Abstract
During systemic infection, Staphylococcus aureus acquires nutrient iron from heme, the cofactor of vertebrate myoglobin and hemoglobin. Upon exposure to heme, S. aureus up-regulates the expression of the heme-regulated transporter, HrtAB. Strains lacking hrtAB exhibit increased sensitivity to heme toxicity, and upon heme exposure they elaborate a secreted protein response that interferes with the recruitment of neutrophils to the site of infection. Taken together, these results have led to the suggestion that hrtAB encodes an efflux system responsible for relieving the toxic effects of accumulated heme. Here we extend these observations by demonstrating that HrtA is the ATPase component of the HrtAB transport system. We show that HrtA is an Mn2+/Mg2+-dependent ATPase that functions at an optimal pH of 7.5 and exhibits in vitro temperature dependence uncommon to ABC transporter ATPases. Furthermore, we identify conserved residues within HrtA that are required for in vitro ATPase activity and are essential for the functionality of HrtA in vivo. Finally, we show that heme induces an alteration in the gene expression pattern of S. aureus ΔhrtA, implying the presence of a novel transcriptional regulatory mechanism responsible for the previously described immunomodulatory characteristics of hrtA mutants exposed to heme.
The important human pathogen Staphylococcus aureus is the cause of significant morbidity and mortality worldwide (13). A commensal of the skin and anterior nares, S. aureus is able to breach sites of colonization, resulting in infection of a number of anatomical locations (9, 49). These pathologies range in severity and invasiveness from skin and soft tissue infections to endocarditis, osteomyelitis, pneumonia, and meningitis. The importance of S. aureus as a pathogen is highlighted by the emergence of strains resistant to a number of commonly used antibiotics (15, 16).
Essential to the ability of bacterial pathogens to proliferate and cause disease is their capacity to acquire nutrients from the host in the context of infection. Equally essential to the growth and viability of such microorganisms is their ability to export toxic metabolic by-products of nutrient acquisition into the surrounding milieu. In gram-positive bacteria, these import and export processes are accomplished through cell wall- and membrane-localized transport systems. The functions of these systems often hinge on a conserved class of ATP-cleaving transport factors known as ATP-binding cassette (ABC) transporters. Typically composed of an integral membrane permease which forms a transport substrate channel and a permease-bound peripheral membrane ATPase, ABC transporters couple the cleavage of ATP to the translocation of small molecules, ions, and proteins across the cell membrane (4, 32, 51).
One nutrient which S. aureus must acquire is iron, the cofactor for a number of essential metabolic enzymes. In the context of infection, the low concentration of free iron within mammalian tissues limits the growth of S. aureus (5). However, iron is abundant in the host in the form of the metalloporphyrin heme, the cofactor of hemoproteins such as hemoglobin and myoglobin. To satisfy its iron requirement during infection, S. aureus utilizes heme from hemoglobin as an iron source. S. aureus is hypothesized to access hemoglobin through the hemolysin-mediated lysis of erythrocytes and is known to import host heme from hemoglobin into the staphylococcal cytoplasm through the actions of cell wall, membrane, and cytoplasmic members of the Isd and Hts systems (33, 40). The central conduits for heme import into the staphylococcal cytoplasm are the IsdDEF and HtsABC complexes, both of which are ABC transporter heme import systems. S. aureus strains with mutations in either system display decreased virulence in animal models of infection, highlighting the importance of heme iron acquisition during infection (14, 33, 40).
Although heme acquisition satisfies the iron requirement of invasive S. aureus, heme is toxic to S. aureus due to its ability to generate reactive oxygen species which damage lipids, proteins, and nucleic acids (12). S. aureus is sensitive to heme toxicity but is able to adapt to growth in the presence of toxic heme concentrations by sensing heme through the HssRS two-component system (42, 45). Upon sensing heme, HssRS induces the expression of HrtAB, a predicted ABC transporter efflux system essential for adaptation to heme toxicity (14). Importantly, S. aureus ΔhrtA displays a liver-specific virulence phenotype in a murine abscess model of infection, a phenotype which is associated with heme-induced overexpression and secretion of virulence factors in this heme-sensitive strain (45). These observations further underscore the importance of proper heme metabolism to staphylococcal physiology and virulence.
Heme acquisition and subsequent protection against heme toxicity are dependent on ABC transporter systems. In particular, the HrtAB system appears to play a critical role at the point of transition between the use of heme as an iron source and the avoidance of heme toxicity. Although the importance of HrtAB in staphylococcal pathogenesis and in the response of S. aureus to heme has been explored, neither the functional details of this system nor a role for its catalytic activity in the response of S. aureus to heme has been demonstrated. In addition, although the transcripts for a few important virulence factors are known to be altered in S. aureus ΔhrtA exposed to heme, the global transcriptional response of S. aureus ΔhrtA to heme has not been reported. Here, we show that the predicted ABC transporter ATPase HrtA is capable of catalyzing the cleavage of ATP in vitro, an activity which is dependent on pH, substrate concentration, temperature, and divalent metal cation. We also show that HrtA ATPase activity is dependent on a number of conserved residues predicted to be essential for nucleotide binding and hydrolysis. In addition, we demonstrate that the catalytic activity of HrtA is essential for the ability of S. aureus to proliferate in the presence of toxic concentrations of heme. Finally, we show that upon exposure to heme, S. aureus ΔhrtA induces a dramatic change in the levels of over 500 transcripts, including those for an array of virulence factors, characterized and hypothetical transcriptional regulators, and a variety of stress response systems.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Staphylococcus aureus strain Newman (11) and its derivatives were used in all experiments (Table 1). Roswell Park Memorial Institute (RPMI) medium plus 1% Casamino Acids (for samples for microarray analysis) or tryptic soy broth (for all other experiments and all genetic manipulations) was used for the growth of S. aureus; for plasmid selection in S. aureus, chloramphenicol was used at a concentration of 10 μg/ml. Luria broth (for genetic manipulations) and Terrific broth (for protein expression) were used for the growth of Escherichia coli; for plasmid selection in E. coli, ampicillin was used at a concentration of 100 μg/ml (34).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Reference or source | Comment |
|---|---|---|
| S. aureus strains | ||
| RN4220 | 30 | Restriction-deficient strain that accepts DNA from E. coli |
| Newman | 11 | S. aureus clinical isolate |
| Newman ΔhrtA | 45 | Isogenic Newman hrtA mutant |
| E. coli strains | ||
| DH5α | Strain for use in all cloning steps | |
| BL21(DE3) | Novagen | Recombinant protein expression strain |
| E. coli plasmids | ||
| pET15b | Novagen | E. coli expression construct with N-terminal His6 tag |
| pET15b.hrtA | This study | Expression construct for N-terminally His6-tagged full-length HrtA |
| pET15b.hrtA:K45A | This study | pET15b.hrtA derivative with K45A mutation |
| pET15b.hrtA:R76A | This study | pET15b.hrtA derivative with R76A mutation |
| pET15b.hrtA:G145A | This study | pET15b.hrtA derivative with G145A mutation |
| pET15b.hrtA:G145T | This study | pET15b.hrtA derivative with G145T mutation |
| pET15b.hrtA:E167Q | This study | pET15b.hrtA derivative with E167Q mutation |
| S. aureus plasmids | ||
| pOS1 | 36 | Empty E. coli/S. aureus shuttle vector |
| phrtA | 45 | ΔhrtA complementation construct containing hrtA under control of its native promoter |
| phrtA-myc | This study | ΔhrtA complementation construct containing c-myc-tagged hrtA under control of its native promoter |
| phrtA-myc:K45A | This study | phrtA-myc derivative with K45A mutation |
| phrtA-myc:R76A | This study | phrtA-myc derivative with R76A mutation |
| phrtA-myc:G145A | This study | phrtA-myc derivative with G145A mutation |
| phrtAmyc:G145T | This study | phrtA-myc derivative with G145T mutation |
| phrtAmyc:E167Q | This study | phrtA-myc derivative with E167Q mutation |
Genetic manipulations in S. aureus.
All plasmids were first electroporated into the restriction-deficient primary recipient RN4220 (30), after which they were electroporated into appropriate electrocompetent secondary recipients (35).
S. aureus microarray analysis.
Fifteen-hour cultures of bacteria were subcultured 1:100 into 15 ml of fresh RPMI plus 1% Casamino Acids with or without 1 μM heme (Fluka BioChemika) in 50-ml conical tubes. Cultures were grown at 37°C with shaking at 180 rpm until the bacterial density reached an A600 of 0.25. RNA was reverse transcribed, cDNA fragmented, 3′ biotinylated, and hybridized to commercially available S. aureus GeneChips following the manufacturer's recommendations for antisense prokaryotic arrays (Affymetrix, Santa Clara, CA). GeneChips were washed, stained, and scanned as previously described (3).
Purification of HrtA and HrtA mutants.
The full-length hrtA open reading frame was inserted between the NdeI and BamHI sites of pET15b, creating pET15b.hrtA for the expression of an N-terminal hexahistidine-tagged HrtA. Pfu mutagenesis (48) was used to create expression constructs for the mutants HrtA:K45A, HrtA:R76A, HrtA:G145A, HrtA:G145T, and HrtA:E167Q. All mutants were verified by sequencing (Vanderbilt University DNA sequencing facility). For all protein expression, E. coli BL-21(DE3) harboring each plasmid was subcultured 1:100 from 15-h cultures into Terrific broth at 37°C with shaking at 225 rpm until the A600 of the culture reached 0.4. The growth temperature was then switched to 16°C for 1 h, and expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside (IPTG) (0.25 mM). After an additional 20 h of growth at 16°C, bacteria were harvested and recombinant proteins were purified by nickel affinity chromatography using Ni-nitrilotriacetic acid Superflow (Qiagen), following the manufacturer's recommendations. Purified proteins were dialyzed against dialysis buffer (10 mM Tris [pH 7.5], 200 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 50% glycerol) overnight and stored at −20°C. The protein concentration was determined by the Bio-Rad protein assay (Bio-Rad). For removal of the N-terminal hexahistidine tag from HrtA, restriction grade thrombin protease (Novagen) was utilized according to the manufacturer's instructions, and cleaved HrtA was stored at −20°C.
ATPase activity assays.
ATPase activities were determined using an Innova Biosciences colorimetric ATPase assay system according to the manufacturer's recommendations. Briefly, recombinant HrtA was diluted to a final concentration of 0.5 μM in 80 μl of 50 mM Tris (pH 7.5, or another pH as indicated), 2.5 mM MgCl2 (or another divalent cation if indicated), and 0.5 mM ATP (or another concentration as indicated). All assays except the temperature dependence assay were carried out at 20°C for maximal ATPase activity. At 3, 10, and 20 min after the addition of ATP (or at other time points as indicated), 20 μl of Innova Biosciences Gold Mix (1:100 dilution of Accelerator into PiColorLock Gold) was added to the HrtA reaction mixture. Quenched reaction mixtures were incubated for 2 min, after which 10 μl of Stabilizer reagent was added. A650 readings were taken on a Cary 50 MPR microplate reader coupled to a Cary 50 Bio UV-visible spectrophotometer (Varian, Inc.).
Complementation constructs.
A plasmid containing a copy of wild-type hrtA under the control of its native promoter (phrtA) was created as described previously (45). A C-terminal c-myc-tagged hrtA under the control of its native promoter was created by PCR, amplifying hrtA from phrtA using a 5′ primer within the hrtA promoter and a 3′ primer matching the 3′ end of hrtA and including the coding sequence for the c-myc epitope (EQKLISEEDL). Amplified DNA was inserted into the E. coli/S. aureus shuttle vector pOS1 (36), creating phrtA-myc. The hrtA-myc K45A, R76A, G145A, G145T, and E167Q mutations were introduced as described above for the generation of E. coli HrtA mutant expression constructs, generating phrtA-myc:K45A, phrtA-myc:R76A, phrtA-myc:G145A, phrtA-myc:G145T, and phrtA-myc:E167Q. Mutations were confirmed and secondary mutations were ruled out by sequencing. Expression of tagged HrtA and HrtA mutants in S. aureus was tested by preparing bacterial extracts and immunoblotting as follows. Fifteen-milliliter cultures of bacteria grown in tryptic soy broth plus 5 μM heme for 15 h were centrifuged, and pellets were washed with wash buffer (50 mM Tris [pH 7.5], 150 mM NaCl). Bacteria were centrifuged again and resuspended in TSM (100 mM Tris [pH 7.0], 500 mM sucrose, 10 mM MgCl2) with 20 μg/ml lysostaphin and incubated at 37°C for 30 min, and protoplasts were harvested by centrifugation. Protoplasts were resuspended in 800 μl of lysis buffer (wash buffer containing one tablet of complete EDTA-free protease inhibitor [Roche] per 10 ml) and were sonicated. Insoluble material was removed by centrifugation at 16,000 × g for 20 min, and the supernatant was analyzed. Thirty micrograms of supernatant material was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and blotted with sc-789 polyclonal rabbit anti-c-myc primary (Santa Cruz Biotechnology, Santa Cruz, CA) and AlexaFluor-680-conjugated anti-rabbit secondary (Invitrogen) antibodies. Membranes were dried and scanned using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
S. aureus growth kinetics.
Fifteen-hour cultures of S. aureus grown in the presence of 2 μM heme were diluted 1:75 into 150 μl of fresh medium containing 10 μM heme (Fluka BioChemika) in triplicate on a 96-well round-bottom cell culture plate. Cells were grown at 37°C with shaking at 180 rpm, and absorbance values were determined at the indicated times after inoculation. All spectrophotometry was performed using a Cary 50 MPR microplate reader coupled to a Cary 50 Bio UV-visible spectrophotometer (Varian, Inc.).
RESULTS
HrtA is a staphylococcal ATPase.
We have previously identified HrtA as a cytoplasmic protein which is increased in abundance 45-fold upon exposure of S. aureus to high concentrations of heme (14). Although previous studies have demonstrated a critical role for HrtA in the protection of S. aureus from heme toxicity and have elucidated the mechanism by which expression of HrtA is controlled, no enzymatic activity has been shown for this protein (42, 45). HrtA is annotated as an ABC transporter ATPase, and we hypothesized that HrtA, cooperating with the membrane-localized permease HrtB, couples the cleavage of ATP to the export of a molecule responsible for the toxicity of heme.
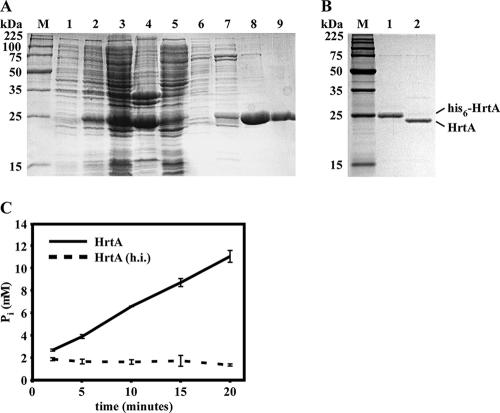
Consistent with its assignment as an ABC transporter ATPase, purified recombinant HrtA catalyzed the liberation of inorganic phosphate from ATP (Fig. 1). Importantly, heat-inactivated HrtA failed to induce detectable ATPase activity, indicating that inorganic phosphate accumulation was the result of the catalytic activity of HrtA and not phosphate contamination in our HrtA preparations (Fig. 1C). Based on these findings, we conclude that HrtA is capable of catalyzing the cleavage of ATP to ADP and PO43−.
FIG. 1.
HrtA purification and ATPase activity. (A) Coomassie blue-stained SDS-polyacrylamide gel showing purification of S. aureus His6-HrtA expressed in E. coli. Lanes: M, molecular weight ladder; 1, E. coli before IPTG induction; 2, E. coli after IPTG induction; 3, total cell lysate; 4, insoluble pellet obtained after French press treatment and centrifugation; 5, soluble extract used for purification; 6 to 9, fractions obtained by elution of material bound to Ni-nitrilotriacetic acid with imidazole (10 mM, 50 mM, 100 mM, and 500 mM). (B) SDS-PAGE showing removal of the hexahistidine tag from His6-HrtA by thrombin protease. Lanes: M, molecular weight ladder; 1, 1 μg purified His6-HrtA; 2, 1 μg HrtA with hexahistidine tag removed by thrombin cleavage. (C) Time course analysis of ATP hydrolysis by HrtA. HrtA or heat-inactivated HrtA [HrtA (h.i.)] was incubated at 20°C in the presence of ATP, and release of inorganic phosphate was measured at the indicated time points. Error bars indicate standard deviations.
HrtA ATPase activity is influenced by ATP concentration, temperature, pH, and metal cofactors.
ABC transporter ATPases have been shown to have enzymatic activities which are influenced by multiple physicochemical conditions (19, 37). In order to assess the conditions under which HrtA displays maximum enzymatic activity and as a means of testing whether the phosphate-liberating activity of our HrtA preparations is sensitive to perturbations which would be expected to alter the activity of an ATPase, we tested the effects of ATP concentration, temperature, pH, and metal cofactor on the activity of HrtA.
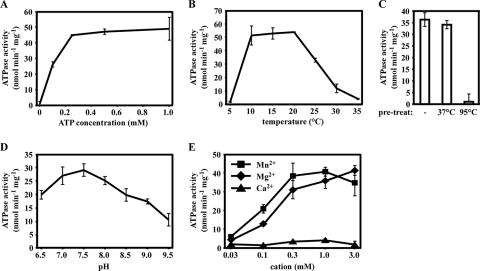
HrtA ATPase activity reached a plateau at 0.25 mM ATP in vitro (Fig. 2A). Interestingly and unexpectedly, the enzymatic activity of HrtA reached a peak at 10 to 20°C and not 37°C (Fig. 2B). This observation is not likely a result of enzyme instability at this temperature, as preincubation of HrtA at 37°C for 20 min failed to alter the activity of this ATPase at 20°C (Fig. 2C). Therefore, it is possible that HrtA displays an unusual temperature dependence in vitro that is not common to ABC transporter ATPases. HrtA reached maximal catalytic activity within the range of pH 7.0 to 8.0 (Fig. 2D), a range consistent with the near-neutral pH of the gram-positive bacterial cytoplasm (8). Divalent metal cations are usually critical for the catalytic activity of ATPases. We found that magnesium (Mg2+) and manganese (Mn2+) both supported the catalytic activity of HrtA at a number of tested concentrations, with Mn2+ supporting slightly elevated HrtA activity over Mg2+ at low concentrations (Fig. 2E). In contrast, calcium (Ca2+) did not support ATPase activity at any concentration tested (Fig. 2E). These results demonstrate that HrtA is an Mn2+/Mg2+-dependent ATPase which functions at an optimal pH of 7.0 to 8.0 and displays an unusual in vitro temperature sensitivity uncommon to ABC transporter ATPases.
FIG. 2.
ATPase activity of HrtA is influenced by various physicochemical conditions. (A) Concentration of ATP required for saturation of enzymatic activity. HrtA (0.5 μM) was incubated with increasing concentrations of ATP at 20°C, and ATPase activity was measured. (B) ATPase activity determination at different temperatures. (C) Effect of temperature pretreatment on HrtA activity. HrtA was incubated at the indicated temperature for 20 min, and ATPase activity was determined at 20°C. (D) Influence of pH on HrtA ATPase activity. (E) Effect of divalent metal cations on HrtA activity. HrtA ATPase activity was determined with the indicated concentration of MnCl2, MgCl2, or CaCl2. For all experiments, the average of triplicate ATPase activities is indicated; error bars represent one standard deviation from the mean.
Identification of ATP binding and cleavage residues required for in vitro HrtA ATPase activity.
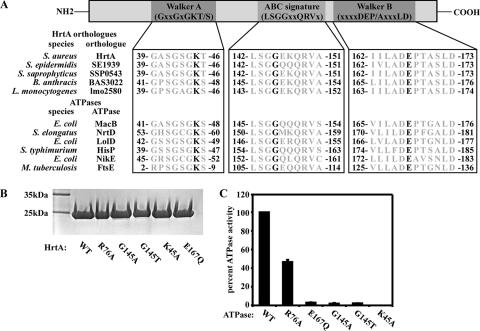
ABC transporter ATPases are characterized by the presence of highly conserved sequence motifs involved in ATP binding and hydrolysis (19, 37). These include the Walker A motif (GxxGxGKS/T), the Walker B motif (DEP/AxxxLD), and the ABC signature sequence (LSGGxxQRV) (47). The signature motif is specific to nucleotide-binding domains of ABC-type transport ATPases, while the Walker A and B motifs are common to most ATPases involved in the binding and hydrolysis of nucleotides (25). Walker A motifs bind ATP through a conserved lysine residue, while Walker B motifs employ a conserved glutamic acid residue to hydrolyze ATP. In addition, the signature motif contains an absolutely conserved LSGGQ motif required for ATPase activity (25). In HrtA, all three sequence motifs can be detected (Fig. 3A). To determine whether these sequences are critical for the ability of HrtA to bind and cleave ATP, highly conserved residues within each motif (K45 of Walker A, G145 of ABC signature, E167 of Walker B) as well as a residue outside of these motifs (R76) were individually mutated, and HrtA mutants were purified and tested for ATPase activity (Fig. 3B and C). Importantly, mutation of R76 reduced the rate of ATP cleavage by HrtA to about 50% of that observed for wild-type HrtA, while mutation of residues K45, G145, or E167 eliminated the ability of HrtA to cleave ATP (Fig. 3C). These data indicate that conserved residues within nucleotide-binding and hydrolysis motifs of HrtA are essential for ATP cleavage. The fact that mutation of a residue outside of the conserved nucleotide-binding motif also altered the rate of ATP hydrolysis by HrtA suggests that nonconserved regions of HrtA are involved in catalysis in vitro.
FIG. 3.
Residues essential for HrtA ATPase activity. (A) Representation of a prototypical ABC transporter ATPase subunit and sequence alignment of conserved domains. Walker A, ABC signature, and Walker B motifs can be detected within HrtA orthologues and other characterized bacterial ABC transporter ATPases. Conserved residues selected for mutagenesis (Lys 45, Gly 145, and Glu 167 of S. aureus HrtA) are shown in black. (B) Coomassie blue-stained SDS-polyacrylamide gel showing recombinant purified wild-type HrtA (WT) and HrtA mutants with mutations in residues within motifs predicted to be involved in nucleotide binding or hydrolysis (G145A, G145T, K45A, E167Q) or a residue outside of such motifs (R76A). (C) Relative ATPase activities of HrtA mutants compared to the wild type. ATPase activities of recombinant HrtA and HrtA site mutants described for panel B were determined and are expressed as the percentage of ATPase activity exhibited by wild-type HrtA. The average percentage of triplicate ATPase activities is indicated; error bars represent one standard deviation from the mean.
Residues required by HrtA for in vitro ATPase activity are required for the adaptation of S. aureus to heme toxicity.
We next tested whether HrtA residues which are required for ATPase activity in vitro are required for the functionality of HrtAB in vivo. To this end, we tested the ability of HrtA point mutants provided in trans to rescue the heme-sensitive phenotype of S. aureus ΔhrtA. Full-length hrtA under the control of the hrtAB promoter was tagged with a C-terminal c-myc epitope tag, inserted into an S. aureus complementation vector, and independently mutated at the four residues analyzed for in vitro ATPase activity (Fig. 3C). S. aureus ΔhrtA was then transformed with these plasmids, and the resulting strains were analyzed for their ability to adapt to heme toxicity by growth curve analysis following subculture into 10 μM heme.
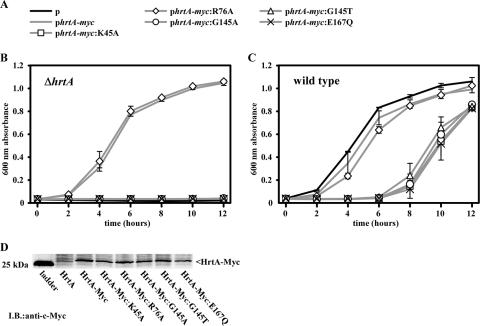
While S. aureus ΔhrtA cannot proliferate in the presence of 10 μM heme, S. aureus ΔhrtA containing a copy of wild-type hrtA-myc provided in trans can grow in 10 μM heme (Fig. 4A and B). Significantly, none of the strains containing hrtA point mutants which were unable to catalyze the cleavage of ATP in vitro were able to rescue the heme-sensitive phenotype of S. aureus ΔhrtA (Fig. 4B). Furthermore, hrtA-myc:R76A, which contains a mutation outside of the conserved nucleotide-binding and hydrolysis motifs, was able to complement S. aureus lacking hrtA (Fig. 4B). Notably, this mutant was also able to cleave ATP in vitro, although at a reduced rate compared to the wild type (Fig. 3C). The fact that full complementation occurred in vivo with a protein that exhibits half the rate of wild-type HrtA ATPase activity in vitro suggests that either this reduction is not great enough to significantly reduce the function of HrtAB in vivo or that mutation of R76 does not alter the ATPase activity of HrtA in vivo. Importantly, all of the analyzed HrtA-myc point mutants were expressed by S. aureus to a similar extent as wild-type HrtA-myc, as analyzed by immunoblotting against the c-Myc epitope tag of these proteins (Fig. 4D).
FIG. 4.
HrtA residues essential for adaptation of S. aureus to heme toxicity. (A) Symbols for plasmids used to transform strains for panels B and C. Plasmids include an empty vector (black line), a plasmid encoding wild-type hrtA-myc (gray line with no symbol), or plasmids encoding hrtA-myc mutants (gray lines with symbols). (B) S. aureus ΔhrtA harboring the plasmids described in panel A was grown overnight in medium containing 2 μM heme and were subcultured into medium containing 10 μM heme. Bacterial growth was tracked by measuring the A600 of triplicate cultures. Data points represent averages of triplicate measurements; error bars represent one standard deviation from the mean. (C) Wild-type S. aureus harboring the plasmids shown in panel A were grown and analyzed as for panel B. (D) Immunoblot (I.B.) against cytoplasmic extracts of S. aureus ΔhrtA expressing C-terminally c-Myc-tagged wild-type HrtA-myc or HrtA-myc mutants. Results are representative of triplicate independent experiments.
Last, we analyzed the ability of catalytically inactive HrtA-myc point mutants to interfere with the normal functioning of HrtAB in vivo. We transformed wild-type S. aureus with the plasmids described above encoding HrtA-myc and its respective point mutants and analyzed the ability of these strains to adapt to heme toxicity. While HrtA-myc and HrtA-myc:R76A had a minimal impact on the ability of wild-type S. aureus to adapt to heme toxicity, all of the HrtA-myc mutants which were unable to catalyze the cleavage of ATP in vitro displayed statistically significant dominant-negative activity in vivo (Fig. 4C). This indicates that HrtA mutants which are unable to cleave ATP interfere with the functioning of HrtAB in vivo.
Staphylococci lacking hrtA induce a dramatic transcriptional response to heme.
HrtAB protects S. aureus from the toxic effects of high concentrations of heme (14, 45). Accordingly, strains of S. aureus lacking the ability to either sense heme (through mutation of the hrtAB promoter-activating response regulator hssR) or respond to growth in heme (through mutation of hrtA) display enhanced sensitivity to heme toxicity (14, 42, 45). Furthermore, S. aureus ΔhrtA increases the expression of a number of immunomodulatory toxins in response to heme (45). In an effort to understand the gene-regulatory response of an hrtA mutant to growth in heme, we performed genome-wide microarray analysis on wild-type and ΔhrtA S. aureus grown in the presence and absence of a subinhibitory concentration of heme to identify transcripts with altered abundances. These experiments were performed at 1 μM heme to permit growth of S. aureus ΔhrtA.
While wild-type S. aureus did not exhibit a significant transcriptional response to 1 μM heme, over 500 transcripts were changed in abundance greater than twofold in S. aureus ΔhrtA exposed to this same concentration of heme (see Tables S1 and S2 in the supplemental material). Comparison of the global expression profile of heme-exposed S. aureus ΔhrtA with the global expression profile of S. aureus upon cold shock, heat shock, SOS induction, stringent response induction, acid shock, alkaline shock, Mn2+ starvation, or Zn2+ starvation indicated that the stringent response most closely resembles the heme response of S. aureus ΔhrtA (37.4% similarity) (Table 2) (1, 7). The bacterial stringent response is induced under carbon or amino acid insufficiency through ribosome stalling, which leads to an increase in the intracellular concentration of (p)ppGpp (6). This occurs through the induction of the stringent response regulatory factor RelA (SACOL1689), the transcript of which is up-regulated upon treatment of S. aureus with the stringent response-inducing antimicrobial agent mupirocin and is increased 9.2-fold in S. aureus ΔhrtA exposed to heme (see Table S1 in the supplemental material) (1). A number of other stringent-response-induced transcripts are increased in S. aureus ΔhrtA upon growth with heme, including those encoding virulence factors such as the transpeptidase sortase A (7.4-fold in stringent response and 9.5-fold in S. aureus ΔhrtA plus heme) and fibrinogen-binding protein (7.9-fold in stringent response and 32.3-fold in S. aureus ΔhrtA plus heme) (see Table S1 in the supplemental material) (1). These results indicate that in S. aureus ΔhrtA, heme induces a response that displays some similarities to the S. aureus stringent response.
TABLE 2.
Correlation between heme-regulated genes in S. aureus ΔhrtA and stress responses
Consistent with previous findings, a dramatic increase in expression of a number of transcripts coding for virulence factors was observed in S. aureus ΔhrtA exposed to heme (Table 3). These include the transcripts for 8 of the 11 staphylococcal superantigen-like exotoxins encoded within the SaPI pathogenicity island in strain Newman (NWMN_0388, NWMN_0389, NWMN_0392, NWMN_0393, NWMN_0394, NWMN_0395, NWMN_0396, and NWMN_0397) as well as two predicted exotoxins located outside of this locus (NWMN_1077 and NWMN_1503) (Table 3) (2). In addition, the expression of a number of pore-forming toxins was dramatically repressed in S. aureus ΔhrtA exposed to heme. These include the transcripts encoding all three of the gamma-hemolysin subunits (SACOL2419, SACOL2421, and SACOL2422) as well as the transcript for a leukotoxin (SACOL1637) (Table 3). Only one characterized class of cell wall-anchored virulence factors, the fibronectin- and fibrinogen-binding proteins (fnbA and efb), were differentially expressed within mutant cells. This transcriptional response appears to be specific to S. aureus ΔhrtA under heme stress, as the wild type grown in up to 20 μM heme did not exhibit a change in expression of secreted proteins as judged by SDS-PAGE analyses (data not shown). Together, these data indicate that upon exposure to heme, S. aureus ΔhrtA undergoes a shift in toxin expression from cell-lytic factors to immunomodulatory factors.
TABLE 3.
Virulence gene transcripts regulated by heme in S. aureus ΔhrtA
| Locusa | Fold change in hemeb | Gene namec | Description of product |
|---|---|---|---|
| NWMN_0388 | 11.3 | set1nm | Staphylococcal superantigen-like toxin NM1 |
| NWMN_0389 | 6.9 | set2nm | Staphylococcal superantigen-like toxin NM2 |
| NWMN_0392 | 16.1 | set5nm | Staphylococcal superantigen-like toxin NM5 |
| NWMN_0393 | 8.9 | set6nm | Staphylococcal superantigen-like toxin NM6 |
| NWMN_0394 | 14.0 | set7nm | Staphylococcal superantigen-like toxin NM7 |
| NWMN_0395 | 6.7 | set8nm | Staphylococcal superantigen-like toxin NM8 |
| NWMN_0396 | 9.4 | set9nm | Staphylococcal superantigen-like toxin NM9 |
| NWMN_0397 | 6.9 | set10nm | Staphylococcal superantigen-like toxin NM10 |
| NWMN_1077 | 6.0 | Staphylococcal superantigen-like toxin | |
| NWMN_1503 | 3.7 | Enterotoxin type A, putative | |
| SACOL0857 | 20.4 | Staphylocoagulase precursor, putative | |
| SACOL1168 | 32.3 | fbp | Fibrinogen-binding protein |
| SACOL1169 | 29.3 | Fibrinogen-binding protein precursor-related protein | |
| SACOL1924 | 2.4 | Toxin-exporting ABC transporter, permease/ATP-binding protein | |
| SACOL2511 | 4.8 | fnbA | Fibronectin-binding protein A |
| SACOL2584 | 23.7 | isaA | Immunodominant antigen A |
| SACOL0137 | −11.5 | cap5B | Capsular polysaccharide biosynthesis protein |
| SACOL0138 | −11.2 | cap5C | Capsular polysaccharide biosynthesis protein |
| SACOL0143 | −8.3 | cap5H | Capsular polysaccharide biosynthesis protein |
| SACOL1637 | −42.1 | lukD | Leukotoxin LukD |
| SACOL1865 | −20.8 | splE | Serine protease SplE |
| SACOL1866 | −22.2 | splD | Serine protease SplD |
| SACOL1867 | −9.1 | splC | Serine protease SplC |
| SACOL1872 | −12.3 | epiE | Epidermin immunity protein F |
| SACOL1873 | −17.3 | epiF | Epidermin immunity protein F |
| SACOL2007 | −8.7 | Peptidase, M20/M25/M40 family | |
| SACOL2419 | −52.8 | hlgA | Gamma hemolysin, component A |
| SACOL2421 | −80.9 | hlgC | Gamma hemolysin, component C |
| SACOL2422 | −42.4 | hlgB | Gamma hemolysin, component B |
All genes with SACOL designation are from S. aureus strain COL; because of strain specificity exhibited by toxins, all S. aureus superantigen-like toxins are referenced with S. aureus strain Newman locus designations (NWMN).
All virulence gene transcripts with at least twofold changes in S. aureus ΔhrtA exposed to heme compared to S. aureus ΔhrtA alone which are statistically significant are shown. Underlining indicates that in S. aureus ΔhrtA alone, the transcript abundance approaches the lower limit of sensitivity and thus values may not be accurate. Bold indicates that in S. aureus ΔhrtA alone, no transcript was detected and thus values may not be accurate.
Transcripts with no gene name have no designated name in the S. aureus COL genome annotation.
In addition to toxins, a wide variety of other transcripts are differentially expressed in S. aureus ΔhrtA exposed to heme (see Tables S1 and S2 in the supplemental material). These include but are not limited to transcripts for metabolic enzymes, transport proteins, and hypothetical factors. Furthermore, a number of transcripts encoding transcriptional regulatory factors are dramatically altered in expression in S. aureus ΔhrtA exposed to heme, providing a possible mechanistic explanation for the broad changes in gene transcription seen in heme-exposed S. aureus ΔhrtA. Regulatory systems with increased expression in S. aureus ΔhrtA exposed to heme include a number of characterized and uncharacterized two-component systems (including hssS, 4.2-fold; lytS, 8.1-fold; lytR, 20.5-fold; vraS, 10.4-fold; SACOL1354, 6.6-fold; SACOL1905, 9.1-fold; SACOL2645-6, 7.1-fold), known transcriptional repressors (fur, 9.5-fold; czrA, 42.9-fold; sirR, 5.2-fold), and other, uncharacterized putative transcriptional regulators, some of which display dramatic alterations in expression (SACOL0403, 21.5-fold decrease; SACOL1904, 24.7-fold increase) (see Tables S1 and S2 in the supplemental material). These data along with previously published findings indicate that heme induces a profound alteration in the gene expression pattern of S. aureus ΔhrtA, possibly through novel transcriptional regulatory mechanisms. This expression change leads to an alteration in virulence gene expression and the hypervirulence of this mutant in a mouse model of systemic infection (45).
DISCUSSION
The import of molecules and ions into the cytoplasm and the export of wastes or toxic compounds across the plasma membrane constitute a tightly controlled process which is essential to the viability of bacteria (19, 22, 31). Many transport pathways in bacteria depend on ABC transporters, which couple the cleavage of ATP to the transport of solutes across the membrane. The gram-positive human pathogen S. aureus encodes over 50 characterized or hypothetical ABC transporters (data not shown), an indication of the importance of this particular transport scheme in this organism. Staphylococcal ABC transporters with described functions are involved in critical processes as diverse as fluoroquinolone resistance (NorA [46, 50]), antimicrobial peptide and/or glycopeptide resistance (GraRS/ApsRS and VraFG [17, 24, 28]), metal ion acquisition (FhuC [41], SirABC [10], and MntABC [20]), molybdate transport (ModABC [29]), and oligopeptide import (Opp-3 [18]). Heme iron acquisition, a process important to the outcome of infection, also depends on ABC transporters (IsdDEF and HtsABC [27, 40]). In addition, the proper metabolism of heme at high concentrations is critically dependent on the ABC transporter HrtAB, a putative efflux pump that is dramatically increased in expression in S. aureus grown in the presence of heme (14, 42, 45).
Here, we have provided insight into the biochemical details of HrtA function and into the gene-regulatory response of S. aureus ΔhrtA to growth in the presence of heme. We demonstrate that HrtA is capable of cleaving ATP in vitro, an activity which is dependent on temperature, pH, substrate concentration, and metal cofactor. We further show that HrtA ATPase activity is sensitive to perturbations in predicted nucleotide-binding residues and that these residues are critical for the ability of S. aureus to adapt to the toxicity of heme. The latter observations demonstrate that the catalytic activity of HrtA and thus the functionality of HrtAB are essential for the survival and growth of S. aureus in the presence of high concentrations of heme (5 μM). The fact that HrtA mutants which are unable to cleave ATP exhibit dominant-negative activity against wild-type HrtAB in vivo suggests that these mutants are capable of associating with endogenous HrtA, HrtB, or both in order to block the normal functioning of HrtAB. ABC transporter permeases typically function as dimers, with one associated ATPase per monomer (19, 37). The dominant-negative activity of HrtA point mutants that we observe is consistent with the known associations that occur between ABC transporter ATPases and their cognate permeases. It is also consistent with the finding that in many ABC transporters, a single functioning ATPase per transporter multimer is insufficient for proper function, but that both ATPases need to be functional (21, 26, 38).
Expression analysis revealed that S. aureus ΔhrtA induces changes in the abundance of over 500 transcripts when exposed to heme, many of which encode important virulence factors, gene-regulatory systems, and as-yet-uncharacterized systems. Interestingly, many (37.4%) S. aureus ΔhrtA heme-induced transcripts are components of the staphylococcal stringent response (Table 2). Consistent with this observation, RelA, the major activator of the stringent response, was up-regulated >9-fold within heme-challenged mutant cells. This observation suggests that heme accumulation redirects transcript synthesis in a manner that promotes up-regulation of alternative transport systems which are major components of the stringent response; this may represent an attempt to rid the cell of heme. Taken together, these observations connect the catalytic activity of HrtAB to the heme toxicity response of S. aureus and provide clues concerning the mechanism of hypervirulence of a strain lacking hrtA.
The toxin transcript expression profile of S. aureus ΔhrtA exposed to heme may reveal new regulatory mechanisms or signals which S. aureus recognizes in order to coordinate a specific toxin expression profile upon exposure to distinct host environments. It is important to note that not only does the total quantity of toxins secreted by S. aureus ΔhrtA exposed to heme appear to be altered (14), but the profile of toxin secretion is dramatically changed to decrease the expression of host-cell-lytic toxins and increase the expression of immunomodulatory toxins such as the staphylococcal superantigen-like toxins. It may be that certain types of stress, such as that experienced by S. aureus ΔhrtA grown in heme, may mimic a physiologically relevant cue which S. aureus senses in order to adjust its toxin expression and secretion profile accordingly. It is tempting to speculate that this stressor is one which represents a signal for invading staphylococci to switch from a cytolytic toxin secretion profile to an immunoevasive response.
The identity of the transport substrate for HrtAB is unknown. We have postulated that HrtAB is capable of exporting heme which accumulates to toxic levels within the cytoplasm of S. aureus, thereby alleviating heme toxicity (45). However, inductively coupled plasma mass spectrometry-based tracking experiments using isotopically labeled heme have not revealed a role for HrtAB in heme efflux from staphylococcal cells (data not shown). Furthermore, it is not clear that heme export would alleviate heme toxicity, especially if the Isd and Hts systems are continually and efficiently importing heme into the cytoplasm and if the cytoplasm is not the compartment in which heme exerts its toxic effects. Biochemical fractionation experiments suggest that most exogenous nondegraded heme accumulates in the S. aureus membrane fraction and not the cytoplasm (39). Based on these observations and speculations, we favor the idea that heme is somehow damaging the S. aureus membrane. Heme is known to damage lipid bilayers through lipid peroxidation and disruption of membrane fluidity (12, 23, 33, 43, 44). We hypothesize that, rather than exporting heme directly, HrtAB may protect against heme-mediated cell damage through the export of as-yet-unidentified toxic compounds which accumulate in S. aureus when this organism encounters heme.
Taken together, these results have established HrtA as the ATPase component of the HrtAB transport system. These observations strengthen our previous hypothesis that HrtAB protects S. aureus from the toxic side effects of heme by connecting the catalytic activity of HrtA to the survival of staphylococci in heme. Based on the observation that strains lacking hrtA exhibit significant transcriptional reprogramming upon exposure to heme, it is possible that small-molecule modifiers of HrtAB or its regulatory system HssRS that work synergistically with host heme during the bloodstream component of staphylococcal infections to attenuate bacterial virulence can be identified.
Supplementary Material
Acknowledgments
We thank members of the Skaar laboratory for critical reading of the manuscript.
This work was supported by Vanderbilt University Medical Center development funds, the Searle Scholars Program, and Public Health Service grant AI69233 from the National Institute of Allergy and Infections Diseases. E.P.S. holds an Investigator in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. D.L.S. was supported by grant T32 HL069765 from the National Institute of Allergy and Infectious Diseases, and V.J.T. was supported by Ruth L. Kirschstein postdoctoral fellowship NRSA AI071487. P.M.D. is supported by University of Nebraska Medical Center development funds and American Heart Association award 0535037N. K.L.A. is supported by American Heart Association predoctoral fellowship 0715547Z.
Footnotes
Published ahead of print on 7 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 1886739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, P., and R. O. Elferink. 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71537-592. [DOI] [PubMed] [Google Scholar]
- 5.Bullen, J. J., and E. Griffiths. 1999. Iron and infection: molecular, physiological and clinical aspects, vol. John Wiley and Sons, New York, NY.
- 6.Cassels, R., B. Oliva, and D. Knowles. 1995. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 1775161-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbin, B. D., E. H. Seeley, A. Raab, J. Feldmann, M. R. Miller, V. J. Torres, K. L. Anderson, B. M. Dattilo, P. M. Dunman, R. Gerads, R. M. Caprioli, W. Nacken, W. J. Chazin, and E. P. Skaar. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319962-965. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creech, C. B., II, T. R. Talbot, and W. Schaffner. 2006. Community-associated methicillin-resistant Staphylococcus aureus: the way to the wound is through the nose. J. Infect. Dis. 193169-171. [DOI] [PubMed] [Google Scholar]
- 10.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 7229-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 695-107. [DOI] [PubMed] [Google Scholar]
- 12.Everse, J., and N. Hsia. 1997. The toxicities of native and modified hemoglobins. Free Radic. Biol. Med. 221075-1099. [DOI] [PubMed] [Google Scholar]
- 13.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 3521436-1444. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 16.Hassan, K. A., R. A. Skurray, and M. H. Brown. 2007. Active export proteins mediating drug resistance in staphylococci. J. Mol. Microbiol. Biotechnol. 12180-196. [DOI] [PubMed] [Google Scholar]
- 17.Herbert, S., A. Bera, C. Nerz, D. Kraus, A. Peschel, C. Goerke, M. Meehl, A. Cheung, and F. Gotz. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiron, A., E. Borezee-Durant, J. C. Piard, and V. Juillard. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 1895119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293381-399. [DOI] [PubMed] [Google Scholar]
- 20.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 441269-1286. [DOI] [PubMed] [Google Scholar]
- 21.Hrycyna, C. A., M. Ramachandra, S. V. Ambudkar, Y. H. Ko, P. L. Pedersen, I. Pastan, and M. M. Gottesman. 1998. Mechanism of action of human P-glycoprotein ATPase activity. Photochemical cleavage during a catalytic transition state using orthovanadate reveals cross-talk between the two ATP sites. J. Biol. Chem. 27316631-16634. [DOI] [PubMed] [Google Scholar]
- 22.Krulwich, T. A., O. Lewinson, E. Padan, and E. Bibi. 2005. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat Rev. Microbiol. 3566-572. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., and U. Bandyopadhyay. 2005. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157175-188. [DOI] [PubMed] [Google Scholar]
- 24.Li, M., Y. Lai, A. E. Villaruz, D. J. Cha, D. E. Sturdevant, and M. Otto. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 1049469-9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomovskaya, O., H. I. Zgurskaya, M. Totrov, and W. J. Watkins. 2007. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 656-65. [DOI] [PubMed] [Google Scholar]
- 26.Loo, T. W., and D. M. Clarke. 1994. Reconstitution of drug-stimulated ATPase activity following co-expression of each half of human P-glycoprotein as separate polypeptides. J. Biol. Chem. 2697750-7755. [PubMed] [Google Scholar]
- 27.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299906-909. [DOI] [PubMed] [Google Scholar]
- 28.Meehl, M., S. Herbert, F. Gotz, and A. Cheung. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 512679-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neubauer, H., I. Pantel, P. E. Lindgren, and F. Gotz. 1999. Characterization of the molybdate transport system ModABC of Staphylococcus carnosus. Arch. Microbiol. 172109-115. [DOI] [PubMed] [Google Scholar]
- 30.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204587-636. [DOI] [PubMed] [Google Scholar]
- 31.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 32.Pohl, A., P. F. Devaux, and A. Herrmann. 2005. Function of prokaryotic and eukaryotic ABC proteins in lipid transport. Biochim. Biophys. Acta 173329-52. [DOI] [PubMed] [Google Scholar]
- 33.Reniere, M. L., V. J. Torres, and E. P. Skaar. 2007. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20333-345. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73133-138. [DOI] [PubMed] [Google Scholar]
- 36.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70267-281. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 221-20. [DOI] [PubMed] [Google Scholar]
- 38.Senior, A. E., M. K. al-Shawi, and I. L. Urbatsch. 1995. The catalytic cycle of P-glycoprotein. FEBS Lett. 377285-289. [DOI] [PubMed] [Google Scholar]
- 39.Skaar, E. P., M. Humayun, T. Bae, K. L. DeBord, and O. Schneewind. 2004. Iron-source preference of Staphylococcus aureus infections. Science 3051626-1628. [DOI] [PubMed] [Google Scholar]
- 40.Skaar, E. P., and O. Schneewind. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6390-397. [DOI] [PubMed] [Google Scholar]
- 41.Speziali, C. D., S. E. Dale, J. A. Henderson, E. D. Vines, and D. E. Heinrichs. 2006. Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 1882048-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stauff, D. L., V. J. Torres, and E. P. Skaar. 2007. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 28226111-26121. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic, I., B. D. Evavold, and V. Kumar. 2001. Antimicrobial properties of porphyrins. Expert Opin. Investig. Drugs 10309-320. [DOI] [PubMed] [Google Scholar]
- 44.Stojiljkovic, I., V. Kumar, and N. Srinivasan. 1999. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31429-442. [DOI] [PubMed] [Google Scholar]
- 45.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ubukata, K., N. Itoh-Yamashita, and M. Konno. 1989. Cloning and expression of the norA gene for fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 331535-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151119-123. [DOI] [PubMed] [Google Scholar]
- 49.Wertheim, H. F., D. C. Melles, M. C. Vos, W. van Leeuwen, A. van Belkum, H. A. Verbrugh, and J. L. Nouwen. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5751-762. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 1726942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters—revisited five years on. Biochim. Biophys. Acta 1461177-200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.