Abstract
Plasmid pSW100 is 1 of the 13 plasmids from Pantoea stewartii subsp. stewartii SW2 which has a replicon that resembles that of ColE1. This work uses a pSW100 derivative, pSW140K, to study how the pSW100 replicon is stably maintained in its hosts. Our results indicate that although pSW140K is stable in Escherichia coli HB101, the plasmid is rapidly lost in another E. coli strain, DH5α, indicating that the genetic background of an E. coli strain affects the stability of pSW140K. Mutagenesis of E. coli HB101 with EZ::TN <DHFR-1> revealed that mutations in traC, traF, traG, traN, and traV, which encode the components of the sex pilus assembly, reduce plasmid stability. Furthermore, this work identified that a 38-bp region located immediately upstream of the RNAII promoter is critical to the maintenance of plasmid stability in E. coli HB101. TraC binds to the region, and in addition, deleting the region destabilizes the plasmid. Furthermore, inserting this 38-bp fragment into a plasmid that contains the minimal replicon from pSW200 stabilizes the plasmid in E. coli HB101. Fluorescence in situ hybridization and immunofluorescence staining also revealed that derivatives of pSW100, pSW128A, and TraC are colocalized in cells, suggesting that pSW100 may use the sex pilus assembly as a partition apparatus to ensure the even distribution of the plasmid during cell division, which may thus maintain the plasmid's stability.
Pantoea stewartii subsp. stewartii SW2 has 13 plasmids ranging in size from 4 to 320 kb (8). Although the total length of these plasmids exceeds 870 kb (8), no function has yet been assigned to them, except for those involved in plasmid replication, mobilization, and conjugation (8, 13-16). Plasmid pSW100 is the smallest plasmid in P. stewartii subsp. stewartii SW2, with a copy number of approximately 10 per cell (14). The plasmid (4,272 bp) is circular and found in almost all P. stewartii subsp. stewartii isolates (8, 14). Although this plasmid has a replicon that is similar to that of ColE1, it is compatible with ColE1 and another ColE1-like plasmid in P. stewartii subsp. stewartii SW2, pSW200 (15). Like ColE1 and pSW200, pSW100 contains a set of mob genes and a bom sequence that are required for plasmid mobilization (Fig. 1A) (14, 15). Since the mobA gene in pSW100 contains a frameshift mutation, the plasmid cannot be mobilized unless pSW200, which contains functional mobA, is present (15). The plasmid also contains a 249-bp and a 1,569-bp open reading frame; the 1,569-bp open reading frame contains five copies of 132-bp direct repeats (Fig. 1A). The functions of these two open reading frames and direct repeats are currently unknown. An earlier work demonstrated that the pSW100 replicon is located in a 702-bp region (Fig. 1A) (14). Unlike the ColE1 plasmid, pSW100 does not contain a rop gene to maintain the plasmid copy number (14). The replicon transcribes a preprimer RNA, RNAII, and an antisense RNA, RNAI, which controls the copy number and the incompatibility of the plasmid (6). Additionally, pSW100 does not seem to carry a gene that is essential to the survival of its host because curing the plasmid does not influence the growth rate, nutritional requirement, or biochemical characteristics of P. stewartii subsp. stewartii SW2 (14). Apart from pSW100 and pSW200, the replicons from three other plasmids in P. stewartii subsp. stewartii SW2, i.e., pSW500 (13), pSW800 (47), and pSW1200 (16), have also been characterized.
FIG. 1.
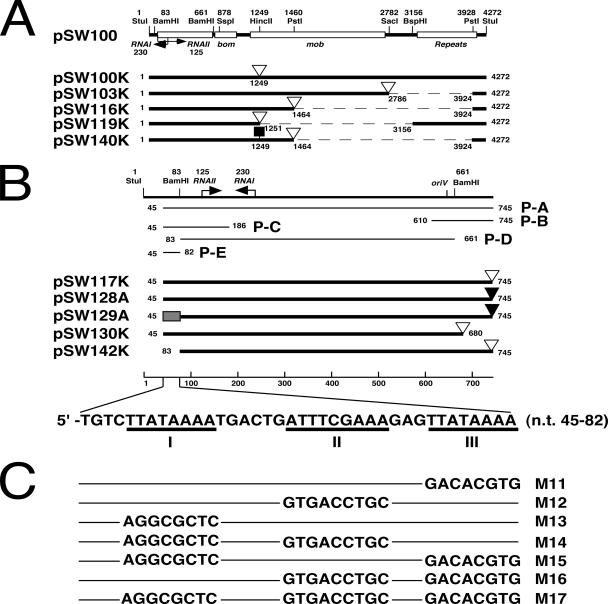
Plasmids and DNA probes. (A) Plasmid pSW100 has a replicon that is homologous to that of ColE1. The plasmid also contains a bom region, four mobilization (mob) genes, and five 132-bp perfect repeats. Numbers denote the nucleotide positions from the StuI site in pSW100 (14). Plasmid pSW100K contains the entire pSW100 sequence and a Km resistance gene (open triangle) that is inserted into the mob region. Plasmids pSW103K, pSW116K, pSW119K, and pSW140K are deletion derivatives of pSW100 that contain a Km resistance gene. Plasmid pSW140K was constructed by inserting a carotenoid synthesis operon (filled square) in pSW116K. Dashed lines represent deletion. (B) Plasmid pSW117K and pSW128A contain the region between nt 45 and nt 745 in pSW100. The region between nt 45 to 82 (filled box) in pSW128A was replaced with the M17 sequence (C) to yield pSW129A. Plasmids pSW130K and pSW142K are deletion derivatives of pSW117K. P-A, P-B, P-C, P-D, and P-E are the DNA fragments that were used in DNA-binding studies. P-E region contains three A-T-rich sequences: regions I, II, and III. (C) The sequence of regions I, II, and III of the P-E fragment in pSW117K was mutated to produce mutants M11 to M17. Filled triangles, Ap resistance gene.
Plasmids with a low copy number frequently use dedicated partition systems to maintain their stability. For instance, P1 and F use the Par and Sop systems, respectively, to ensure that replicated plasmids are uniformly distributed into daughter cells following cell division to prevent plasmid loss (25). Although the proteins and the cis elements differ between these two systems, the proteins that participate in the partition of these plasmids have similar functions and conserved sequences (2, 39). In P1, the cis element that is required for plasmid partition, parS, contains two sets of repeat sequences, which are recognized by a partition protein, ParB (45). Between the repeats is an integration host factor-binding site (17). Although the parS sequence is complex, only a 22-bp region is necessary for P1 segregation (31). Furthermore, ParA, an ATPase that contains Walker A, Walker B ATP-binding motifs, a helix-turn-helix domain, and a motif called “motif 3,” also interacts with ParB to participate in the partition process (39). Unlike the partition systems in P1 and F, pSC101, although containing a par site, does not encode a partition protein. Rather, the partition of pSC101 depends on the binding of host gyrase to the par site (46). Plasmids may also use postsegregational killing systems, as well as partition systems, to maintain their stability (24). For example, F plasmid encodes a potent gyrase inhibitor, CcdB, and an antidote, CcdA; following cell division, CcdB kills the daughter cells without F (9). The plasmid ColE1 is known not to use either a partition system or a postsegregational killing system to maintain its stability (19, 42). Since ColE1 has about 20 times more copies than P1 or F per cell, a random distribution of the plasmid into two daughter cells is thought to suffice to maintain the plasmid's stability (33, 43). However, an immunofluorescence study indicated that the stability of ColE1 may not completely depend on random distribution. A ColE1 derivative, pUC19, was found to aggregate in midcell or near-quarter-cell positions during cell division (35), indicating that the plasmid may use an unknown partition system to maintain its stability. This work indicates that pSW100 uses the sex pilus assembly as a partition tool to maintain its stability.
MATERIALS AND METHODS
Bacterial strains and media.
P. stewartii subsp. stewartii SW2 is a wild-type strain (8). E. coli HB101 (5), E. coli DH5α (22), E. coli DH5α(F), E. coli MG1063 (F+ recA56) (21), E. coli ATCC 23744 (pro thr leu thi Strr F-), E. coli ATCC 23846 (pro trp his lac Strr F-), E. coli ATCC 25257 (thi-1 Hfr), and E. coli ATCC 25256 (metB Hfr) were used to test the stability of pSW140K. E. coli BL21(DE3) (41) was used as a host to express His-tagged TraC. E. coli BW25113 is a host strain for PCR targeting (10, 20). E. coli DT-5, a traC mutant strain of E. coli HB101, was generated using EZ::TN <DHFR-1> (Epicentre, Madison, WI). LB medium (32) was used to culture E. coli cells. Ampicillin (Ap) (100 μg/ml), apramycin (Am) (25 μg/ml), kanamycin (Km) (50 μg/ml), and trimethoprim (Tp) (10 μg/ml) were added to LB medium to select the cells that were resistant to these antibiotics.
Plasmids.
Plasmid pSW100K was constructed by inserting a Km resistance gene that was isolated from pUC4-KIXX (4) by SmaI digestion into the HincII site of pSW100 (Fig. 1A). Plasmid pSW103K was constructed by replacing a 1,137-bp SacI-PstI fragment in pSW100 with a Km resistance gene (Fig. 1A). Plasmids pSW116K and pSW119K were generated by replacing a 2,459-bp PstI fragment and a 1,904-bp HincII-BspHI fragment in pSW100 with a Km resistance gene, respectively (Fig. 1A). A carotenoid synthesis operon, which was isolated from pSL525 (29, 44) by EcoRI digestion, was inserted into the HincII site in pSW116K to generate pSW140K (Fig. 1A). DNA fragments that contained sequences from nucleotides (nt) 45 to 745, 45 to 680, and 83 to 745 of pSW100 (14) were amplified by PCR and ligated with a Km resistance gene to generate pSW117K, pSW130K, and pSW142K, respectively (Fig. 1B). Plasmid pSW128A was constructed by replacing the Km resistance gene in pSW117K with an Ap resistance gene (Fig. 1B). Plasmid pSW129A is identical to pSW128A except that the region between nt 45 and 82 was replaced with an M17 sequence (Fig. 1B and C). Plasmid pML12 was generated by inserting traC, which was amplified by PCR with primers CF1 (5′-CTCTGTGAAGCATGCGGAA) and CR1 (5′-CATGTTGAAGGCGACAGTCACCGGTACCATGAATAACCCACTTGAGGCCGTC) using E. coli HB101 DNA as the template, into the SphI-SmaI sites in pGEM-7Z (Promega Corp., Madison, WI). Plasmid pML13 was constructed by inserting traC, which was amplified by PCR with primers F1 (5′-CCGGTACCATGAATAACCCACTTGAGGCCGTC) and R1 (5′-GGGGAGCTCCATGTTGAAGGCGACAGTCA) using E. coli HB101 DNA as a template, into the KpnI-SacI sites in pET-30b (Novagen, San Diego, CA). Plasmid pF101, constructed using a PCR targeting method (10, 20), contains an Am resistance cassette that was inserted into the tnpA gene in Tn1000 of F plasmid. Plasmid pSW245 contains the minimal replicon of pSW200, from nt 380 to 998 (15), and a Km resistance gene. Near the junction between the upstream region of the RNAII promoter and the Km resistance gene in pSW245, the plasmid contains a BglII site. A 38-bp P-E sequence from pSW100 (Fig. 1B) was inserted into the BglII site to construct pSW246.
Expression and purification of TraC.
E. coli BL21(DE3)(pML13) was cultured overnight in 1 liter of LB-Km medium. Following centrifugation at 10,000 × g for 15 min, a cell pellet was suspended in ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1.5% Triton X-100, pH 8.0) and homogenized using a French press (Thermo Spectronic, Rochester, NY) at 10,000 lb/in2. Cell lysate was then centrifuged at 17,000 × g for 30 min at 4°C. The supernatant was applied to a 0.5-ml Ni-nitrilotriacetic acid agarose column (Qiagen, Valencia, CA) which had been preequilibrated with 10 column volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). His-TraC was finally eluted from the column with a buffer that contained 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0. Eluted His-TraC was concentrated with Amicon-Ultra4 centrifugal filters (Millipore, Billerica, MA) and stored at −70°C until use.
Transposon mutagenesis.
E. coli HB101 was mutagenized with EZ::TN <DHFR-1> (Epicentre) on the basis of the manufacturer's method. Mutants with an EZ::TN <DHFR-1> insertion were selected on LB-Tp agar. Chromosomal DNA from the mutants was purified (34), digested with NheI and PvuII, and then inserted into the NheI-PvuII sites in pBR322. Plasmids that contain EZ::TN <DHFR-1> were transformed into E. coli HB101 and selected on LB agar that contained Ap and Tp. Chromosomal fragments adjacent to the transposon insertions were sequenced using two EZ::TN <DHFR-1> primers, 5′-GGCGGAAACATTGGATGCGG and 5′-GACACTCTGTTATTACAAATCG (Epicentre).
Plasmid stability test.
E. coli strains that contained a plasmid with the pSW100 replicon were initially cultured in LB-Km broth overnight. The culture was then used to inoculate LB broth with an inoculum size of 1%. At the inoculation time, the culture was estimated to have a cell density of about 5 × 107 CFU/ml. The cells were subsequently cultured for 12 h at 37°C with constant shaking and subcultured for another 12 h in LB broth with the same inoculum size. This subculturing procedure was continued for 6 days. Under such subculturing conditions, cells were made to reach the stationary phase and grown for about seven generations in each subculturing period. After each subculturing, cells were plated on LB agar; the number of the colonies that had lost the plasmid during subculturing was determined by replica plating about 1,000 colonies from LB agar to LB-Km agar. Since plasmid pSW140K contains a carotenoid synthesis operon (29), the stability of pSW140K was determined by calculating the percentage of the colonies that could produce orange-colored β-carotene. Each stability test was repeated at least twice, and the numbers of colonies on three LB-agar plates were averaged. Standard deviations were calculated and are shown in each figure.
DNA affinity precipitation assay.
Five DNA probes, P-A, P-B, P-C, P-D, and P-E (Fig. 1B), were labeled with biotin using a 3′-end DNA labeling kit (Pierce, Rockford, IL). His-TraC (100 μg in 10 μl) and a biotinylated DNA probe (25 ng) were added to a binding buffer that contained 60 mM KCl, 12 mM HEPES, pH 7.9, 4 mM Tris-HCl, 5% glycerol, 0.5 mM EDTA, and 1 mM dithiothreitol. The reaction mixture (25 μl) was incubated on ice for 45 min. M280 streptavidin beads (30 mg; Dynal Biotech, Norway), which had been preequilibrated in the binding buffer, were added to the mixture. The beads were then captured with a magnet and washed five times in the binding buffer (7). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (15 μl) was added to the beads, and the solution was boiled for 5 min to dissociate the proteins from the beads. Proteins were then separated in a 7.5% SDS-polyacrylamide gel and detected by immunoblotting with anti-His antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Electrophoretic mobility shift assay (EMSA).
The double-stranded DNA probes P-E, which contains a sequence from nt 45 to 82 in pSW100 (5′-TGTCTTATAAAATGACTGATTTCGAAAGAGTTATAAAA) (Fig. 1B), and mP-E (5′-TGTCAGGCGCTCTGACTGGTGACCTGCGAGGACACGTG), which contains the M17 sequence (Fig. 1C), were labeled at their 3′ ends with biotin. The probe (25 ng) was then mixed with 6 μg of His-TraC in 20 μl of a reaction mixture that contained 1 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 2.5% glycerol, 5 mM MgCl2, 1 μg poly(dI-dC), and 0.05% NP-40. The mixture was incubated at room temperature for 20 min. Electrophoresis was performed with a 10% polyacrylamide gel at 100 V for 70 min. Following electrophoresis, DNA was detected using horseradish peroxidase-conjugated streptavidin (Pierce Biotechnology, Inc.).
Preparation of DNA probes for FISH.
DNA probes for fluorescence in situ hybridization (FISH) were amplified by PCR with the primers ori-F (5′-TGTCTTATAAAATGACTG) and ori-R (5′-GTGATACTGCGGCGGGCGTTA), using pSW128A as a template. DNA was labeled using Ulysis nucleic acid labeling kits (Molecular Probes) according to the manufacturer's instructions. Briefly, 1 μg of probe DNA was denatured at 95°C for 5 min. Alexa Fluor 546 ULS (Universal Linkage System) labeling reagent (5 μl; Molecular Probes, Inc., Eugene, OR) was then added. The reaction mixture was incubated at 80°C for 15 min and then at 0°C to stop the reaction. Labeled DNA was finally precipitated by ethanol.
FISH and immunofluorescence staining.
Cells that had been cultured overnight were washed three times in phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) and immobilized on poly-l-lysine-coated coverslips in a six-well plate. The cells were fixed with an acetone-methanol (8:1) solution for 30 s and washed with PBS three times for 10 min. The fixed cells were subsequently treated with 1 mg/ml lysozyme for 30 min at 37°C and 4% Triton X-100 in PBS for 5 min at room temperature. Prehybridization solution (2 ml) that contained 1 mg/ml sonicated salmon sperm DNA, 70% formamide, and 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0) was added to each well, and the plates were incubated at 42°C for 1 h. DNA was denatured by placing the coverslips on a 95°C heat block for 2 min. A solution that contained 200 ng/μl of a labeled DNA probe, 50% formamide, and 2× SSC was then added to each well. After incubating at 42°C for 3 h, the cells were washed twice in 2 ml of 50% formamide in 2× SSC at 42°C for 30 min, once in 2 ml of 50% formamide in 2× SSC at room temperature for 10 min, three times in 2 ml of 0.1% Tween 20 in 2× SSC at room temperature for 10 min, and twice in PBS. To detect the location of TraC, blocking solution that contained 5% bovine serum albumin in PBS was added to each well. Cells were incubated for 1 h and then treated with anti-His polyclonal antibody for 1 h. Following antibody binding, proteins were stained with Alexa Fluor 488-conjugated anti-rabbit secondary antibody (Molecular Probes, Inc.). Chromosomal DNA was finally stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO). Cells were observed under a Zeiss confocal laser scanning microscope (model LSM 510 META).
Southern blot hybridization.
Chromosomal DNAs from E. coli DH5α and HB101 were digested with BglII and NotI. Plasmids from P. stewartii subsp. stewartii SW2 were isolated using an alkaline lysis method (26). DNA fragments were separated by agarose gel electrophoresis and transferred onto a Hybond-N+ membrane (Amersham) by following a standard procedure (36). The traC gene was amplified by PCR, digested with MseII, end labeled with biotin (Pierce, Rockford, IL), and used as a probe. Hybridization was performed using a North2South chemiluminescence hybridization and detection kit (Pierce, Rockford, IL).
RESULTS
Determining the region in pSW100 critical to the maintenance of plasmid stability.
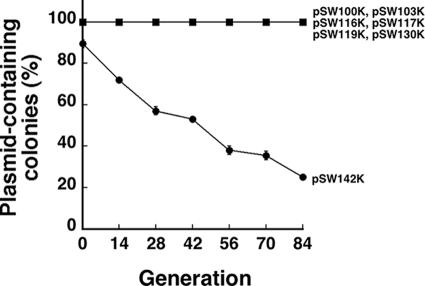
Plasmid pSW100K, which contains a Km resistance gene inserted into the mob region in pSW100 (Fig. 1A), was deleted to delineate the regions that are required to maintain plasmid stability. Plasmid pSW100K was stable in E. coli HB101 during 84 generations of culturing in LB broth (Fig. 2). Additionally, deleting the mob gene and the repeat region in pSW100, including pSW103K, pSW116K, pSW117K, and pSW119K (Fig. 1), influenced neither the plasmid copy number nor the plasmid's stability. These plasmids remained stable throughout the culturing period (Fig. 2). Of these plasmids, pSW117K had the smallest size, with a pSW100 sequence of 701 bp. The DNA sequence that is required to maintain the plasmid must therefore be in this 701-bp region (Fig. 1B). In this respect, a 65-bp region, which runs from nt 681 to 745 of pSW117K and downstream to the RNAII region, was deleted to yield pSW130K (Fig. 1B). This deletion did not appear to influence the plasmid's stability, as no plasmid loss was observed during 84 generations of culturing (Fig. 2). When a 38-bp P-E region, which contains the sequence between nt 45 and 82 upstream of the RNAII promoter, was deleted (pSW142K) (Fig. 1B), the plasmid became unstable. After the first 14 generations of culturing in LB broth, 28% of the population had lost pSW142K. The proportion of the population that contained pSW142K further declined to 25% upon 84 generations of culturing (Fig. 2).
FIG. 2.
Analysis of the region in pSW100 that affects plasmid stability. E. coli HB101(pSW100K), E. coli HB101(pSW103K), E. coli HB101(pSW116K), E. coli HB101(pSW117K), E. coli HB101(pSW119K), E. coli HB101(pSW130K), and E. coli HB101(pSW142K) were cultured in LB-Km broth overnight and used to inoculate LB broth. Cells were subcultured every 12 h (seven generations), plated on LB agar, and then replica plated on LB-Km agar to determine the numbers of the colonies that did not contain the plasmid.
Stability of pSW140K in E. coli strains.
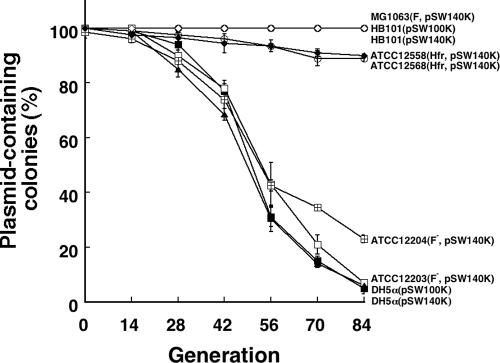
The stability of the pSW100 replicon was studied in seven E. coli strains to determine whether the genetic background of the host affects plasmid stability. To facilitate this study, a carotenoid synthesis operon (29) was inserted into pSW116K to yield pSW140K (Fig. 1A). The colonies that contained pSW140K synthesized β-carotene and, therefore, were orange; those colonies that could not stably maintain pSW140K were nonpigmented. The cells that contained pSW140K were initially cultured in LB-Km broth and then subcultured in LB broth for 84 generations. The instability of pSW140K in these strains was examined every 14 generations by plating the cells on LB agar and calculating the percentage of the nonpigmented colonies on the plates. After colonies from each strain were plated, pSW140K was found to be stable in E. coli HB101 and E. coli MG1063. No plasmid loss was observed during 84 generations of culturing (Fig. 3). Compared to these two strains, pSW140K was relatively unstable in E. coli ATCC 25257 and E. coli ATCC 25256; 10% and 11% of the population lost pSW140K after 84 generations of culturing, respectively (Fig. 3). However, pSW140K was extremely unstable in E. coli DH5α, E. coli ATCC 23744, and E. coli ATCC 23846; 95%, 94%, and 77% of the population, respectively, had lost the plasmid after 84 generations of culturing (Fig. 3). The stability pattern of pSW100K, which was determined by replica plating the colonies on LB-Km plates, was found to be similar to that of pSW140K in E. coli HB101 and E. coli DH5α (Fig. 3). The copy numbers of pSW100K and pSW140K in strain HB101 were also similar to each other. Although pSW140K is unstable in E. coli DH5α, the copy number of the plasmid per cell in the strain was close to that in E. coli HB101 (data not shown).
FIG. 3.
Stability of the pSW100 replicon in E. coli strains. E. coli strains HB101(pSW100K), HB101(pSW140K), and MG1063(pSW140K) (circles with an empty diamond), DH5α(pSW100K) (filled triangles), DH5α(pSW140K) (filled squares), ATCC 25257(pSW140K) (filled diamonds), ATCC 25256(pSW140K) (empty circles), ATCC 23744(pSW140K) (empty squares), and ATCC 23846(pSW140K) (squares with a cross) were cultured in LB-Km broth overnight and subcultured in LB broth with an inoculation size of 1% for 84 generations. Cells were plated on LB agar after every 14 generations of culturing. The percentages of the colonies that had lost their orange color, indicating the loss of pSW140K, were calculated. The stability of pSW100K was examined by replica plating the cells on LB and LB-Km agar.
Identifying the genes in E. coli HB101 that are critical to the stability of pSW140K.
The fact that pSW140K is stable in certain E. coli strains indicates that the genetic background of the hosts affects the stability of pSW140K. To elucidate the genes that are crucial to the stability of pSW140K, E. coli HB101 cells were mutagenized with EZ::TN <DHFR-1> to screen the mutants that could not stably maintain pSW140K. This mutagenesis was achieved by selecting the mutants that contained a transposon insertion in the chromosome by culturing the cells in LB-Tp broth. These cells, as a pool, were transformed with pSW140K. After the transformants had been subcultured for 84 generations in LB broth, the cells were plated on LB agar to select the colonies that had no pigmentation. In this selection, 25 mutants were isolated. These mutants were retransformed with pSW140K for another round of stability tests to verify the instability of pSW140K in these mutants. The transposon and its adjacent chromosomal fragments of these mutants were then cloned. DNA sequencing indicated that the transposon was inserted into 10 genes on the chromosome; 5 of these genes, traC, traF, traG, traN, and traV, are encoded by the F plasmid. This result also showed that E. coli HB101, although an F− strain, may contain a segment of F on the chromosome.
Genetic complementation.
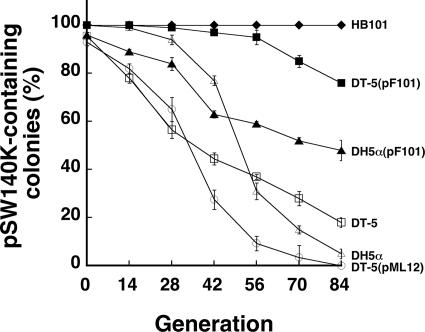
The proteins encoded by traC, traF, traG, traN, and traV are the components of the sex pilus assembly (11, 12). Of these proteins, TraF, TraG, TraN, and TraV are either associated with the membranes or present in the periplasm (28); only TraC is present in the cytoplasm and at the base of the pilus assembly (38). Accordingly, TraC became a candidate for interacting with pSW140K to maintain plasmid stability, and so traC was studied further. A complementation study was performed using traC to determine whether the plasmid stability in a traC mutant could be recovered. A mutant E. coli HB101 strain with an EZ::TN <DHFR-1> insertion in traC, DT-5, was transformed with pSW140K. Plasmid pSW140K was unstable in this mutant strain. By the 14th generation, 22% of the cells had lost pSW140K (Fig. 4). The proportion of the population that contained pSW140K further decreased to 18% at the 84th generation (Fig. 4). When DT-5(pSW140K) was transformed with pF101, an F plasmid with an Am resistance gene inserted into Tn1000; after the transformants were cultured in the presence of Am, the stability of pSW140K improved. No plasmid loss occurred during the first 28 generations. When the population reached the 42nd generation, only 3% had lost pSW140K (Fig. 4). The proportion of the population was increased to 24% by the 84th generation (Fig. 4). However, when the mutant was transformed with a high-copy-number plasmid, pML12 (which carries traC), an adverse effect on the stability of pSW140K was observed. Plasmid pML12 caused a rapid loss of pSW140K from the DT-5 mutant. The loss was 18% at the 14th generation and 63% at the 42nd generation (Fig. 4). When the cells reached the 84th generation, all of the colonies that were examined had lost pSW140K (Fig. 4). Meanwhile, the same experiment was performed to study whether pF101 could stabilize pSW140K in E. coli DH5α. The results indicated that 68% and 95% of the E. coli DH5α cells lost pSW140K after 56 and 84 generations of culturing, respectively (Fig. 4). However, introducing pF101 into the strain increased the stability of pSW140K; only 39% and 50% of the population lost the plasmid after 56 and 84 generations of culturing, respectively (Fig. 4). However, in E. coli DH5α, pF101 could not restore the stability of pSW140K as it did in strain HB101, indicating that factors other than F may also be required for the stable maintenance of pSW140K in E. coli DH5α.
FIG. 4.
Stabilizing pSW140K by plasmids that carry traC. E. coli DT-5(pSW140K), an E. coli HB101 mutant that contains mutated traC (empty squares), was transformed with pF101 (filled squares) and pML12 (empty circles) to determine how traC affected the stability of pSW140K, which was also tested in E. coli HB101 (filled diamonds), E. coli DH5α (empty triangles), and E. coli DH5α(pF101) (filled triangles). Cells were initially cultured in LB-Km broth and then subcultured in LB or LB-Am broth for 84 generations. Nonpigmented colonies on LB or LB-Am agar, indicating the loss of pSW140K, were counted.
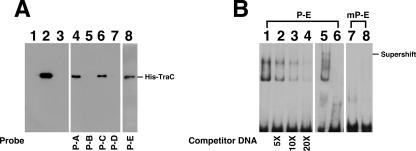
Binding of TraC to pSW100.
A DNA affinity precipitation assay was performed using biotinylated probes to identify the region in pSW100 (Fig. 1B) that interacts with TraC. The analysis revealed that His-TraC, purified from E. coli BL21 (DE3)(pML13) (Fig. 5A, lane 2), was retained by the probe P-A (Fig. 5A, lane 4), indicating the binding of His-TraC to the replicon region of pSW100. However, His-TraC did not bind to probes P-B and P-D (Fig. 5, lane 5, 7), which contain sequences from nt 610 to 745 and nt 83 to 661 (Fig. 1B), respectively, indicating that His-TraC did not bind to the region between nt 83 and 745. Yet, His-TraC bound to probe P-C (Fig. 1B), which contains a sequence from nt 45 to 186 (Fig. 5A, lane 6), and probe P-E (Fig. 5A, lane 8), which contains a sequence of nt 45 to 82 from pSW117K (Fig. 1B), indicating that His-TraC binds to the region between nt 45 and 82. EMSA with probe P-E and a mutant probe, mP-E, was also conducted. PAGE revealed a shift in the P-E band by His-TraC (Fig. 5B, lanes 1). As expected, unlabeled probe P-E competed the binding (Fig. 5B, lanes 2 to 4). The analysis also demonstrated that anti-His antibody supershifted the probe (Fig. 5B, lane 5), further indicating the binding of TraC to the probe. Meanwhile, His-TraC did not shift a probe with a mutated P-E sequence, mP-E (Fig. 5B, lane 8), in which the P-E sequence was replaced with the M17 sequence (Fig. 1C).
FIG. 5.
Binding of TraC to pSW100. (A) Purified histidine-tagged TraC (lane 2) was added to a binding mixture that contained a biotinylated DNA probe, P-A (lane 4), P-B (lane 5), P-C (lane 6), P-D (lane 7), or P-E (lane 8). A DNA-protein complex was then captured using streptavidin-coated magnetic beads. Proteins bound to the beads were separated by SDS-PAGE and detected by immunoblotting using an anti-histidine tag antibody. In a negative control, a P-A probe was incubated with protein extract from E. coli BL21(DE3)(pET-30b) (lane 1). Lane 3 was loaded with the P-A probe. (B) The P-E probe (lanes 1 to 6) and a mutant P-E probe (mP-E) (lanes 7 and 8) were used to analyze the binding of His-TraC to the P-E region by EMSA. Anti-His antibody was utilized to demonstrate the supershifting of the probe (lane 5). Lanes 6 and 7 were loaded with the P-E and mP-E probes, respectively. Each binding reaction involved 5 ng of biotinylated probe, 6 μg of TraC, and 1 μg of poly(dI-dC). Unlabeled P-E DNA was used to compete the binding (lanes 2 to 4). The protein-DNA complex was separated with a 7% polyacrylamide gel and detected using a LightShift chemiluminescence EMSA kit (Pierce).
Colocalization of TraC with pSW128A.
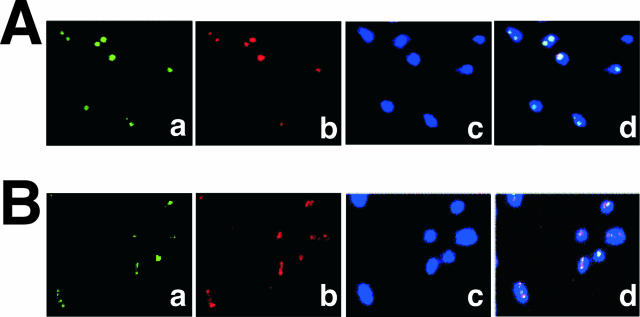
FISH analysis coupled with immunofluorescence staining was performed to visualize intracellular TraC and the pSW100 replicon. The work was performed using two plasmids, pSW128A and pSW129A. Plasmid pSW128A has a sequence identical to that of pSW117K, except that pSW128A contains an Ap resistance gene (Fig. 1B). In pSW129A (Fig. 1B), the P-E sequence in pSW128A is replaced with that of M17 (Fig. 1C). In this experiment, TraC was stained green by indirect immunofluorescence staining, plasmid DNA was stained red by FISH, and chromosomes were stained blue by DAPI (Fig. 6). Confocal microscopy demonstrated that TraC colocalized with pSW128A but not pSW129A (Fig. 6A and B, panels d), suggesting that TraC interacted with the P-E region in pSW128A.
FIG. 6.
Intracellular localization of pSW128A and TraC. E. coli BL21(DE3)(pSW128A, pML13) (A) or E. coli BL21(DE3)(pSW129A, pML13) (B) was cultured at 37°C overnight. (a) TraC protein stained by indirect immunofluorescence staining using anti-His polyclonal antibody and Alexa Fluor 488-conjugated anti-rabbit secondary antibody (green); (b) plasmid DNA visualized by FISH using an Alexa Fluor 546 ULS (Universal Linkage System)-labeled probe (red); (c) chromosome stained by DAPI (blue); (d) merged image.
Mutational analysis of the P-E region.
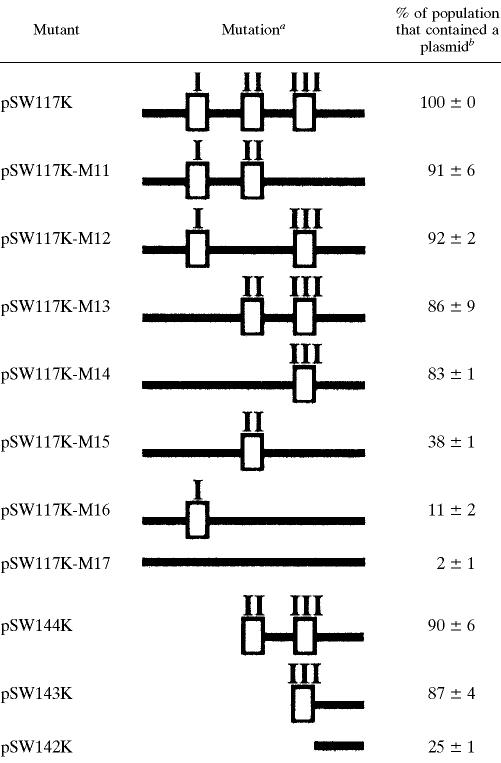
The 38-bp P-E fragment was deleted from pSW117K (Fig. 1B; Table 1) to analyze the sequence in the P-E region that is required to maintain the stability of pSW100. Deleting the regions from nt 45 to 62 and nt 45 to 74, which generated pSW144K and pSW143K, respectively, reduced the plasmid stability by 10 to 13% in E. coli HB101 after 84 generations of culturing (Table 1). However, the plasmid became extremely unstable when the deletion was extended to nt 82 (pSW142K), 75% of the population lost the plasmid after 84 generations of culturing (Table 1). Since the 38-bp PE DNA fragment contained three A-T-rich regions, regions I, II, and III (Fig. 1B), this work further investigated how these regions affect the plasmid's stability. These sequences were mutated, and three mutants, pSW117K-M11, pSW117K-M12, and pSW117K-M13 (Fig. 1C; Table 1), were generated. These mutant plasmids were relatively stable during culturing: 91% of pSW117K-M11, 92% of pSW117K-M12, and 86% of the pSW117K-M13 population contained the plasmid after 84 generations of culturing (Table 1). When both regions I and II were mutated (mutant pSW117K-M14), the population that contained the plasmid declined to 83% after 84 generations of culturing (Table 1). Moreover, only 38% of the population contained the plasmid after both regions I and III had been deleted (mutant pSW117K-M15). When regions II and III were mutated (mutant pSW117K-M16), only 11% of the population contained the plasmid (Table 1). The proportion of the population that contained the plasmid decreased to 2% when all three regions had mutated (mutant pSW117K-M17). Although mutations in the P-E region affected the plasmid's stability, the copy numbers of these mutants were all similar in E. coli HB101 (data not shown).
TABLE 1.
Mutations in the P-E region and the stability of pSW117K derivatives
The region deleted from each plasmid is shown schematically. Empty boxes denote three A-T-rich sequences in the P-E region labeled I, II, and III (Fig. 1B).
Percentage of the population that contained a plasmid after 84 generations of culturing in LB broth.
Detecting traC in P. stewartii subsp. stewartii SW2.
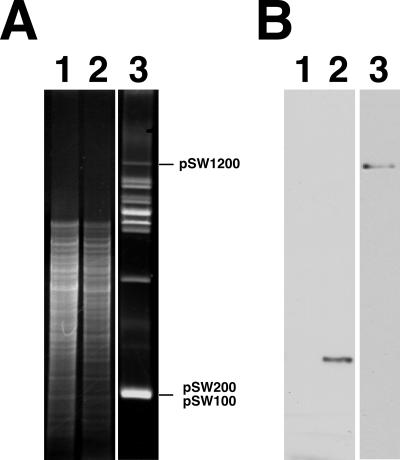
Southern blot analysis using a biotinylated traC probe was adopted to detect traC in P. stewartii subsp. stewartii SW2. As expected, E. coli HB101 DNA had only one BglII/NotI fragment that hybridized with the probe (Fig. 7B, lane 2). Conversely, no hybridization signal was observed when the E. coli DH5α DNA was used (Fig. 7B, lane 1). In P. stewartii subsp. stewartii SW2, pSW1200, which contains an IncY replicon (16), hybridized with the probe (Fig. 7B, lane 3), suggesting that pSW1200 contains traC.
FIG. 7.
Southern blot analysis of traC in E. coli and P. stewartii subsp. stewartii. (A) Five micrograms of DNA from E. coli DH5α (lane 1) and E. coli HB101 (lane 2) was digested with BglII/NotI. Lane 3 was loaded with P. stewartii subsp. stewartii SW2 plasmids that had been purified by an alkaline lysis method (26). (B) Southern hybridization was performed using a biotin-labeled traC probe.
Stabilizing the minimal replicon of pSW200 by the P-E sequence.
Plasmid pSW200, another ColE1-like plasmid in P. stewartii subsp. stewartii SW2, is stably maintained in P. stewartii subsp. stewartii SW2 and E. coli HB101. However, pSW245, which contains the minimal replicon and a Km resistance gene, is unstable in E. coli HB101; about 40% of the population lost the plasmid after 84 generations of culturing (Fig. 8). This work inserted the P-E fragment to a BglII site in pSW245, which is located at the position that is 43 nt upstream of the +1 site of the RNAII promoter (15), and generated pSW246. This plasmid became stable in E. coli HB101; no plasmid loss was observed after 84 generations of culturing.
FIG. 8.
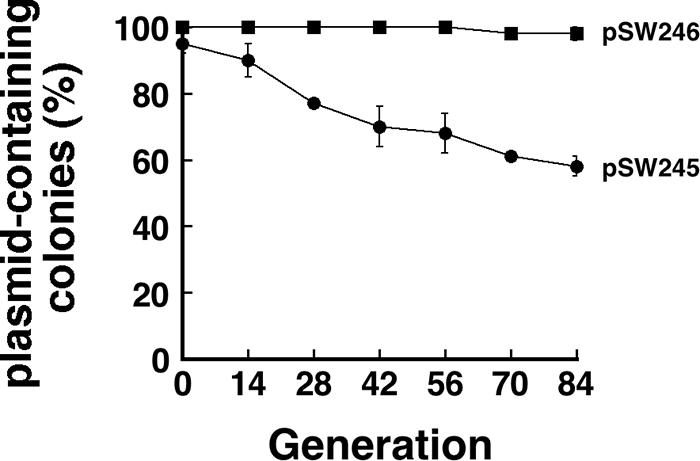
Stabilizing pSW245 with the P-E sequence. E. coli HB101(pSW245) and E. coli HB101(pSW246) were initially cultured in LB-Km broth and then subcultured in LB broth for 84 generations. The stability of the plasmids was determined by replica plating on LB-Km plates.
DISCUSSION
Plasmid pSW100 from P. stewartii subsp. stewartii SW2 can be stably maintained in its natural host and E. coli HB101. Since the plasmid does not appear to encode any function that is involved in plasmid partition or postsegregational killing (14), how this plasmid maintains its stability in its hosts is unknown. This study examined the stability of pSW140K, a pSW100 derivative, in several E. coli strains and found that pSW140K is stable in E. coli HB101 but unstable in E. coli DH5α over 84 generations of culturing in LB broth (Fig. 3). This observation indicates that the genetic background of the strains is critical to the stability of the plasmid. A study of mutagenized E. coli HB101 revealed that pSW140K is unstable in traC, traF, traG, traN, and traV mutants. These five genes encode the proteins that are components of the sex pilus assembly (12). This finding is somewhat surprising since E. coli HB101 is not known to be an F+ or an Hfr strain. In fact, we had previously demonstrated that E. coli HB101 can be used as a recipient strain for F conjugation (29). E. coli HB101 was originally obtained by mating an E. coli B with an E. coli K-12 strain (5, 37). Therefore, a segment of F may have remained on the chromosome, allowing the cells to synthesize the sex pilus assembly and function as a recipient during conjugation.
Among the five Tra proteins, TraC and TraF, which are present in the cytoplasm and periplasm, respectively, participate in pilus assembly (38, 48). Additionally, TraG is an inner membrane protein with a C-terminal periplasmic domain that is required for pilus assembly and mating pair stabilization (1). TraN, however, is an outer membrane protein that is associated with OmpA and is involved in mating pair stabilization (27). TraV is an outer membrane lipoprotein; it serves as an anchor that enables TraK and TraB to produce a transenvelope structure that is necessary for F-pilus assembly (23). Since TraC is present in the cytoplasm, it is the protein that most likely interacts with pSW100. In fact, the DNA-binding study established that TraC interacts with the 38-bp P-E fragment that is located immediately upstream of the RNAII promoter (Fig. 1B and 5). Confocal microscopy using FISH and immunofluorescence staining also demonstrated that, in cells (Fig. 6), TraC colocalizes with pSW128A but not with pSW129A, a plasmid that contains a mutated P-E sequence, suggesting that TraC binds to the P-E region in vivo. Furthermore, the fact that pSW140K became unstable when any of the five tra genes was mutated indicates the importance of an intact sex pilus assembly in maintaining the stability of pSW140K. The interaction between TraC and the P-E region suggests that pSW100 may become attached to the base of the sex pilus assembly during plasmid segregation.
To determine if the stability of pSW140K depends on traC, a complementation study was performed by introducing pF101 into E. coli strains. The results indicated that pSW140K was unstable in the E. coli HB101 traC mutant, DT-5. After 84 generations of culturing, more than 80% of the population had lost the plasmid. When pF101, a plasmid that carries traC, was transformed, pSW140K was stabilized, although not completely back to the wild-type level, and at least within the first 28 generations, all the examined colonies contained pSW140K. Even toward the 84th generation, pSW140K was present in almost 80% of the population (Fig. 4). Additionally, pSW140K was unstable in DT-5(pML12) (Fig. 4) because pML12 is a high-copy-number plasmid; the amount of TraC expressed from this plasmid probably exceeded that from F. The presence of an excess of TraC in the cell may prevent pSW140K from binding to the TraC that is attached to the sex pilus assembly. Additionally, pSW140K appeared more stable in F+ and Hfr strains than in F- strains (Fig. 3), once again supporting the claim that the sex pilus assembly participates in stabilizing pSW140K.
The results did not demonstrate a complete restoration of the stability of pSW140K after the introduction of pF101, an F plasmid that contains an Am resistance gene, into E. coli DT-5(pSW140K). The lack of complete restoration is mysterious. The DT-5 strain may contain a second mutation that influences the stability of pSW140K, or the F fragment on the chromosome may cause aberrant expression of an adjacent gene that might be important to the stability of pSW140K. Moreover, in DT-5(pF101), the numbers of copies of traF, traG, traN, and traV are twice as many as those of traC. A precise amount of Tra proteins expressed by F may be critical to the formation of the sex pilus assembly. A relatively low level of TraC in the cells may influence the structure of the sex pilus assembly and the stability of pSW140K, explaining why complete complementation of pSW140K stability cannot be achieved. Moreover, complete restoration of pSW140K by pF101 was not achieved in E. coli strain DH5α (Fig. 4), suggesting that pSW140K may depend on factors other than the sex pilus assembly to maintain its stability in E. coli DH5α. In fact, the results of our mutagenesis study indicated that mutations in genes apart from the five tra genes may also affect the stability of pSW140K (data not shown). These genes include rfbC, which encodes dTDP-4-deoxyrhamnose-3,5-epimerase, which participates in O-antigen synthesis (30), and mrcA, which encodes penicillin-binding protein 1a (18). How these genes influence the stability of pSW140K is now being studied.
This work addressed how pSW100 is stabilized in E. coli and found that TraC can bind to a 38-bp region (P-E region) that is located immediately upstream of the −35 site of the RNAII promoter (Fig. 1B, 5, and 6). Sequence analysis indicated that TraC contains a sequence that resembles that of a DNA-binding motif, NUMOD3 (40), in the region between amino acid residues 312 and 325, which may participate in binding to pSW140K. The 38-bp region not only is bound by TraC but also is critical to plasmid stability, since deleting this region in pSW117K (pSW142K) significantly reduced the stability of the plasmid in E. coli HB101 (Fig. 2). Further analysis of the P-E region by deleting the regions from nt 45 to 62 (pSW144K) and 45 to 74 (pSW143K) showed that plasmid stability was only slightly reduced (Table 1). However, the plasmid became extremely unstable when the deletion was extended to nt 82 (pSW142K) (Table 1; Fig. 2). Mutational analysis also indicated that although mutating the three A-T-rich sequences in the P-E region individually only slightly decreased the plasmid's stability, simultaneously mutating any two of these sequences, especially regions II and III, substantially reduced the plasmid's stability (Table 1), indicating that these A-T-rich sequences may cooperate in maintaining plasmid stability.
Plasmid ColE1 and its derivatives are generally believed to segregate at random into daughter cells (33, 42). The copy numbers of these plasmids are relatively high, so a random distribution of the plasmids is unlikely to produce a daughter cell that does not contain a plasmid (42, 43). However, as is generally known, many cloning vectors that are derived from ColE1 may have as many as 200 copies (35). If ColE1 plasmids indeed segregate randomly, then these plasmids should be extremely stable. Yet, the instability of these plasmids is frequently observed (3), indicating that mechanisms other than random distribution may be involved in maintaining plasmid stability. In fact, a recent study indicated that pUC19 aggregates in the mid- or quarter-cell position during cell division (35), suggesting that a specific segregation mechanism may control the stability of the plasmid with a ColE1 replicon. The results of this work demonstrated that pSW140K may use the sex pilus assembly as a partition apparatus to maintain its stability. Indeed, P. stewartii subsp. stewartii SW2 contains a conjugatable IncY plasmid, pSW1200 (16), that contains the traC sequence (Fig. 7). Accordingly, in its natural host, plasmid pSW100 may use this plasmid, pSW1200 (16), to maintain its stability.
Acknowledgments
We thank Jen-Fen Fu, Cheng-Yeu Wu, and Chyi-Liang Chen for their valuable suggestions.
This work was supported by grants from the Ministry of Education, Taiwan, Republic of China (to Chang Gung University); the Chang-Gung Memorial Hospital (grant CMRPD160111); the Chang-Gung Molecular Medicine Research Center (grant CMRPD140014); and the National Science Council of the Republic of China (grant NSC 96-3112-B-182-002).
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Audette, G. F., J. Manchak, P. Beatty, W. A. Klimke, and L. S. Frost. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153442-451. [DOI] [PubMed] [Google Scholar]
- 2.Austin, S., and A. Abeles. 1983. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J. Mol. Biol. 169353-372. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10411-421. [DOI] [PubMed] [Google Scholar]
- 4.Barany, F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37111-123. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41459-472. [DOI] [PubMed] [Google Scholar]
- 6.Cesareni, G., M. Helmer-Citterich, and L. Castagnoli. 1991. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 7230-235. [DOI] [PubMed] [Google Scholar]
- 7.Chang, L. K., J. Y. Chung, Y. R. Hong, T. Ichimura, M. Nakao, and S. T. Liu. 2005. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 336528-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coplin, D. L., R. G. Rowan, D. A. Chisholm, and R. E. Whitmoyer. 1981. Characterization of plasmids in Erwinia stewartii. Appl. Environ. Microbiol. 42599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dao-Thi, M. H., D. Charlier, R. Loris, D. Maes, J. Messens, L. Wyns, and J. Backmann. 2002. Intricate interactions within the ccd plasmid addiction system. J. Biol. Chem. 2773733-3742. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firth, N., K. Ippen-Ihler, and R. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. Neidhardt (ed.), Escherichia coli and Salmonella, 2nd ed. ASM Press, Washington, DC.
- 12.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, J. F., H. C. Chang, Y. M. Chen, Y. S. Chang, and S. T. Liu. 1996. Characterization of the replicon of plasmid pSW500 of Erwinia stewartii. Mol. Gen. Genet. 250699-704. [DOI] [PubMed] [Google Scholar]
- 14.Fu, J. F., H. C. Chang, Y. M. Chen, Y. S. Chang, and S. T. Liu. 1995. Sequence analysis of an Erwinia stewartii plasmid, pSW100. Plasmid 3475-84. [DOI] [PubMed] [Google Scholar]
- 15.Fu, J. F., J. M. Hu, Y. S. Chang, and S. T. Liu. 1998. Isolation and characterization of plasmid pSW200 from Erwinia stewartii. Plasmid 40100-112. [DOI] [PubMed] [Google Scholar]
- 16.Fu, J. F., S. W. Ying, and S. T. Liu. 1997. Cloning and characterization of the ori region of pSW1200 of Erwinia stewartii: similarity with plasmid P1. Plasmid 38141-147. [DOI] [PubMed] [Google Scholar]
- 17.Funnell, B. E. 1988. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc. Natl. Acad. Sci. USA 856657-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghuysen, J. M. 1991. Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 4537-67. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54681-708. [DOI] [PubMed] [Google Scholar]
- 20.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 1001541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyer, M. S. 1978. The gamma delta sequence of F is an insertion sequence. J. Mol. Biol. 126347-365. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 20463-113. [DOI] [PubMed] [Google Scholar]
- 23.Harris, R. L., V. Hombs, and P. M. Silverman. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42757-766. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 3011496-1499. [DOI] [PubMed] [Google Scholar]
- 25.Hiraga, S. 1992. Chromosome and plasmid partition in Escherichia coli. Annu. Rev. Biochem. 61283-306. [DOI] [PubMed] [Google Scholar]
- 26.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1451365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klimke, W. A., C. D. Rypien, B. Klinger, R. A. Kennedy, J. M. Rodriguez-Maillard, and L. S. Frost. 2005. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology 1513527-3540. [DOI] [PubMed] [Google Scholar]
- 28.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2241-15. [DOI] [PubMed] [Google Scholar]
- 29.Lee, L. Y., and S. T. Liu. 1991. Characterization of the yellow-pigment genes of Erwinia herbicola. Mol. Microbiol. 5217-224. [DOI] [PubMed] [Google Scholar]
- 30.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 1775539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, K. A., M. A. Davis, and S. Austin. 1991. Fine-structure analysis of the P1 plasmid partition site. J. Bacteriol. 1733630-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Nordstrom, K., and K. Gerdes. 2003. Clustering versus random segregation of plasmids lacking a partitioning function: a plasmid paradox? Plasmid 5095-101. [DOI] [PubMed] [Google Scholar]
- 34.Perry, K. L., T. A. Simonitch, K. J. Harrison-Lavoie, and S. T. Liu. 1986. Cloning and regulation of Erwinia herbicola pigment genes. J. Bacteriol. 168607-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 984486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J. F., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 6.33. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sambrook, J. F., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. A3.7. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schandel, K. A., M. M. Muller, and R. E. Webster. 1992. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J. Bacteriol. 1743800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumacher, M. A. 2007. Structural biology of plasmid segregation proteins. Curr. Opin. Struct. Biol. 17103-109. [DOI] [PubMed] [Google Scholar]
- 40.Sitbon, E., and S. Pietrokovski. 2003. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 28473-477. [DOI] [PubMed] [Google Scholar]
- 41.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 42.Summers, D. 1998. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 291137-1145. [DOI] [PubMed] [Google Scholar]
- 43.Summers, D. K., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell 361097-1103. [DOI] [PubMed] [Google Scholar]
- 44.To, K. Y., E. M. Lai, L. Y. Lee, T. P. Lin, C. H. Hung, C. L. Chen, Y. S. Chang, and S. T. Liu. 1994. Analysis of the gene cluster encoding carotenoid biosynthesis in Erwinia herbicola Eho13. Microbiology 140331-339. [DOI] [PubMed] [Google Scholar]
- 45.Vecchiarelli, A. G., M. A. Schumacher, and B. E. Funnell. 2007. P1 partition complex assembly involves several modes of protein-DNA recognition. J. Biol. Chem. 28210944-10952. [DOI] [PubMed] [Google Scholar]
- 46.Wahle, E., and A. Kornberg. 1988. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 71889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, C. Y., J. F. Fu, and S. T. Liu. 2001. The replicon of pSW800 from Pantoea stewartii. Microbiology 1472757-2767. [DOI] [PubMed] [Google Scholar]
- 48.Wu, J. H., P. Kathir, and K. Ippen-Ihler. 1988. The product of the F plasmid transfer operon gene, traF, is a periplasmic protein. J. Bacteriol. 1703633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]