHo1 insertions and of the lacZY cassette located in accF. The horizontal top bars indicate the direction of transcription reported by the lacZ fusions. (B) Constructs containing the lacZ reporter genes from insertions 153, 476, and the lacZY cassette located in accF. The restriction map and genetic organization of each insert fragment is shown below the name of the corresponding plasmid. The direction of expression of the lacZ gene on the Tn3HoHo1 insertions and of the lacZY fusion to accG are shown as red arrows. The open vertical arrows located under the restriction maps of the inserts indicate locations of the candidate operator sequences, while the closed arrows indicate the location of the pho box-like elements. Fragments that contain the same enzyme sites at both ends were obtained from pHK153 or pTHB476 by digesting the plasmids either with BamHI or with HindIII. The resulting fragments of insert DNAs were ligated into the vector pLAFR6, which had been digested with the appropriate enzyme, resulting in clones, pB153, pBH153, pB476, and pBH476. The reporter insert in pLFZY was obtained from pTHH206F (Table 1) by double digesting the plasmid with PstI and HindIII. This 2.5-kb fragment containing the reporter fusion was ligated into pLAFR6 digested with the same enzyme. The insert of pBHS476 was obtained as follows. The insert of pBH476 was cloned into pUC18 as a HindIII fragment, and an internal SalI fragment, which spans from a site in the insert to a site in the multiple cloning site of the vector, was deleted. The accA::lacZ fragment was recovered from this clone as a BamHI fragment, which spans from a site in the insert fragment to a site in the MCS of pUC18. This fragment was ligated into pLAFR6, which had been digested with BamHI.
Cell culture, induction, and β-galactosidase assays.
Each strain to be tested was grown in 4.5 ml of ABM medium containing appropriate antibiotics to an optical density at 600 nm of 0.3 to 0.4. A 1-ml volume of each culture was transferred to two sterile culture tubes, and agrocinopines were added to a final concentration of 20 μM to one subculture. These cultures were grown in parallel for an additional 2 h, and the bacteria were harvested by centrifugation and resuspended in 300 μl of 0.9% NaCl. β-Galactosidase activities were quantified as described by Miller (45). To assay for the induction by phosphate starvation, cells were collected by centrifugation from a 2-ml sample of culture as described above, washed twice with 0.9% NaCl, and resuspended in 2 ml of low-phosphate ABM medium. The bacterial suspension was split, and agrocinopines were added to one portion as described above. The subcultures were incubated for 12 h, and the cells were assayed for β-galactosidase activities as described above. β-Galactosidase activities of each strain incubated under all four conditions described above were confirmed in at least two separate experiments.
Primer extension assay.
Primer extension reactions were conducted as described by Piper et al. (55). The primer used was 5′-GTCGCGCCTCGCAGTGGCCACCG-3′. The primers were 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (BRL Life Technologies) as directed by the manufacturer. Dideoxy sequencing reactions were performed using the same primer and pKPS7 (55) as the template. The products of primer extension reactions and sequencing reactions were separated by electrophoresis on 6% polyacrylamide sequencing gels and visualized by exposing the gel to X-ray films.
Construction of His-tagged AccR.
The accR gene was cloned into pET29b (Novagen, Inc.) to add a tag of six histidine residues to the C terminus. The primers used to amplify the accR fragment were 5′-CTCTGGGAAGCGCATATGTTCAACTC-3′ (rightward) and 5′-CTCTCGTCGAACCTCGAGCGCAACAA-3′ (leftward). The underlined bases represent NdeI and XhoI sites incorporated into the primers by changing four bases (rightward primer) or three bases (leftward primer). The boldface ATG represents the translational start codon of the accR gene. Base substitutions in the leftward primer change the last two codons of accR from those for asparagine and termination to those for leucine and glutamic acid. The accR fragment was amplified by PCR using purified pRKW26 (3) as a template. The PCR product was digested with NdeI and XhoI and ligated with pET29b (Novagen, Inc.) linearized by the same set of enzymes. The insert in the resulting construct, pEXPR, was confirmed by DNA sequence analysis, and was used to transform E. coli strain BL21(DE3)(pLysS).
Purification of His-tagged AccR.
Strain BL21(DE3)(pLysS, pEXPR) was cultured in 2 liters of L broth containing kanamycin and chloramphenicol to an optical density at 600 nm of 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and the culture was incubated for an additional 2 h. Bacteria were collected by centrifugation, and the cell pellet was frozen overnight at −20°C. The cells were resuspended in 40 ml of cold buffer A (20 mM Tris-HCl [pH 7.9] and 0.5 M NaCl) containing 5 mM imidazole. The resulting suspension was sonicated on ice until the sample was no longer viscous. Cell debris was removed by centrifugation (39,000 × g for 20 min), and the supernatant was passed through a 0.45-μm-pore-size nitrocellulose membrane filter (Nalgene, Inc.). The filtered supernatant was subjected to Ni2+ affinity column chromatography (1.25-ml bed volume) as described by the supplier (Novagen, Inc.). The column was washed with 25 ml of buffer A containing 5 mM imidazole and then with the same volume of buffer A containing 60 mM imidazole. Bound proteins were eluted by using 20 ml of buffer A containing 1 M imidazole. Samples from each 1-ml fraction were assayed for the presence of proteins (BCA protein assay; Promega). The fractions that showed positive reactions in the protein assays were combined, and a small volume was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel. The elution buffer was exchanged with storage buffer containing 10 mM Tris-HCl (pH 7.8), 0.5 mM EDTA, 10 mM MgCl2, 1 mM dithiothreitol, and 50% glycerol by using a spin column (Microcon YM-10; Millipore). The protein preparation that remained soluble was separated from the insoluble part by centrifugation and was adjusted to a concentration of 200 nM in a total volume of about 100 μl. The sample in storage buffer was analyzed by SDS-PAGE on a 12% gel to determine molecular weight of the His-tagged AccR and was stored at −20°C.
Gel retardation assays.
Complementary pairs of oligonucleotides corresponding to potential cis-acting operator sites were annealed together to make double-stranded DNA molecules. The 3′ ends of the DNA fragments were labeled using digoxigenin-11-ddUTP and terminal transferase as recommended by the supplier (Boehringer Mannheim). Protein-DNA binding reactions in a 20-μl volume contained 0.4 pmol of DNA probe, 0.05% NP-40, and AccR protein at the concentrations indicated in a buffer of 10 mM Tris-HCl (pH 7.6), 1 mM EDTA, 50 mM NaCl, and 5% glycerol provided by the supplier (Boehringer Mannheim). When required, opines (agrocinopines or nopaline) were added as indicated. After 10 min of incubation at room temperature, samples were subjected to electrophoresis in 6% polyacrylamide gels at 4°C in Tris-borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.0]). After electrophoresis, the free DNA fragments and the AccR-DNA complexes were transferred by electrophoresis (Trans-Blot cell; Bio-Rad Co.) onto nylon membranes. The digoxigenin-labeled probes subsequently were detected by autoradiography using an enzyme immunoassay with antidigoxigenin-AP, Fab fragments, and the chemiluminescent substrate, CSPD (Boehringer Mannheim).
Gene induction assays.
Inducibility of accF or accG was tested by a modification of the agrocin 84 susceptibility assay described by Hayman et al. (27). Briefly, reporter assay medium containing the antibiotic was prepared by growing the agrocin 84-producer strain NT1(pAgK84-A1) as a single colony in the center of a plate of Stonier's medium to which X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) had been added. The plates were incubated for 48 h. Indicator strains C58KF or C58K11 that contain lacZ fusions to accF or to accG, respectively, were grown to late exponential phase and spread as a suspension in melted agar (0.7%) buffered with 20 mM potassium phosphate (pH 7.0) over the surface of the Stonier's medium plates prepared as described above. Paper strips containing 10 nmol of agrocinopines, l-arabinose, or sucrose were placed on the surface of the soft-agar overlays to provide a localized source of the substrate being tested for induction activity. The resulting plates were incubated at 28°C for 2 to 3 days before we examined the results.
pho box search in the Agrobacterium genome.
Upstream sequences of 300 bp counted from the start codon, or just as much as available if the neighboring gene is closer than this, of the genes that contain at least 50 bp of untranslated upstream region were retrieved from the genome of A.tumefaciens C58 (Refseq numbers NC_003304, NC_003305, NC_003306, and NC_003308) (73). Target sequences were then scanned for pho box motifs by using a software tool patser available at Regulatory Sequence Analysis Tools (RSAT [http://rsat.ulb.ac.be/rsat/]). A weight matrix used in the analysis was generated by using SEQLOGO (http://ep.ebi.ac.uk/EP/SEQLOGO/) by aligning 27 putative pho box motifs previously identified in A. tumefaciens C58 by Yuan et al. (75). (see Tables S1 and S2 in the supplemental material). Annotated functions of the genes associated with identified pho boxes were looked up from the NCBI database (http://www.ncbi.nlm.nih.gov/) or from the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/).
RESULTS
The acc operon is regulated by agrocinopines and by phosphate limitation.
Previous studies showed that acc is induced in the presence of the agrocinopines or when accR is mutated (3, 28, 34). Further, sequences closely related to pho boxes in E. coli and to the one in A. tumefaciens virG gene (41, 42, 72) were found in the region upstream of accR. This raised the possibility that the expression of acc may also be controlled by the availability of phosphate (34, 75). In fact, the reporter fusions to the acc operon, which do not disrupt the agrocinopine uptake system, i.e., C58K11 and NT1(pHK153), showed increased expression of the operon by agrocinopines and by phosphate limitation (Table 2).
TABLE 2.
β-Galactosidase activities of Agrobacterium strains in response to two environmental cues
| Strain | Reporter fusion | β-Galactosidase activity (Miller units)a at:
|
|||
|---|---|---|---|---|---|
| 25 mM PO4−2
|
0.1 mM PO4−2
|
||||
| − Agrocinopines | + Agrocinopines | − Agrocinopines | + Agrocinopines | ||
| NT1 | 1 | 1 | 1 | 1 | |
| C58K476 | accA::lacZb | 16 | 17 | 272 | 1,222 |
| C58KF | accF::lacZ | 8 | 8 | 36 | 1,119 |
| C58K11 | accG::lacZ | 11 | 603 | 27 | 224 |
| C58KFΔR | accF::lacZ | 168 | 171 | 473 | 527 |
| C58K11ΔR | accG::lacZ | 121 | 113 | 364 | 362 |
| NT1(pHK153) | accR-lacZ | 5 | 30 | 11 | 30 |
| NT1(pTHB476) | accA::lacZ | 104 | 103 | 440 | 2,008 |
| NT1(pTHH206F) | accF::lacZ | 14 | 16 | 64 | 398 |
| NT1(pHK115) | traII::lacZ | 4 | 15 | 5 | 17 |
β-Galactosidase assays were done as described in Materials and Methods.
lacZ is in the intergenic region between accR and accA just outside the accR coding region.
Others with nonfunctional uptake system [i.e., C58K476, C58KF, NT1(pTHB476), and NT1(pTHH206F)] (34), however, failed to show the induction by agrocinopines under phosphate-rich conditions due to the inaccessibility to the inducers. However, these strains demonstrated induction by the opines under phosphate-limiting conditions, suggesting concomitant induction of a secondary means of opine uptake that is not available when phosphate level is high. Strains with a mutation in the regulatory gene, accR (i.e., C58KFΔR and C58K11ΔR), showed derepressed levels of expression of the reporter genes but still exhibited induction by phosphate starvation. The clone reporting the expression of the arc operon (55), pHK115 (for the map, see Fig. 1A), showed induction by opines but not by low phosphate levels.
A promoter located in the region upstream of accR is responsible for the regulated expression of the acc operon by agrocinopines and by phosphate limitation.
To dissect the expression of acc, we cloned fragments from the operon, each with its relevant downstream lacZ reporter fusion, into pLAFR6 (Fig. 1). This vector contains a set of two transcription terminators on either side of the multiple cloning site (31). The resulting constructs were tested for expression of the lacZ reporters in three different strains: NT1, C58, and NT1(pTiC58ΔaccR). Each strain has the same chromosomal background but differs with respect to the status of pTiC58. NT1 is a Ti plasmidless derivative of the wild-type strain C58, C58 has the wild-type pTiC58, and NT1(pTiC58ΔaccR) harbors a derivative of pTiC58 that contains a 5-bp deletion in the 5′ end of the accR coding sequence (3).
C58 and NT1(pTiC58ΔaccR) harboring pB153, which has a reporter to accR, exhibited induction of accR by agrocinopines and by phosphate limitation (Table 3). In contrast, NT1 harboring the same plasmid, which lacks the agrocinopine uptake system coded for by acc, did not show induction under phosphate-rich conditions, but it did when starved for phosphate. pBHS476 contains a lacZ fusion to accA, as well as the intergenic region between accR and accA, but lacks most of accR and all of its 5′ sequences (Fig. 1). All three tester strains harboring this clone expressed the reporter only at a low level (Table 3). Moreover, expression was not affected by the presence or absence of accR and did not increase in response to agrocinopines or to phosphate starvation. This suggests that a weak promoter is located between accR and accA and that expression from this promoter does not respond to AccR, opines, or phosphate limitation.
TABLE 3.
β-Galactosidase activities of Agrobacterium strains harboring subclones of acc in response to two environmental cues
| Strain | Reporter fusion | β-Galactosidase activity (Miller units)a at:
|
|||
|---|---|---|---|---|---|
| 25 mM PO4−2
|
0.1 mM PO4−2
|
||||
| − Agrocinopines | + Agrocinopines | − Agrocinopines | + Agrocinopines | ||
| NT1 | 1 | 1 | 1 | 1 | |
| Clones with both IR | |||||
| NT1(pB153) | accR-lacZb | 5 | 5 | 10 | 132 |
| C58(pB153) | 5 | 32 | 10 | 34 | |
| NT1(pTiC58ΔaccR, pB153) | 5 | 30 | 12 | 32 | |
| NT1(pB476) | accA::lacZ | 56 | 58 | 157 | 1,405 |
| C58(pB476) | 34 | 182 | 169 | 903 | |
| NT1(TiC58ΔaccR, pB476) | 79 | 143 | 334 | 1,336 | |
| Clones with only one IR | |||||
| NT1(pBH153) | accR-lacZ | 16 | 15 | 76 | 279 |
| C58(pBH153) | 22 | 22 | 78 | 100 | |
| NT1(pTiC58ΔaccR, pBH153) | 23 | 23 | 77 | 247 | |
| NT1(pBH476) | accA::lacZ | 295 | 281 | 981 | 2,131 |
| C58(pBH476) | 260 | 270 | 979 | 1,763 | |
| NT1(pTiC58ΔaccR, pBH476) | 346 | 367 | 977 | 2,182 | |
| Clones lacking accR promoter | |||||
| NT1(pBHS476) | accA::lacZ | 4 | 5 | 3 | 4 |
| C58(pBHS476) | 3 | 4 | 3 | 5 | |
| NT1(pTiC58ΔaccR, pBHS476) | 5 | 6 | 4 | 7 | |
| NT1(pLFZY) | accF::lacZ | 4 | 4 | 3 | 4 |
| C58(pLFZY) | 3 | 9 | 4 | 20 | |
| NT1(pTiC58ΔaccR, pLFZY) | 17 | 17 | 18 | 19 | |
β-Galactosidase assays were done as described in Materials and Methods.
lacZ is in the intergenic region between accR and accA just outside the accR coding region.
However, expression of the accA::lacZ reporter responded to opines and to phosphate limitation when tested in a clone pB476, which contains an insert that includes accR and its 5′ upstream sequences (Table 3). This suggests that the regulated expression of accA is mediated by the promoter located before accR. C58 and NT1(pTiC58ΔaccR) harboring pB476 showed significant increases in the levels of expression of the lacZ reporter in response to agrocinopines and to phosphate limitation. Regulated expression in NT1(pTiC58ΔaccR) was expected since the reporter clone provides wild-type AccR. NT1(pB476), which does not express the acc transport system, did not exhibit induction in response to agrocinopines under phosphate-rich conditions but did under low-phosphate levels (Table 3).
Expression of the accF and accG reporters placed in pTiC58 was repressed in the absence of the agrocinopines, but these same fusions in pTiC58ΔaccR were expressed at high levels, even in the absence of the opines (Table 2). This indicates that accF and accG are part of the acc operon. However, pLFZY, which lacks the regions upstream of accR and accA (Fig. 1) showed only low levels of expression of the accF::lacZ reporter in strain NT1 (Table 3). Interestingly, the same reporter in C58 showed moderate increases in gene expression in response to agrocinopines. Also, NT1(pTiC58ΔaccR) harboring pLFZY expressed the lacZ reporter at a level severalfold higher than did NT1 harboring the same plasmid in the absence of agrocinopines (Table 3). This suggests that the weak promoter in the region upstream of accF and accG serves as the secondary promoter that is regulated by agrocinopines. However, none of the strains harboring pLFZY responded to low phosphate levels.
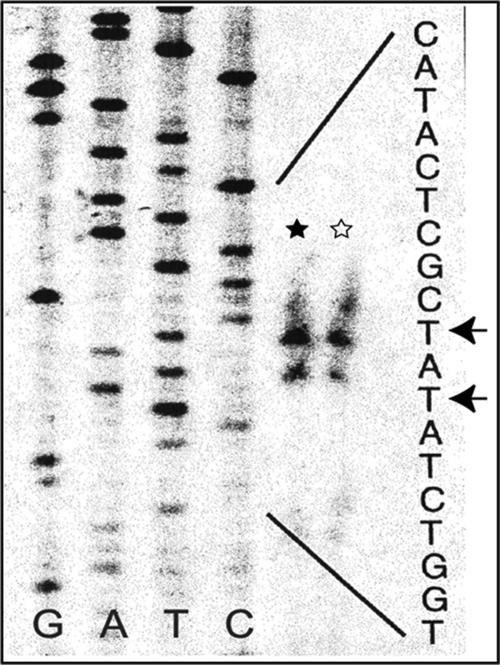
Consistent with the genetic data, primer extension analyses for the expression of the acc operon by agrocinopines and for that by phosphate-limitation mapped the same two transcription start sites in the region upstream of accR (Fig. 2). This verifies that the accR promoter is responsible for the regulated expression of acc by both agrocinopines and by low phosphate levels.
FIG. 2.
Mapping the transcriptional start site for the acc operon. Two transcription start sites mapped by primer extension analyses using total RNA isolated from either NT1(pTiC58ΔaccR) (denoted by ⋆) or C58 under a low phosphate condition (denoted by ★) are marked with two arrows in the gel picture.
Two operators are required for regulated expression of the accR promoter by agrocinopines under phosphate-rich conditions.
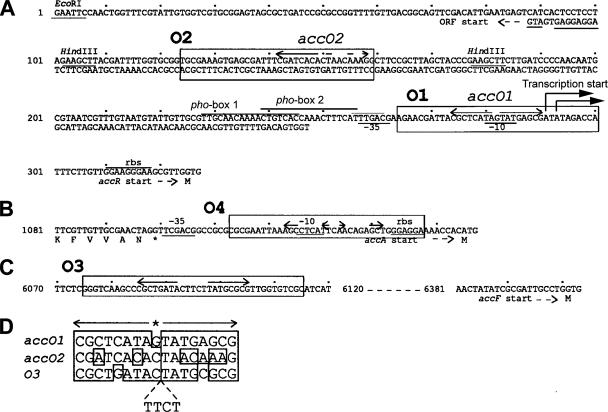
The accR promoter contains a candidate operator in the form of an IR sequence that overlaps the putative −10 element (34). We mapped two consecutive transcription start sites associated with this −10 element (Fig. 2 and 3A). A second, imperfect copy of this IR is located about 110 bp upstream from this sequence. Analysis of deletion derivatives suggests that both IR elements are required for the repression of acc by AccR; pBH153 and pBH476, which truncate at the leftward end of HindIII fragment 31 that contains the imperfect IR (Fig. 1 and 3A), showed severalfold-higher levels of expression of the lacZ reporters compared to the same strains harboring pB153 and pB476, respectively, which contain about 2.3 kb of sequence upstream of this site (Fig. 1 and Table 3). While strains harboring pB153 and pB476 exhibited induction of the lacZ reporters in response to agrocinopines, the levels of expression in the strains harboring pBH153 and pBH476 did not show further increases when supplemented with the opines, indicating that the reporter in this plasmid is constitutively expressed under these conditions. However, strains harboring pBH153 and pBH476 showed induction in response to low phosphate levels, and the expression was further increased when supplemented with the opines.
FIG. 3.
Nucleotide sequence of the promoter regions of acc. (A) Sequence of the 5′-untranslated region of the acc operon. Two sequences homologous to the pho box are overlined. The accR promoter, and the two putative operators, accO1 and accO2, are indicated. Putative −10 and −35 sequences are underlined. The −10-like sequence of the second phosphate-responsive promoter of accR is indicated by a line above the nucleotide sequence. Other lines above or below the nucleotide sequence indicate the positions of potential ribosomal binding sites (rbs). The translational start site of each gene is preceded by its name, a short dashed arrow showing the direction of expression, and by the single-letter code M for methionine. IR sequences are indicated by divergent arrows above the sequences. Boxes labeled O1, O2, O3, and O4 represent the double-stranded probes used for gel retardation assays. (B) The sequence of the region containing the putative promoter of accA. (C) Sequence upstream of accF. (D) Nucleotide sequence similarities among accO1, accO2, and O3. The arrows demark the IR sequences. The asterisk denotes the central nucleotide which defines the axis of symmetry.
AccR interacts with the operators in the absence of agrocinopines.
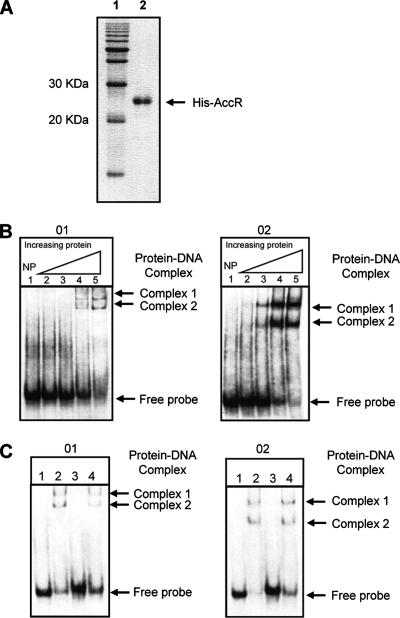
To study AccR-operator interactions, we purified the C-terminal His-tagged protein (see Materials and Methods). The purified protein migrates as band with a size of about 26 kDa in an SDS-polyacrylamide gel (Fig. 4A). The apparent molecular mass differs from the predicted mass (29 kDa) of His-tagged AccR. However, the nucleotide sequence of the accR-His tag fusion fragment was as expected. Visual analysis of the Coomassie blue-stained SDS-PAGE gel indicates that the preparation of AccR is better than 95% pure. We confirmed that the His-tagged AccR has the wild-type-level activity in repressing expression of the lacZ reporter fusion to accG and in decreasing susceptibility to agrocin 84 by repressing acc expression in the strains tested (data not shown).
FIG. 4.
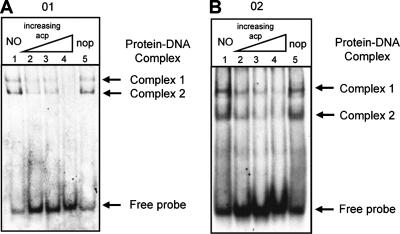
Binding of purified AccR to DNA probes O1 and O2. (A) SDS-PAGE analysis of purified His-tagged AccR protein. A protein preparation of about 1 μg was electrophoresed in a SDS-12% polyacrylamide gel. Lanes contain the following: 1, protein size marker; 2, His-tagged AccR. (B) Probe O1 (in the left panel) is a 37-mer, and probe O2 (in the right panel) is a 36-mer. The two probes correspond to the boxed regions shown in Fig. 2. Binding reactions were carried out as described in Materials and Methods. Lane 1, DNA probe only. The other lanes contain probe and His-tagged AccR at concentrations of 0.04 nM (lane 2), 0.2 nM (lane 3), 1 nM (lane 4), and 5 nM (lane 5). (C) Binding of AccR to DNA probes O1 and O2 is specific. Probes O1 and O2 show the effect of adding homologous or heterologous competitor DNA to reactions. All reactions contain 0.4 pmol of labeled DNA. Lane 1, DNA probe only; lanes 2, 3, and 4, DNA probes mixed with AccR at a final concentration of 1 nM. Lanes 3 and 4 also contain 0.8 pmol of the unlabeled homologous DNA and 3 μg of calf thymus DNA, respectively.
In gel mobility-shift assays, fragments O1 and O2, that contain the perfect and imperfect IRs, respectively, each formed detectable complexes with purified AccR (Fig. 4B and C). The amount of complex formed increased with increasing amounts of protein (Fig. 4B). Binding of AccR to fragments O1 and O2 was specific for the DNA; excess amounts of unlabeled calf thymus DNA did not compete with the labeled probes for binding by AccR (Fig. 4C). However, addition of unlabeled O1 or O2 DNA to the reactions in amounts as little as twice that of the probes significantly reduced the amount of complexes formed between AccR and the labeled fragments. Both fragments yielded two major bands of AccR-DNA complexes in the gel retardation assays. However, the electrophoretic mobilities of the two bands formed with fragment O1 differed from those formed with fragment O2, although the sizes of the two fragments themselves differ by only 1 bp.
We tested the influence of agrocinopines on the binding of AccR to DNA fragments O1 and O2 (Fig. 5). Addition of a mixture of agrocinopines A and B to the reaction decreased the amounts of complexes formed between AccR and fragments O1 or O2. Moreover, the amounts of complexes formed decreased in proportion to the amounts of the opines added. In binding reactions with agrocinopines at 200 μM, virtually no bands corresponding to the AccR-DNA complexes were detected on the gel. On the other hand, nopaline, another opine associated with pTiC58, had no detectable influence, inhibitory or otherwise, on the formation of AccR-DNA complexes at concentrations as high as 500 μM.
FIG. 5.
Inhibition of binding of AccR to O1 and O2 by agrocinopines. Probes O1 and O2 show the effect of adding agrocinopines or nopaline to the reactions. Each reaction contains AccR at a final concentration of 1 nM. Agrocinopines were added to the reaction at final concentrations of 50 μM (lane 2), 100 μM (lane 3), or 200 μM (lane 4). Lane 1, no opines; lane 5, nopaline at a final concentration of 500 μM. acp, agrocinopines; NO, no opines; nop, nopaline.
We also tested the IR sequence present about 300 bp upstream of accF, which is similar to the two IRs from the accR promoter (Fig. 3B and C). Despite the sequence similarity, the mobility of the fragment O3 containing this IR sequence was not affected by His-tagged AccR (data not shown). An imperfect IR sequence unrelated to those present upstream of accR is located in the promoter region of accA (Fig. 3B). Fragment O4, which contains this IR sequence, also did not form detectable complexes with AccR (data not shown).
A. tumefaciens C58 genome contains 191 putative pho boxes associated with 226 genes.
Yuan et al. identified 99 putative pho boxes in the A. tumefaciens strain C58 genome using the weight matrix generated from known Sinorhizobium and E. coli pho boxes (75). We generated an Agrobacterium-specific weight matrix (see Table S2 in the supplemental material) from 27 Agrobacterium pho boxes identified from the present study and reanalyzed the Agrobacterium genome. Using a software tool patser from RSAT with a cutoff score of 8.0, we found 12 putative pho box motifs associated with 12 genes in pTiC58, refinding the two previously identified in the region upstream of accR (34, 75) (Fig. 3A). The same number of motifs and associated genes were found in pAtC58, although it is larger than pTiC58. We also found 121 motifs with 142 genes and 46 motifs with 60 genes in the circular and the linear chromosomes, respectively. Many of the genes associated with the putative pho box motifs have predicted roles in the utilization of phosphate or related compounds. Some of these genes include Atu0419 (phoR, two-component sensor kinase), Atu0420 (pstS, a subunit of ABC transporter for phosphate), Atu0174 (phnC, a subunit of ABC transporter for phosphonate), Atu6108 (phnA, alkylphosphonate uptake protein), Atu0305 (ugpA, a subunit of ABC transporter for sn-glycerol-3-phosphate), and Atu1144 (ppk, polyphosphate kinase). On the other hand, there also are many genes whose known or predicted functions do not appear to be directly connected to phosphate utilization. These include Atu6178 (virG, two-component response regulator), which is involved in plant transformation (72); Atu1657 [nadE, NAD(+) synthase] encoding de novo synthesis of NAD (30); and Atu2085 (lpxC, UDP-3-O-[3-hydroxymyristoryl] N-acetylglucosamine deacetylase), which encodes lipopolysaccharide biosynthesis. For a complete list of genes with putative pho box sequences, see Table S3 in the supplemental material.
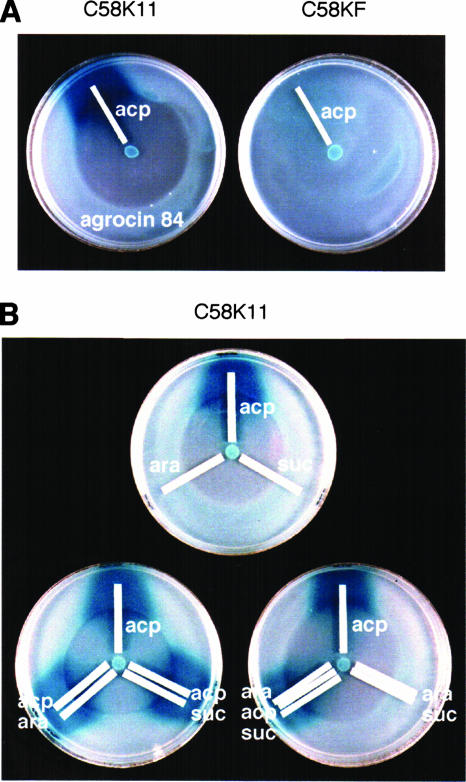
Agrocin 84 and the sugars that constitute agrocinopine A do not serve as inducers of acc.
That Agrocin 84 is transported and processed by functions encoded by the acc operon (27, 34) raises the possibility that the antibiotic may induce the expression of this operon. We tested this using a modification of the agrocin 84 susceptibility assay we previously described (34) (Fig. 6). Strains C58KF and C58K11, containing lacZ reporters to accF and accG, respectively, were spread over the surface of the assay plates as indicator strains. Both strains can take up agrocin 84, whereas only C58K11 can take up agrocinopines (34). Moreover, while able to take up agrocin 84, strain C58KF is resistant to the antibiotic, whereas strain C58K11 remains susceptible. The medium in the assay plate contains agrocin 84, as well as X-Gal, the chromogenic indicator of the expression of the lacZ reporters. A paper strip containing agrocinopines was placed on the surface of the assay plates to assess induction of the expression of the reporters by the opines. In overlays containing strain C58K11, which takes up both agrocinopines and agrocin 84, an intense blue color reaction formed around the paper strip containing the opines (Fig. 6A). This is consistent with the fact that the opines induce the expression of the acc operon. However, no such color reaction was observed at the edge of the inhibition zone caused by agrocin 84, indicating that the reporter is not induced by subinhibitory levels of agrocin 84. Strain C58KF, which takes up agrocin 84 but not agrocinopines, showed no induction of the lacZ reporter either around the paper strip or in the central area of the plate. Thus, even at high levels, the antibiotic does not induce the reporter gene.
FIG. 6.
Inducibility of the acc operon by alternative substrates. Susceptibility to agrocin 84 was determined as described by Kim and Farrand (34). Paper strips containing agrocinopines, l-arabinose, or sucrose were placed on the assay plates containing agrocin 84 and X-Gal as described in Materials and Methods. (A) Agrocin 84 does not induce the expression of the acc operon. Strains C58K11 and C58KF, which contain lacZ reporters located in accF and in accG, respectively, were used as reporters. (B) l-Arabinose and sucrose do not induce expression of the acc operon. Strain C58K11 was used as the reporter. acp, agrocinopines; ara, l-arabinose; suc, sucrose.
Using strain C58K11 as the indicator, we also determined whether l-arabinose and sucrose, the constituents of agrocinopine A (58), can induce the expression of acc (Fig. 6B). Paper strips containing 10 nmol of l-arabinose, sucrose, or agrocinopines were placed separately or in close proximity to each other in various combinations. Neither arabinose nor sucrose, separately or together, induced the accG::lacZ reporter or affected susceptibility of strain C58K11 to agrocin 84. Nor did either of these sugars, alone or in concert, exert any detectable influence on the induction of the acc operon by agrocinopines.
DISCUSSION
Studies with lacZ fusions to accR, accA, accF, and accG showed that expression of each of these genes within the intact acc operon is inducible by agrocinopines and also by phosphate starvation (Table 2). We found that the promoter located in the region upstream of the first gene of the acc operon, accR, is the main promoter responsible for the regulated expression of the operon (Table 3). Thus, accR is autoregulated by its own gene product as part of the acc operon. This results in a high level of expression of AccR in the presence of agrocinopines and also under phosphate-limiting conditions. AccR produced at elevated levels may allow quick repression of the system when the opines become scarce. It also may serve to keep the expression of the acc operon below a certain level under conditions of phosphate limitation when the opines are not available.
Clones pBH153 and pBH476, which contain only the IR located proximal to accR (Fig. 1 and 3A), expressed the reporter at constitutively elevated levels in all strains when tested under phosphate-rich conditions (Table 3). This suggests that both of the IRs are required to repress the expression of the acc operon under these conditions. The arrangement of the two IRs in the operon, one in the promoter of accR and the other about 110 bp upstream from this site, and the requirement of both sequences for repression are not without precedent. Full repression of araBAD, the arabinose operon of E. coli, by AraC requires two operators: araI and araO2 (40, 61). araI is located immediately adjacent to PBAD, the promoter of araBAD, while araO2 is located more than 200 bp upstream from araI. In the absence of arabinose, AraC represses expression from PBAD, through formation of a DNA loop by simultaneously binding to araI and araO2. However, closes pBH153 and pBH476 showed further increase in the expression of the reporter gene under phosphate limitation when agrocinopines are added (Table 3). This suggests that unlike the repression by AccR under phosphate-rich conditions, binding of AccR to the proximal IR alone apparently can exert some repressional effect under phosphate-limiting conditions.
Gel retardation assays with fragments containing IR sequences formed detectable complexes with purified AccR (Fig. 4B and C). Moreover, interaction between AccR and the target fragments is sequence specific; binding was inhibited by unlabeled homologous DNA, but heterologous DNA in vast excess did not detectably compete with the labeled probes. DNA fragments each formed two complexes with purified AccR distinguishable by different mobilities (Fig. 4B and C). DeoR, which is highly homologous to AccR (1), also forms sets of complexes when tested with DNA fragments containing a single copy of its cognate operator in gel retardation assays (46). It is intriguing to note that this protein represses the deo operon by binding simultaneously to multipartite operators, thereby forming DNA loops in the process (1, 13, 26, 61).
While there are extra sequences in the fragments used in gel retardation assays, these IR sequences, that we named accO1 and accO2 (Fig. 3A), most likely comprise the operators that are responsible for the repression of acc by AccR. First, accO1 highly resembles the structure of the experimentally verified E. coli lac operator in that each arm consists of long nucleotides of up to 10, and the arms are separated by a GC base pair (4, 65). Moreover, this IR shows a perfect palindrome structure of as many as 17 nucleotides that overlaps the −10 sequence of the acc operon (Fig. 3A). Second, half of accO1 is highly conserved in accO2, whereas the other half is diverged (left arm, 6/8 match; right arm, 4/8 match). This resembles the organization of the DeoR-regulated dra-nupC-pdp operon in Bacillus subtilis, operators of which consist of one perfect IR and a sequence matching only to one arm of this IR (77). Third, many repressors bend DNA at the center of their operators (17, 37, 52, 53, 62, 78), and DNA molecules containing a bend at the end migrate faster in the gels than those with the same bend in the middle (74). Consistent with this, AccR-O2 complexes, with accO2 at the end of the fragment, migrated faster than the complexes formed with fragment O1, which contains accO1 in the middle (Fig. 4B and C). Lastly, there is an IR sequence similar to accO1 about 300 bp upstream from accF (Fig. 3C). Although fragment O3, which contains this IR, failed to form detectable complexes with AccR in our assays (Fig. 3D and data not shown), the possibility of this IR cooperatively binding AccR with other operators in the operon is not excluded. Even if it simply represents an evolutionary remnant, its presence supports the legitimacy of accO1 and accO2 as operators. Additional IR sequences notable on fragments O1 and O2 include GAACGAT-TACGCTC and GAGCGAT-TTCGATC, which overlap the left arms of accO1 and accO2, respectively. High conservation between these sequences and their particular locations associated with the putative operators suggest possible involvement of these IRs in AccR-operator interactions that warrants future investigations.
That agrocinopines interfere with the formation of AccR-DNA complexes in gel retardation assays (Fig. 5) suggests that the opines, not one of the degraded metabolites, are the true inducers. This is consistent with our observation in that C58KF that cannot catabolize agrocinopines exhibits induction of the acc operon by agrocinopines under phosphate-limiting conditions, whereas C58KFΔR, lacking the intact AccR, shows derepressed levels of expression (Table 2). Under phosphate-rich conditions, however, C58KF does not show such induction due to the inaccessibility to the agrocinopines (34) (see below). Two other opines, octopine and nopaline, also directly interact with their respective activator proteins OccR and NocR (2, 39, 70). On the other hand, agrocin 84 does not induce the acc operon (Fig. 5), although this antibiotic is recognized by the transport and catabolism functions coded for by the operon (27, 34). This suggests that AccR is more discriminatory with respect to its substrates than is the opine transport system (34). l-Arabinose and sucrose, the two sugar components of agrocinopine A, did not induce the acc operon, nor did they inhibit induction of this operon by the opines. The results suggest that these sugars do not compete with agrocinopines for binding to AccR or to AccA, the periplasmic agrocinopine-binding protein coded for by accA (34).
In E. coli and in Sinorhizobium meliloti, which is closely related to Agrobacterium, a low phosphate concentration in the environment is sensed by a two-component sensor kinase, PhoR (38, 41, 75). Then, PhoR activates a cognate response regulator, PhoB, by phosphorylating the protein. Activated PhoB then induces the expression of various genes by binding to pho boxes in their upstream regions. virG of pTi58, a two-component response regulator of the vir operon involved in T-DNA transfer into plant genome, is preceded by a pho box motif in its upstream sequence (the pho box at ca. −148 to −131 [see Table S3 in the supplemental material]). Its close homolog (81% identity at DNA level), virG of octopine-type Ti plasmid pTiA6, was previously shown to be regulated by phosphate starvation (72) and has an upstream pho box sequence that matches 13 of 18 bp from that in pTi58 virG. In this regard, it is interesting that the accR promoter contains two putative 18-bp pho boxes, which overlap each other by 7 bp (Fig. 3A and see Table S3 in the supplemental material) (pho box 1 at ca. −94 to −77 bp from the start codon, pho box 2 at ca. −83 to −66). Since acc indeed is induced by phosphate starvation and phosphate is a constituent of agrocinopines, it appears evident that it is part of the pho regulon in Agrobacterium. A pair of phoR (Atu0419) and phoB (Atu0425) is present in the circular chromosome and also are two additional orphan phoB homologs (Atu5119 and Atu5121) in pAtC58. That phoR (Atu0419) is associated with a putative pho box in the promoter region (see Table S3 in the supplemental material) suggests its role in the pho regulon circuits in A. tumefaciens strain C58.
The reporters in strains that do not contain the intact agrocinopine transport system coded for by acc failed to respond to agrocinopines under phosphate-rich conditions (Tables 2 and 3). However, all of these strains showed induction by the opines under phosphate-limiting conditions (Tables 2 and 3). This indicates that, under conditions of phosphate limitation, agrocinopines are taken up by other transport systems. In this regard, it is noteworthy that many chromosomal genes encoding ABC type transporters for inorganic phosphate (Atu0420), phosphonates (Atu0174, phnC), sn-glycerol 3-phosphate (Atu0305, ugpA), and iron (Atu0202) are preceded by putative pho box motifs (see Table S3 in the supplemental material). In E. coli, the Phn system encoded by phnCDE transports broad range of substrates, such as phosphonates, phosphates, phosphites, and organophosphates (18, 44).
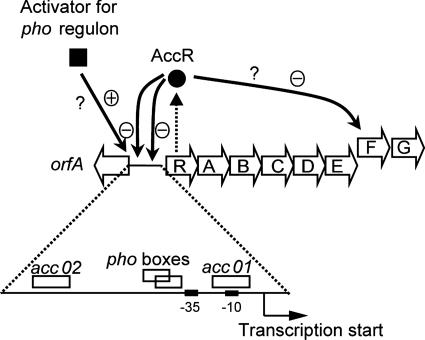
Taking all of the data together, we propose that the expression of acc is regulated as follows (Fig. 7). In the absence of agrocinopines, high-level expression of the acc locus from the accR promoter is repressed by AccR through interactions with the two operators. Under these conditions, we propose that acc is expressed at a low basal level from the promoters present in the regions upstream of accA and accF and perhaps accR. When agrocinopines are present, expression of acc is increased most likely because the promoter is freed from the repression exerted by AccR. On the other hand, under conditions of phosphate limitation, the promoter is activated, most probably by a homolog of E. coli PhoB (e.g., Atu0425, Atu5119, or Atu5121). In the absence of the opines, expression from this promoter also appears to be influenced by the interaction between AccR and the two operator sequences. Presumably, AccR bound to the operators impedes expression from the promoter. However, expression of the acc operon is activated even in the absence of the opines under phosphate-limiting conditions, indicating that interactions between AccR and the operators do not fully block expression from the promoter.
FIG. 7.
Model for the regulation of expression of the acc operon in response to agrocinopines and to phosphate starvation. Genes in acc and orfA, the first gene of the divergently transcribed arc operon, are shown as open arrows. A dashed arrow indicates the production of AccR. Solid arrows indicate the location of the promoters regulated by AccR or by a putative PhoB homolog. The two operator sequences, accO1 and accO2, that bind AccR, −10, and −35 sequences, as well as a pho box-like sequences located in the region upstream of accR, are indicated by boxes. •, AccR; ▪, an unknown transcriptional activator protein for genes of the pho regulon; ⊕, activation of transcription; ⊖, repression of transcription by AccR; ?, uncharacterized regulatory elements.
In the soil environment, where general nutrient sources often are limiting (23), opines produced by plant tumors can be an important source for carbon and energy. While ecological studies await, we can expect that agrocinopines, which also serve as a source for phosphate, may play an important role in survival of A. tumefaciens in the environment. Agrocinopines induce not only their own uptake by and catabolism in strain C58 but also spread of pTiC58 among Ti-plasmidless agrobacteria in the community by relieving repression exerted by AccR on the arc operon (3), where traR, a quorum-sensing regulator for conjugative transfer of pTiC58, is the fourth gene (for the map, see Fig. 1A) (34, 55). This ensures maximum use of agrocinopines as specific nutrients for agrobacteria in the environment. Under phosphate-limiting conditions, coactivation of the acc operon as part of the pho regulon has a significance in that it increases the ability of the bacterium to locate a source of agrocinopines for phosphate, since acc also confers chemotaxis to the opines (35). The opine-phosphate dual regulatory system governing the expression of the acc operon is therefore of a sophisticated design to maximize the survival of A. tumefaciens in the environment.
Supplementary Material
Acknowledgments
This study was supported by grants 2007-E00116-00 from KNIH and K0619251 from Korea University to H.S.K. and by the Korea Foundation for International Cooperation of Science and Technology (KICOS) through a grant provided by the Korean Ministry of Science and Technology (MOST) (K20601000002-07E0100-00240). Portions of this work were supported by grant R01GM52465 from the NIH to S.K.F.
Footnotes
Published ahead of print on 14 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adhya, S. 1989. Multipartite genetic control elements: communication by DNA loop. Annu. Rev. Genet. 23227-250. [DOI] [PubMed] [Google Scholar]
- 2.Akakura, R., and S. C. Winans. 2002. Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J. Biol. Chem. 27715773-15780. [DOI] [PubMed] [Google Scholar]
- 3.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, C. E., and M. Lewis. 2001. The Lac repressor: a second generation of structural and functional studies. Curr. Opin. Struct. Biol. 1119-25. [DOI] [PubMed] [Google Scholar]
- 5.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollaston, and A. W. B. Johnston. 1978. Transfer of the drug-resistance transposon Tn5 to Rhizobium. Nature 276633-634. [Google Scholar]
- 6.Bomhoff, G., P. M. Klapwijk, H. C. M. Kester, R. A. Schilperoort, J. P. Hernalsteens, and J. Schell. 1976. Octopine and nopaline synthesis and breakdown genetically controlled by a plasmid of Agrobacterium tumefaciens. Mol. Gen. Genet. 145177-181. [DOI] [PubMed] [Google Scholar]
- 7.Broer, I., W. Dröge-Laser, R. F. Barker, K. Neumann, W. Klipp, and A. Pühler. 1995. Identification of the Agrobacterium tumefaciens C58 T-DNA genes e and f and their impact on crown gall tumor formation. Plant Mol. Biol. 2741-57. [DOI] [PubMed] [Google Scholar]
- 8.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204384-397. [DOI] [PubMed] [Google Scholar]
- 9.Chilton, M.-D., T. C. Currier, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. USA 713672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chilton, M.-D., R. K. Saiki, N. Yadav, M. P. Gordon, and F. Quétier. 1980. T-DNA from Agrobacterium Ti plasmid is in the nuclear DNA fraction of crown gall tumor cells. Proc. Natl. Acad. Sci. USA 774060-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chilton, M.-D., D. A. Tepfer, A. Petit, C. David, F. Casse-Delbart, and J. Tempé. 1982. Agrobacterium rhizogenes inserts T-DNA into plant roots. Nature 295432-434. [Google Scholar]
- 12.Citovsky, V., S. V. Kozlovsky, B. Lacroix, A. Zaltsman, M. Dafny-Yelin, S. Vyas, A. Tovkach, and T. Tzfira. 2007. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 99-20. [DOI] [PubMed] [Google Scholar]
- 13.Dandanell, G., and K. Hammer. 1985. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 43333-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depicker, A., S. Stachel, P. Dhaese, P. Zambryski, and H. M. Goodman. 1982. Nopaline synthase: transcript mapping and DNA sequence. J. Mol. Appl. Genet. 1561-573. [PubMed] [Google Scholar]
- 15.Dessaux, Y., A. Petit, and J. Tempé. 1992. Opines in Agrobacterium biology. CRC Press, Boca Raton, FL.
- 16.Dische, Z., and E. Borenfreund. 1957. A new color reaction for the determination of aldopentose in the presence of other saccharides. Biochim. Biophys. Acta 23639-642. [DOI] [PubMed] [Google Scholar]
- 17.Economides, A. N., D. Everdeen, and N. Panayotas. 1996. A shared, non-canonical DNA conformation detected at DNA/protein contact sites and bent DNA in the absence of supercoiling or cognate binding. J. Biol. Chem. 27124836-24841. [DOI] [PubMed] [Google Scholar]
- 18.Elashvili, I., J. J. Defrank, and V. C. Culotta. 1998. phnE and glpT genes enhance utilization of organophosphates in Escherichia coli K-12. Appl. Environ. Microbiol. 642601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis, J. G., A. Kerr, M. Van Montagu, and J. Schell. 1979. Agrobacterium: genetic studies on agrocin 84 production and the biological control of crown gall. Physiol. Plant Pathol. 15311-319. [Google Scholar]
- 20.Ellis, J. G., and P. J. Murphy. 1981. Four new opines from crown gall tumors-their detection and properties. Mol. Gen. Genet. 18136-43. [Google Scholar]
- 21.Farrand, S. K., I. Hwang, and D. M. Cook. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 1784233-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrand, S. K., J. E. Slota, J.-S. Shim, and A. Kerr. 1985. Tn5 insertions in the agrocin 84 plasmid: the conjugal nature of pAgK84 and the locations of determinants for transfer and agrocin 84 production. Plasmid 13106-117. [DOI] [PubMed] [Google Scholar]
- 23.Fried, M., and H. Broeshart. 1967. The soil-plant system in relation to inorganic nutrition. Academic Press, Inc., New York, NY.
- 24.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site specific mutagenesis. Cell 27142-153. [DOI] [PubMed] [Google Scholar]
- 25.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 6716-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammer, K., L. Bech, P. Hobolth, and G. Dandanell. 1993. DNA specificity of Escherichia coli deoP1 operator-DeoR repressor recognition. Mol. Gen. Genet. 237129-133. [DOI] [PubMed] [Google Scholar]
- 27.Hayman, G. T., S. Beck von Bodman, H. Kim, P. Jiang, and S. K. Farrand. 1993. Genetic analysis of the agrocinopine catabolic region of Agrobacterium tumefaciens Ti plasmid pTiC58, which encodes genes required for opine and agrocin 84 transport. J. Bacteriol. 1755575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayman, G. T., and S. K. Farrand. 1988. Characterization and mapping of the agrocinopine-agrocin 84 locus on the nopaline Ti plasmid pTiC58. J. Bacteriol. 1701759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holsters, M., B. Silva, F. Van Vliet, C. Genetello, M. De Block, P. Dhaese, A. Depicker, D. Inzé, G. Engler, R. Villarroel, M. Van Montagu, and J. Schell. 1980. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3212-230. [DOI] [PubMed] [Google Scholar]
- 30.Hove-Jensen, B. 1996. Phosphoribosyl diphosphate synthetase-Independent NAD de novo synthesis in Escherichia coli: a new phenotype of phosphate regulon mutants. J. Bacteriol. 178714-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh, T., D. Dahlbeck, and B. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 2451374-1377. [DOI] [PubMed] [Google Scholar]
- 32.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109137-141. [DOI] [PubMed] [Google Scholar]
- 33.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 34.Kim, H., and S. K. Farrand. 1997. Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J. Bacteriol. 1797559-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, H., and S. K. Farrand. 1998. Opine catabolic loci from Agrobacterium plasmids confer chemotaxis to their cognate substrates. Mol. Plant-Microbe Interact. 11131-143. [DOI] [PubMed] [Google Scholar]
- 36.Kim, J.-G., B. K. Park, S.-U. Kim, D. Choi, B. H. Nahm, J. S. Moon, J. S. Reader, S. K. Farrand, and I. Hwang. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. USA 1038846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J., C. Zwieb, C. Wu, and S. Adhya. 1989. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene 8515-23. [DOI] [PubMed] [Google Scholar]
- 38.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Gen. Genomics 2721-17. [DOI] [PubMed] [Google Scholar]
- 39.Lintig, J., D. Kreusch, and J. Schröder. 1994. Opine-regulated promoters and LysR-type regulators in the nopaline (noc) and octopine (occ) catabolic regions of Ti plasmids of Agrobacterium tumefaciens. J. Bacteriol. 176495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobell, R., and R. Schleif. 1990. DNA looping and unlooping by AraC protein. Science 250528-532. [DOI] [PubMed] [Google Scholar]
- 41.Makino, K., M. Amemura, S.-K. Kim, A. Nakata, and H. Shinagawa. 1994. Mechanism of transcriptional activation of the phosphate regulon in Escherichia coli. ASM Press, Washington, DC.
- 42.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli: activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 20385-95. [DOI] [PubMed] [Google Scholar]
- 43.Messens, E., A. Lenaerts, R. W. Hedges, and M. Van Montagu. 1985. Agrocinopine A, a phosphorylated opine is secreted from crown gall cells. EMBO J. 4571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, J. H. 1972. Assay of β-galactosidase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Mortenen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy, P. J., and W. P. Roberts. 1979. A basis for agrocin 84 sensitivity in Agrobacterium radiobacter. J. Gen. Microbiol. 114207-213. [Google Scholar]
- 48.Murphy, P. J., M. E. Tate, and A. Kerr. 1981. Substituents at N6 and C-5′ control sensitive uptake and toxicity of the adenine-nucleotide bacteriocin, agrocin 84, in bacteria. Eur. J. Biochem. 115539-543. [DOI] [PubMed] [Google Scholar]
- 49.Nautiyal, C. S., and P. Dion. 1990. Characterization of the opine-utilizing microflora associated with samples of soil and plants. Appl. Environ. Microbiol. 562576-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oger, P., and S. K. Farrand. 2001. Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol. Microbiol. 411173-1185. [DOI] [PubMed] [Google Scholar]
- 51.Oger, P., and S. K. Farrand. 2002. Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum-sensing activator gene traR. J. Bacteriol. 1841121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Martín, J., and M. Espinosa. 1993. Protein-induced bending as a transcriptional switch. Science 260805-807. [DOI] [PubMed] [Google Scholar]
- 53.Pérez-Martín, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362448-450. [DOI] [PubMed] [Google Scholar]
- 55.Piper, K. R., S. Beck von Bodman, I. Hwang, and S. K. Farrand. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 321077-1089. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, W. P., M. E. Tate, and A. Kerr. 1977. Agrocin 84 is a 6-N-phosphoramidate of an adenine nucleotide analogue. Nature 265379-380. [DOI] [PubMed] [Google Scholar]
- 57.Rossignol, G., and P. Dion. 1985. Octopine, nopaline, and octopinic acid utilization in Pseudomonas. Can. J. Microbiol. 3168-74. [Google Scholar]
- 58.Ryder, M. H., M. E. Tate, and G. P. Jones. 1984. Agrocinopine A, a tumor-inducing plasmid-coded enzyme product, is a phosphodiester of sucrose and l-arabinose. J. Biol. Chem. 2599704-9710. [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Schell, J., M. Van Montague, M. De Beuckeleer, M. De Block, A. Depicker, M. De Wilde, G. Engler, C. Genetello, J.-P. Hernalsteens, M. Holsters, J. Seurinck, B. Silva, F. Van Vliet, and R. Villaroel. 1979. Interaction and DNA transfer between Agrobacterium tumefaciens, the Ti plasmid and the host. Proc. R. Soc. London Ser. B 204251-266. [DOI] [PubMed] [Google Scholar]
- 61.Schleif, R. 1992. DNA looping. Annu. Rev. Biochem. 61199-223. [DOI] [PubMed] [Google Scholar]
- 62.Schumacher, M. A., K. Y. Choi, H. Zalkin, and R. G. Brennan. 1994. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science 266763-770. [DOI] [PubMed] [Google Scholar]
- 63.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in vivo and in vitro manipulations of gram-negative bacteria. Springer-Verlag KG, Berlin, Germany.
- 64.Slota, J. E., and S. K. Farrand. 1982. Genetic isolation and physical characterization of pAgK84, the plasmid responsible for agrocin 84 production. Plasmid 8175-186. [DOI] [PubMed] [Google Scholar]
- 65.Spronk, C. A., G. E. Folkers, A.-M. Noordman, R. Wechselberger, N. van den Brink, R. Boelens, and R. Kaptein. 1999. Hinge-helix formation and DNA bending in various lac repressor-operator complexes. EMBO J. 186472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tate, M. E., J. G. Ellis, A. Kerr, J. Tempé, K. E. Murray, and K. J. Shaw. 1982. Agropine: a revised structure. Carbohydr. Res. 104105-120. [Google Scholar]
- 67.Tate, M. E., P. J. Murphy, W. P. Roberts, and A. Kerr. 1979. Adenine N6-substituent of agrocin 84 determines its bacteriocin-like specificity. Nature 280697-699. [DOI] [PubMed] [Google Scholar]
- 68.Tempé, J., P. Guyon, D. Tepfer, and A. Petit. 1979. The role of opines in the ecology of the Ti plasmids of Agrobacterium. Elsevier/North Holland Biomedical Press, Amsterdam, The Netherlands.
- 69.Tremblay, G., R. Gagliardo, W. S. Chilton, and P. Dion. 1987. Diversity among opine-utilizing bacteria: identification of coryneform isolates. Appl. Environ. Microbiol. 531519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, L., J. D. Helmann, and S. C. Winans. 1992. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell 69659-667. [DOI] [PubMed] [Google Scholar]
- 71.Watson, B., T. C. Currier, M. P. Gordon, M.-D. Chilton, and E. W. Nester. 1975. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winans, S. C. 1990. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J. Bacteriol. 1722433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, L. Chen, G. E. Wood, Y. Chen, L. Woo, J. P. Kitajima, V. K. Okura, et al. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
- 74.Wu, H.-M., and D. M. Crothers. 1984. The locus of sequence-directed and protein induced DNA bending. Nature 308509-513. [DOI] [PubMed] [Google Scholar]
- 75.Yuan, Z.-C., R. Zaheer, R. Morton, and T. M. Finan. 2006. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 342686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zanker, H., J. von Lintig, and J. Schröder. 1992. Opine transport genes in the octopine (occ) and nopaline (noc) catabolic region in Ti plasmids of Agrobacterium tumefaciens. J. Bacteriol. 174841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng, X., and H. H. Saxild. 1999. Identification and characterization of a DeoR-specific operator sequence essential for induction of dna-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 1811719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zwieb, C., J. Kim, and S. Adhya. 1989. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 3606-611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.