Abstract
Osmotic stress is known to increase the thermotolerance and oxidative-stress resistance of bacteria by a mechanism that is not adequately understood. We probed the cross-regulation of continuous osmotic and heat stress responses by characterizing the effects of external osmolarity (0.3 M versus 0.0 M NaCl) and temperature (43°C versus 30°C) on the transcriptome of Escherichia coli K-12. Our most important discovery was that a number of genes in the SoxRS and OxyR oxidative-stress regulons were up-regulated by high osmolarity, high temperature, or a combination of both stresses. This result can explain the previously noted cross-protection of osmotic stress against oxidative and heat stresses. Most of the genes shown in previous studies to be induced during the early phase of adaptation to hyperosmotic shock were found to be also overexpressed under continuous osmotic stress. However, there was a poorer overlap between the heat shock genes that are induced transiently after high temperature shifts and the genes that we found to be chronically up-regulated at 43°C. Supplementation of the high-osmolarity medium with the osmoprotectant glycine betaine, which reduces the cytoplasmic K+ pool, did not lead to a universal reduction in the expression of osmotically induced genes. This finding does not support the hypothesis that K+ is the central osmoregulatory signal in Enterobacteriaceae.
Bacteria possess ensembles of transcriptionally regulated genes, commonly called stress or shock response systems, that enable them to adapt very rapidly to changes in the chemical or physical aspects of their environment, including water activity (osmolarity), pH, temperature, and oxygen concentration (65). Adaptation to most, if not all, environmental shifts generally involves two stages: a transient or acute phase (shock response) that consists of rapid responses needed to initiate the adaptation to the new conditions, and a continuous or chronic phase (stress response) that consists of responses that are needed to support exponential growth, possibly at a new growth rate, in the altered environment.
To adapt to a reduction in external water activity, cells accumulate low-molecular-weight solutes to maintain the proper intracellular osmotic balance. Major osmoregulatory solutes in Enterobacteriaceae in minimal medium are K+, glutamate, and trehalose (12). It has been proposed that the increased concentration of cytoplasmic K+ is the central regulatory signal that turns on most of the other osmotically controlled responses (22, 56). A class of compounds called osmoprotectants can be accumulated to high concentrations inside the cells by transport from the medium and thereby can alleviate the inhibitory effects of osmotic stress (66). These include glycine betaine (GB), proline, and about a dozen other structurally related zwitterionic molecules (17), of which GB is one of the most potent. Osmoprotectants can mitigate the adverse effects of high osmolarity by increasing the free water content of the cytoplasm (12), and they may also increase the stability of macromolecules in solutions of low water activity (2, 34). In Enterobacteriaceae, the accumulation of K+, glutamate, and trehalose is suppressed by GB and to a lesser extent by proline, if these are present in the medium (12, 56).
The response of bacteria to high temperature has often been studied by applying a short temperature upshift and thus is called the heat shock response (3, 45). Although originally defined as a reaction to elevated temperature, this response is also used in other adverse conditions that lead to the accumulation of unfolded and damaged proteins, such as exposure to harmful chemicals (antibiotics, solvents) or overproduction of endogenous and recombinant proteins (65). In Escherichia coli, the heat shock response consists of the induction of more than 20 different heat shock proteins (HSPs) (29, 54), the majority of which are either molecular chaperones that assist the refolding of denatured proteins or proteases that degrade misfolded and abnormal proteins (16, 65, 67).
Exposure of cells to one type of stress can also condition them against other, seemingly unrelated, stresses. For example, when bacteria are challenged with high osmolarity, they acquire increased resistance to high temperature and oxidative stresses (10, 24, 32, 57). The high-osmolarity-dependent increase in thermotolerance has two manifestations: elevation of the upper limit of the growth temperature and enhanced survival at otherwise lethal high temperatures (24, 32, 57). Although osmoprotectants can confer a striking growth rate improvement at high osmolarity, they can have adverse consequences for other aspects of cell physiology (25). GB blocks the high-osmolarity-dependent acquisition of increased resistance against high temperature and H2O2 in Salmonella enterica serovar Typhimurium (24, 25), S. enterica serovar Enteritidis, E. coli K-12, and E coli O157:H7 (L. N. Csonka, unpublished data).
Traditionally, various forms of high-temperature treatment have been effective, inexpensive means of food preparation and preservation. Treatment with high concentrations of salt or sugar is another widely used practice of food preservation (66). Therefore, the mechanism of osmotic regulation of thermotolerance is an important concern for food microbiology. In order to probe transcriptional changes in bacteria subjected to prolonged osmotic and heat stresses and to evaluate the cross-regulation of these stress responses, we determined the global transcriptome of E. coli K-12 during exponential growth at two different temperatures (30 and 43°C) and at low and moderately high osmolarities (0.0 and 0.3 M NaCl). Because GB reduces the accumulation of K+ in media of high osmolarity (12, 56), we determined the effect of this osmoprotectant on the transcriptome in order to test the hypothesis that K+ is the regulatory signal for all other osmotically controlled responses (22, 56).
MATERIALS AND METHODS
Bacterial growth.
E. coli K-12 strain NCM3722 was used for all experiments. This strain is a prototroph that comes as close as possible to reconstructing the ancestral wild-type E. coli K-12 strain (51). NCM3722 has been successfully used previously in microarray studies (30, 31, 51), and with the exception of the fnr region (b1332 to b1344), galactitol operon (b2096 to b2090), and flagellar genes, it shows a gene expression profile similar to that of the sequenced MG1655 strain (51). Cultures were grown with aeration at the indicated temperatures in K medium containing 10 or 20 mM glucose. This medium has an osmotic strength of 0.15 osmol/kg H2O (4). Osmotic stress was imposed by increasing the osmolarity of the medium to 0.64 osmol/kg H2O by 0.3 M NaCl (4). Because E. coli is a methionine auxotroph above 42°C (46), K medium was supplemented with 0.5 mM methionine. For the isolation of RNA, cells from single colonies on LB agar were inoculated into 1 ml liquid LB and grown to saturation at 37°C. Fifty microliters was then inoculated into 5 ml of K medium-0.5 mM methionine-10 mM glucose or this medium containing 0.3 M NaCl with or without 1 mM GB. The cultures were grown to saturation at 30°C and 43°C and then subcultured at a 1:8 dilution into 16 ml of the same medium and incubated at the same temperatures, respectively, as before. After one doubling (∼1 h), exponentially growing cells from these cultures were inoculated to a target optical density at 600 nm (OD600) of 0.05 into 40 ml of medium containing the same NaCl concentrations as in the previous step, and the glucose concentration was increased to 20 mM. The cultures were incubated in 250-ml Erlenmeyer flasks at the same temperatures as previously. These steps ensured that the cells had been adapted to the desired salinity and temperature during the first growth cycle in K medium. For all cultures, doubling times were 0.79 to 0.96 h−1 (Table 1). When the cells reached an OD600 of 0.4 to 0.5 (about three doublings), 25 ml was added to a 2.5-ml mixture of cold 95% ethanol and 5% phenol to preserve RNA (51). The cells were cooled rapidly on ice and immediately harvested by centrifugation (4°C), and the pellet was frozen on dry ice. The six different growth conditions used were as follows, C1, 30°C, no NaCl; C2, 30°C, 0.3 M NaCl; C3, 30°C, 0.3 M NaCl, 1 mM GB; C4, 43°C, no NaCl; C5, 43°C, 0.3 M NaCl; C6, 43°C, 0.3 M NaCl, 1 mM GB.
TABLE 1.
Growth conditions and culture doubling times
| Sample | Growth conditions | Doubling time (h−1)a |
|---|---|---|
| C1 | 30°C, no NaCl | 0.91 ± 0.01 |
| C2 | 30°C, 0.3 M NaCl | 0.92 ± 0.03 |
| C3 | 30°C, 0.3 M NaCl, 1 mM GB | 0.89 ± 0.02 |
| C4 | 43°C, no NaCl | 0.93 ± 0.05 |
| C5 | 43°C, 0.3 M NaCl | 0.82 ± 0.03 |
| C6 | 43°C, 0.3 M NaCl, 1 mM GB | 0.89 ± 0.07 |
Data are shown as the arithmetic mean ± the standard deviation (n = 4).
Microarray sample preparation.
Total RNA was extracted by the hot phenol-chloroform method (30). cDNA synthesis, fragmentation, and labeling and washing and scanning of E. coli GeneChip arrays were performed according to the instructions of the manufacturer (Affymetrix, Inc.). Labeled cDNA was hybridized to an E. coli Genome 2 array (Affymetrix, Inc.). Independent array hybridizations were carried out for three biological replicates (independent cultures) of each condition (C1 to C6).
Microarray data analysis.
Hybridization intensity data were obtained from the scanned array images, and all chips were normalized to the same average target intensity (1,500) in Affymetrix gene chip operating software. All nine possible interchip comparisons were carried out for each pair of conditions by using default gene chip operating software statistical parameters (γ1L = γ1H = 0.002, γ2L = γ2H = 0.002667, perturbation = 1.1). For each set of nine pairwise comparisons, a weighted mean of the signal log ratio (SLR, represents the log2 of the ratio of probe signal intensities between two chips) was computed by using the one-step Tukey biweight estimate (33). A similar approach was used to derive a consensus “detection P value.” Default E. coli array P value cutoff parameters (α1 = 0.05, α2 = 0.065) were applied to these consensus values to estimate the transcript presence under each condition. We eliminated from further consideration genes that were not present under any of the six conditions studied, as well as genes that were always expressed below 1/10 of the average array signal (1,500). These procedures produced a data set with 15 pairwise comparisons for 3,922 E. coli genes.
The filtered data set was imported into the Genesis microarray analysis software (version 1.6.2; [55]) as previously described (30). Euclidean distance was used as a measurement of gene profile similarity. K-means clustering and principal-component analysis were used to analyze the microarray data. Detailed explanations of these clustering algorithms can be found in references 47 and 48. Since in K-means clustering the number of clusters (K) into which the data set is partitioned needs to be user defined, we considered cluster partitions with K = 15, 18, 20, and 25. We chose K = 18 for further analysis because increasing the number of clusters did not result in any new cluster profiles (as judged by visual inspection of SLR averages); lowering the number of clusters below 18 led to the merging of two different cluster profiles into a single cluster. Since the K-means algorithm is not deterministic (i.e., it can produce slightly different results from the same data set in different runs due to random initialization of the clustering algorithm [48]), we confirmed that the cluster profiles and the total number of genes in each cluster either remained unchanged (profiles) or changed little (cluster size) for multiple K-means runs.
Each of the 18 clusters was then classified depending on the profile of its response to heat and osmotic stresses and to the addition of GB. Designation of K clusters assigned as heat, osmolarity, or GB regulated was based on the average SLR of the genes in that particular cluster exceeding a minimum cutoff threshold of 1.5-fold for the corresponding effect. This cutoff value was chosen after manual examination of the average SLR values in each cluster and provided the best overall discrimination between clusters containing previously recognized “responsive” and “nonresponsive” genes. Note that we use a more stringent 2-fold cutoff criterion for the consideration of individual genes (instead of the 1.5-fold cutoff used for cluster averages) as being differentially expressed (18). Cluster contents were exported into Excel and incorporated into an E. coli annotation file (15) for further biological interpretation. The complete data set has been deposited at the NCBI GEO database (record GSE7656) and is also available at http://www.med.wright.edu/bmb/op/papers/Osmo_Temp/.
Cell motility assays.
Cell motility assays of E. coli K-12 NCM3722 were performed with M9 minimal medium supplemented with 0.2% glycerol and 0.2% agar. Bacteria were grown in M9 medium, and cells were diluted to an OD600 of 0.2. A 5-μl aliquot was subsequently spotted in the middle of each agar plate containing a different salt concentration (0.0 M, 0.3 M, or 0.6 M NaCl). Plates were incubated for 72 h at different temperatures (30°C, 41°C, 42°C, and 43°C) and then inspected for cell migration. The incubator temperature was confirmed with a second thermometer; to avoid temperature shifts during experiments, the incubator was not opened until the plates were ready for inspection.
E. coli oxidative-stress regulon.
A list of oxidative-stress response genes was compiled by using the corresponding gene lists from (i) the EcoCyc database (http://ecocyc.org/) and (ii) the GenProtEC MultiFun E. coli protein functional classification (http://genprotec.mbl.edu/) and (iii) a hydrogen peroxide-responsive gene list from Zheng et al. (68).
RESULTS
Growth conditions and data analysis.
We determined the global transcriptome of E. coli strain K-12 growing at low osmolarity (in K medium) or at moderately high osmolarity (in K medium containing 0.3 M NaCl) at moderately low (30°C) and moderately high (43°C) temperatures. We also investigated the effects of the osmoprotectant compound GB on the E. coli transcriptome in K medium augmented with 0.3 M NaCl at both temperatures. These combinations of osmolarity and temperature were used because the cells grew at similar rates under all conditions (Table 1) and because 0.3 M NaCl is sufficient for maximal induction of thermotolerance (24). The uniformity of growth rates was important to ensure that the transcriptional responses we observed were due to osmotic or temperature-dependent regulation of gene expression rather than to growth rate-dependent effects (30). It should be emphasized that our experiments were carried out with exponentially growing cells that had been adapted to high or low osmolarity and high or low temperature for at least 12 generations, and thus the cells experienced continuous osmotic and/or heat stresses. Other studies addressed the acute transcriptional responses of E. coli 9 min (63) or 20 min (64) after osmotic shock or 7 min after high-temperature shock (45). Consequently, the changes in gene expression that were observed in these other studies could include (i) acute responses to the environmental shifts that may not persist after the cells have adapted to the new conditions and (ii) responses to the growth rate transition during shock that may be absent under steady-state conditions (4, 44).
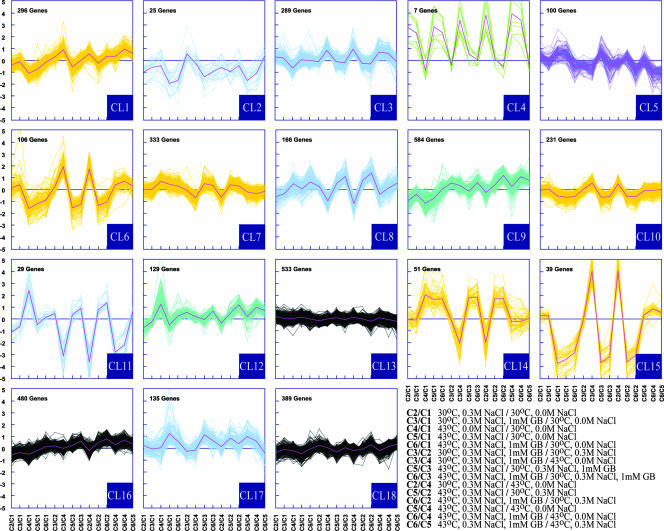
We used Affymetrix E. coli Genome v2 arrays to obtain gene expression profiles for all samples. Cluster analysis (30, 31) was used to identify groups of differentially expressed genes with similar expression profiles. Figure 1 shows a distribution of all genes into 18 clusters by a K-means algorithm (31, 55). Each cluster displays the expression profiles of all of the genes in that cluster across 15 pairwise comparisons as described in the figure legend. The clusters were color coded depending on their expression profiles; the designation of clusters and the response of cluster genes to osmotic and heat stresses and to the presence of GB are shown in Table 2. For example, cluster 4 (CL4) was designated as “overexpressed under osmotic stress only” because the genes in this cluster were consistently up-regulated in all pairwise comparisons in which the osmolarity of the medium was a variable (0.3 versus 0.0 M NaCl), irrespective of the temperature or the presence or absence of GB. Because the genes in this cluster, on average, showed little differences in expression at different temperatures or in the presence or absence of GB at constant osmolarity, they were not considered to consistently respond to the differences in temperature or to the presence of GB. A similar approach was used to assign responses to all other clusters (Table 2).
FIG. 1.
K-means clustering of the complete data set (K = 18). Each box shows a transcriptional response of genes partitioned into a separate cluster. A cluster number is shown in the bottom right corner of each box. The total number of genes in each cluster is shown in the top left corner of each cluster. The 15 pairwise comparisons obtained among six different growth conditions are indicated on the x axis; comparisons are listed in the bottom right corner. The SLRs of expression are indicated on the y axis. The lines in each cluster represent the data for individual genes. Lines are colored as follows, according to the response of the genes in each cluster to osmotic and/or heat stresses, and/or to the presence of GB in the medium: yellow, genes responding to heat stress only; green, osmotic stress only; blue, osmotic and heat stresses; purple, presence of GB; teal, osmotic and heat stresses and presence of GB; black, no difference. The designation of each cluster is also shown in Table 2. The red/pink line in each cluster designates the centroid. Comparisons represent growth condition differences as follows (from left to right): 1, salt concentration; 2, salt and GB; 3, temperature; 4, salt and temperature; 5, salt, temperature, and GB; 6, presence of GB; 7, salt, temperature, and GB; 8, temperature and GB; 9, temperature; 10, salt and temperature; 11, temperature; 12, temperature and GB; 13, salt; 14, salt and GB; 15, presence of GB.
TABLE 2.
Classification of K-means clusters based on gene transcriptional responses to heat and osmotic stresses and to the addition of GBa
| Cluster | Heat stress | Osmotic stress | GB | Effect(s) | Further analysis |
|---|---|---|---|---|---|
| CL1 | ↓ | — | — | Heat | |
| CL2 | ↓ | ↓↓↓ | — | Heat, osmolarity | Yes |
| CL3 | ↓ | ↑ | — | Heat, osmolarity | |
| CL4 | — | ↑↑↑ | — | Osmolarity | Yes |
| CL5 | — | — | ↓ | GB | |
| CL6 | ↓↓↓ | — | — | Heat | Yes |
| CL7 | ↑ | — | — | Heat | |
| CL8 | ↑↑ | ↓ | — | Heat, osmolarity | |
| CL9 | ↓ | ↓ | ↑ | Heat, osmolarity, GB | |
| CL10 | ↓ | — | — | Heat | |
| CL11 | ↑↑↑ | ↓↓↓ | — | Heat, osmolarity | Yes |
| CL12 | ↑↑ | ↓ | ↑ | Heat, osmolarity, GB | |
| CL13 | — | — | — | No change | |
| CL14 | ↑↑↑ | — | — | Heat | Yes |
| CL15 | ↓↓↓ | — | — | Heat | Yes |
| CL16 | — | — | — | No change | |
| CL17 | ↑ | ↑ | — | Heat, osmolarity | |
| CL18 | — | — | — | No change |
The numbering of K-means clusters corresponds to that in Fig. 1. Vertical arrows represent up- or down-regulation of genes in a particular cluster due to heat stress, osmotic stress, or addition of GB. To assign transcriptional responses to each cluster, numerical criteria were used as described in Materials and Methods. Numbers of arrows approximate the magnitude of up- or down-regulation as follows: three arrows, at least a 3-fold average difference; two arrows, 2- to 3-fold difference; one arrow, 1.5- to 2-fold difference. A dash represents no difference in the transcriptional response.
Overall, 3 out 18 clusters, which accounted for 36% of the genome, contained genes that did not show significant variation in their expression level in any of the comparisons (Fig. 1). Included among these were the potassium transporter genes trkG (b1363), trkA (b3290), trkH (b3849), and kup (= trkD, b3747), which have been shown previously to be constitutively expressed in E. coli (7). As expected, the expression of a considerable number of genes was influenced by temperature and/or osmolarity; the magnitude (n-fold change) of the transcriptional responses shown by the thermoregulated genes was similar to that exhibited by the osmotically controlled genes. However, there was a substantially larger number of genes responding specifically to heat stress (six clusters) than exclusively to osmotic stress (one cluster) (Fig. 1). A number of clusters contained genes that responded to both stresses. We confirmed general partitioning of the genes into K-means clusters by visualizing the positioning of the genes in the PCA 3D space (see supplemental figure at http://www.med.wright.edu/bmb/op/papers/Osmo_Temp/). We chose six of the K clusters with the largest expression changes for further analysis of their genes (Table 2).
Transcriptional response to osmotic stress.
Cluster 4 (CL4, Fig. 1) represented seven genes that were highly up-regulated by osmotic stress but did not respond to heat stress alone. Members of two operons participating in the osmotic-stress-stimulated uptake of GB/proline (proU, b2677 to b2679) and K+ (kdpFABC, b4513 and b0696 to b0698) were in this cluster. All three genes of the proU operon (proV, proW, and proX) were in this cluster and showed a ≥16-fold average increase in expression in response to 0.3 M NaCl at 30 or 43°C. The expression of kdpA, the only gene of the kdpFABC operon in this cluster, increased about sevenfold when cells were grown in the presence of 0.3 M NaCl. The other genes of this operon, kdpB and kdpC (the array contains no probes for the kdpF [b4513] gene), were also up-regulated by salt stress (>2.5-fold) but were partitioned into cluster 17. ProP (b4111), a second transporter for proline/GB, was only slightly overexpressed under osmotic stress (up to 1.6-fold). In addition, a hypothetical protein (encoded by yrbL [b3207], identified as a member of the Mg2+ stimulon [40]), a putative ATP-binding protein (bax, b3570), and a small RNA gene (rybA, b4416) were part of cluster 4 and can be considered candidates for new osmotic-stress-regulated genes. It is notable that bax overexpression was also a part of the early response to osmotic upshift (63).
Transcriptional response to heat stress.
We considered three clusters of genes showing the largest responses to continuous heat stress (CL6, CL14, CL15; Fig. 1 and Table 2). Cluster 14 comprised 51 genes that were highly up-regulated under heat stress (at least fourfold induction). This cluster contained several known chaperones and HSPs, such as ClpB and IbpB. Other well-studied heat shock proteins, DnaK, DnaJ, GrpE, GroEL, and GroES, were also up-regulated by the continuous heat stress. However, the magnitude of such up-regulation was not as great as when cells were subjected to a transient heat shock (45) and therefore these genes were partitioned into cluster 7 (DnaJ was in cluster 17), representing genes modestly up-regulated (1.6-fold) by heat stress (Fig. 1). In addition, high temperature up-regulated (about fivefold) several genes involved in sulfur metabolism, including the cysHIJ (b2762 to b2764), cysND (b2751 and b2752), cysA (b2422), and cysP (b2425) genes. The cysD and cysN genes encode sulfate adenylyltransferase, which converts intracellular sulfate to adenosine 5′-phosphosulfate and 3′-phosphoadenylyl-sulfate. Two genes in the cysteine and methionine degradation pathways (metC [b3008] and tnaA [b3708]) were down-regulated at high temperature. Both 5′-phosphosulfate and 3′-phosphoadenylyl-sulfate are known to play important roles in the thermotolerance of Saccharomyces cerevisiae (36), as is methionine in E. coli (26). Cysteine is also a precursor of glutathione and is an important constituent of the antioxidant defense systems (8, 62). Nineteen genes in cluster 14 (Fig. 1) encoded proteins of unknown function.
Clusters 6 and 15 (CL6 and CL15, Fig. 1) contained genes that were considerably down-regulated by high temperature and showed little or no response to continuous osmotic stress. Cluster 15 contained 39 genes that were the most highly down-regulated genes (10-fold on average) in response to heat stress. Almost all of the genes in this cluster (a total of 35) are involved in flagellar biosynthesis or chemotaxis. The fliA gene, which encodes the transcriptional activator for the class III flagellar genes, was down-regulated >150-fold by heat stress. Most of the flagellar and chemotaxis genes missing from cluster 15 were in cluster 6 (CL6, Fig. 1; heat stress down-regulated by about 4.5-fold).
Genes responding to both stresses.
Two clusters (CL2 and CL11, Fig. 1 and Table 2) showed transcriptional responses to both persistent stresses. Cluster 11 (CL11, Fig. 1) contained 29 genes whose expression was down-regulated by continuous osmotic stress but was up-regulated under continuous heat stress. Several acid resistance genes (gadA [b3517], gadB [b1493], hdeAB [b3509 and b3510], and hdeD [b3511]) were in this cluster. Cluster 2 comprised 25 genes that were down-regulated under both stresses. Genes coding for the catabolism of the polyamine putrescine (puuR, puuB, puuC, puuD, and puuE [b1298 to b1302]) were in this cluster. The putrescine transporter gene puuP (b1296) did not show any changes in its expression in any of the comparisons. Putrescine was implicated in the regulation of RpoS protein translation and stability (61), in the early phase of adaptation to hyperosmotic shock (60), and in the response to temperature stress (59). It was also shown recently that putrescine protects E. coli against oxidative stress (58). In E. coli, this divalent cation, which is a major counterion for the negatively charged nucleic acids in cells grown at low osmolarity, is excreted rapidly upon a hyperosmotic shock; the exchange of one molecule of putrescine for two K+ ions enables the cells to balance the anionic charge of macromolecules and at the same time to increase the internal cellular osmolarity (43). The decrease in the internal putrescine pool at high osmolarity might be the regulatory signal for the down-regulation of the puu genes.
The cusCF genes (b0572 and b0573) of the cusCFBA operon, which encodes a copper-transporting efflux system, were down-regulated sixfold under both continuous stresses. The cusB and cusA genes, which were also down-regulated at least twofold by both stress conditions, are part of cluster 18 (CL18, Fig. 1). Copper is a cofactor for several enzymes, including copper-zinc superoxide dismutase (encoded by sodC [b1646]), which protects cells against free radicals generated from oxidative stress (28). In our comparisons, copper-zinc superoxide dismutase increased over twofold due to heat and osmotic stress, in agreement with the down-regulation of the copper efflux system.
Cross-regulation of osmotic and temperature adaptation.
In order to assess whether the increase in thermotolerance that is conferred by high osmolarity might be due to an osmotic control of genes that have thermoprotective functions, we examined E. coli gene expression in the presence of continuous osmotic stress at low and high temperatures and at high temperature in the presence of low and high salt concentrations. A vast number of genes showing mRNA expression differences in these comparisons were of unknown or putative function, but there was also a number of interesting genes with known function in this category. The otsAB genes (b1896 and b1897), which encode enzymes of biosynthesis of trehalose from glucose-6-phosphate and UDP-glucose, were induced under both stresses. Induction of these two genes was higher when cells were exposed to both continuous stresses simultaneously than when they were exposed to one stress, in accord with similar observations made with an otsA-lacZ reporter fusion in S. enterica serovar Typhimurium (10). The transcription of the sufABCDSE (b1679 to b1684) operon, which encodes the alternative Fe-S cluster assembly system in E. coli that is known to protect cells from oxidative damage (37), was up-regulated under combined stress conditions but not when cells were exposed to a single continuous stress.
We found that the betIBA genes (b0311 to b0313), which specify the enzymes for the conversion of choline to GB, were induced by heat stress (two- to fourfold) but not by high osmolarity, whereas the GB transport system (ProU) was induced only under osmotic stress (see above). The expression of the betI and betB genes was found to be down-regulated about fivefold at 16°C compared to 37°C in a previous study (23). Both the betIBA and proU systems were up-regulated when cells were subjected to both stresses simultaneously. Note that choline cannot be synthesized by E. coli and thus is only potentially available to cells from the environment (choline was not added to K medium). This difference in the regulation of betAB and proU may represent an interesting case of two different strategies cells seem to use to acquire GB (9). The preference of transport over synthesis under osmotic-stress conditions might relate to the fact that GB is present in the environment of high osmolarity that E. coli cells are likely to encounter in vivo—human and animal urine (with an osmolarity of up to 1,400 mosmol/kg [13]).
Cell motility under heat and osmotic stresses.
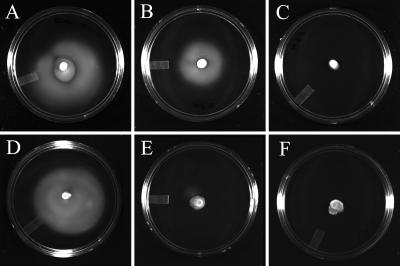
As described above, we observed substantial down-regulation of the genes of the motility and chemotaxis regulon (MCR) under continuous heat stress. There was no significant change in the expression of these genes when cells were subjected to continuous osmotic stress. We showed previously that MCR genes are repressed under poor growth conditions and upon nitrogen and sulfur downshifts (31). Down-regulation of most of the genes of this regulon was also observed for Bacillus subtilis growing with 1.2 M NaCl (52), although only several genes in this set were found to be down-regulated in E. coli during the initial phase of adaptation to heat (45) and osmotic shocks (14). Therefore, we sought to corroborate the apparent differences in MCR gene expression under continuous heat and osmotic stresses by agar motility assays (Fig. 2). The motility of E. coli was strongly impaired at 43°C (Fig. 2F) compared to 30°C (Fig. 2A), consistent with the results of the microarray experiments. Cell growth, as judged by colony size on agar plates and by growth rates of cultures in liquid medium, was not affected at 43°C (data not shown). Motility on soft agar plates was only partially inhibited by 0.3 M NaCl (Fig. 2B). Cell motility in medium with 0.6 M NaCl was completely abolished (Fig. 2C); cell proliferation was also impaired (data not shown).
FIG. 2.
Cell motility of E. coli NCM3722 at different temperatures and osmotic contents of the medium. A, 30°C, no NaCl; B, 30°C, 0.3 M NaCl; C, 30°C, 0.6 M NaCl; D, 41°C, no NaCl; E, 42°C, no NaCl; F, 43°C, no NaCl.
Comparison of the acute and chronic responses of E. coli to heat and osmotic stresses.
The transient transcriptional responses of E. coli to osmotic and heat shocks have been addressed in several previous studies. Weber and Jung (63) profiled E. coli gene expression 9 min after osmotic shock imposed with 0.4 M NaCl in minimal medium; Richmond and colleagues (45) examined the effects of heat shock on E. coli 7 min after a temperature shift from 37 to 50°C. There was only a weak concordance between the results of the previous acute heat shock studies and our chronic heat stress experiments. The majority of the proteins that were induced >30-fold after the heat shock were only slightly overexpressed (up to 3-fold) when cells were continuously grown at the elevated temperature (43°C). Some heat shock regulon genes, such as clpA (b0882), clpP (b0437), clpX (b0438), and lon (b0439), were not induced at all under the long-term heat stress. These results are in agreement with the original work carried out by Straus et al. (54), who found that mRNA levels for the E. coli HSPs increased rapidly for the first 4 min following heat shock and then decreased and settled at a new steady state. One exception to this trend was the hchA gene (= yedU; b1967) coding for the Hsp31 chaperone. The mRNA level for this gene was more than 30-fold higher under heat shock (45) and was up-regulated by about 19-fold in cells growing continuously at 43°C. The Hsp31 protein was proposed to be a holding chaperone that captures early unfolding intermediates under conditions of severe stress and releases them when cells return to physiological conditions (41). Among the genes down-regulated by heat, we noted that only a few MCR members were repressed in the study of Richmond et al., whereas the complete regulon was down-regulated in our experiments. As detailed in Materials and Methods, there are genetic differences between E. coli K-12 strains NCM3722 (motile; used in this study) and MG1655 (expresses flagella poorly; used by Richmond et al.) that could account for the differences between our experimental observations (31, 51).
Several genes known to be involved in the adaptation to high osmolarity (proU operon, proP, otsAB, osmC, osmY) were up-regulated both by the acute osmotic shock in the studies of Weber and Jung and by chronic osmotic stress in our experiments. As mentioned earlier, bax (b3570) was also overexpressed as part of both the transient and long-term responses to osmotic stress. However, a total of 11 genes of unknown and putative function that were classified as being overexpressed during the initial phase of adaptation to osmotic shock were not differentially expressed under continuous osmotic stress. There was also poor agreement between genes that were found to be down-regulated 9 min after the osmotic shock and those that were repressed during the long-term osmotic stress. Many of the transiently down-regulated genes are genes that encode ribosomal proteins, proteins that participate in energy metabolism (nuo, atp, sdhCD), and genes responsible for cell proliferation (fts, prfB, priB) (63). It is possible that their down-regulation was the consequence of the temporary growth arrest after the osmotic shock, whereas there was little difference in the growth rates of E. coli cultures growing exponentially in K medium and K medium containing 0.3 M NaCl (Table 1).
Effect of GB on gene expression under continuous osmotic stress.
GB is one of the most potent osmoprotectants that can alleviate the inhibitory effects of high osmolarity. However, surprisingly, this compound blocks the ability of high osmolarity to increase thermotolerance and oxidative-stress resistance in S. enterica serovar Typhimurium (25) and other pathogenic or laboratory strains of Enterobacteriaceae (L. N. Csonka, unpublished data). It has been proposed that K+ is the intracellular signal that triggers the induction of most of the osmotically controlled operons (22, 56), and because GB reduces the accumulation of K+, glutamate, and trehalose (12), it has been suggested that augmentation with GB would dampen the induction of the osmoregulatory response. In order to test the latter prediction, we determined the effect of GB on gene expression under continuous osmotic stress. Supplementation of the high-osmolarity medium with GB repressed a total of 84 genes: 11 at 30°C and 80 at 43°C, of which 7 were down-regulated at both temperatures (with a twofold change as the cutoff for these comparisons; see gene lists at http://www.med.wright.edu/bmb/op/papers/Osmo_Temp/). Supplementation of the high-osmolarity medium with GB also induced a total of 157 genes: 38 at 30°C and 119 at 43°C, with 10 being up-regulated at both temperatures.
The genes of the proU operon (proVWX, b2677 to b2679) and the otsAB operon (b1896 and b1897), which encode a transport system for the osmoprotectant GB and the biosynthetic enzymes for trehalose, respectively, were down-regulated in the presence of GB; however, the effect was not strong (1.2- to 2.1-fold). Genes of the osmotically inducible potassium transport system kdpABC (b0696 to b0698) were only slightly (1.2- to 1.6-fold) down-regulated in GB-supplemented medium. The levels of the proVWX and kdpABC genes (but not those of otsAB) were still higher in K medium with 0.3 M NaCl and 1 mM GB than when cells were grown in unsupplemented K medium.
The katE gene (b1732), which encodes catalase III, was one of the genes repressed by GB in the high-osmolarity medium. This response could contribute to the reversal of the protection afforded by high osmolarity against oxidative stress (24, 25). The expression of the katE gene is mediated by RpoS, but other genes of the RpoS regulon were not repressed by GB. Surprisingly, supplementation of the high-osmolarity medium with GB at high temperature elevated the expression of several DNA repair genes, including umuDC (b1183 and b1184) (mutagenic repair pathway), dinI (b1061) and dinD (b3645) (encode damage-inducible proteins), and ykfG (b0247) (encodes a DNA damage repair protein). These results suggest that the combination of high temperature, high osmolarity, and GB might increase DNA damage. The stimulus for this response could be the accumulation of reactive oxygen species (ROS) and the resultant increase in DNA damage, which in turn might be due to the down-regulation of katE that we observed.
Induction of oxidative-stress-responsive genes under continuous osmotic and heat stresses.
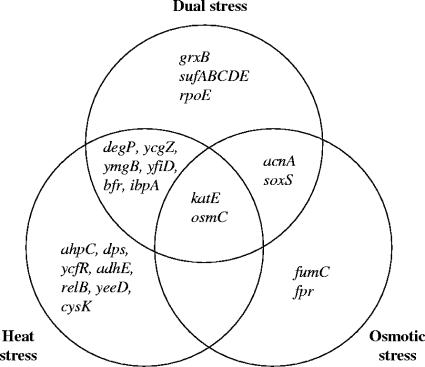
We found that a number of genes participating in a defense against oxidative damage responded to continuous osmotic and/or heat stresses. These included sodC (superoxide dismutase), katE (catalase), osmC (b1482; peroxidase), puu genes (putrescine catabolism), and cusCFBA (copper efflux system). To investigate this further, we compiled a list of 83 known and predicted oxidative-stress-responsive genes by using several sources of information as described in Materials and Methods, and we considered the responses of these genes to osmotic stress, heat stress, or both stresses combined. Genes of the oxidative-stress regulon that showed at least a twofold change in expression upon continuous heat and/or osmotic stress were considered to be cross-regulated by one or both of these latter stresses (Fig. 3). The set of genes that showed elevated expression under our two stress conditions included SoxRS-regulated, as well as OxyR-regulated, genes (53). SoxS, which is the direct transcriptional activator of most of the genes in the SoxRS regulon (19), was induced under each stress (though only 1.9-fold by heat stress); OxyR, which is activated at the protein level by H2O2-mediated oxidation (53), was not transcriptionally induced. Two genes, katE and osmC, were induced under all three conditions (osmotic stress, heat stress, and a combination of both stresses). The grxB gene (b1064) had a 1.8-fold induction under each stress and was up-regulated 2.7-fold by a combination of both stresses; soxS (b4062) was induced 3-fold by osmotic stress, 1.9-fold by heat stress, and 2.4-fold by a combination of the two stresses. Nine genes were induced by one stress and by the two stresses combined; these included the acnA (aconitase A; b1276), bfr (bacterioferritin; b3336), ibpA (heat shock chaperone; b3687), and degP (heat shock serine protease; b0161) genes (49). Proteomic studies confirmed that the Dps, KatE, AcnA, and OsmC proteins were induced during the initial phase of osmotic adaptation (64). A number of genes, including the sufABCDSE operon (b1679 to b1684; encodes components of a secondary pathway of iron-sulfur cluster assembly), were induced when both stresses were applied but not by one stress only (Fig. 3). In summary, a total of 26 genes of the oxidative regulon were up-regulated by at least one of the stresses or by their combination.
FIG. 3.
Responses of genes of the oxidative-stress regulon to osmotic and heat stresses. The Venn diagram indicates genes that showed at least twofold overexpression under a single stress or under a combination of both stresses. There were no genes that were up-regulated under both individual osmotic and heat stresses but not when both stresses were applied simultaneously.
DISCUSSION
In order to probe the mechanism of the osmotic stimulation of thermotolerance and oxidative-stress resistance in E. coli, we determined the effects of moderately high osmolarity and moderately high temperature on the transcriptional profiles of E. coli. Our most important novel finding was the discovery that osmotic stress and high temperature, individually or in combination, induced at least 26 genes of the oxidative-stress regulon. The induction of the members of this regulon could directly account for the increase in the oxidative-stress tolerance of E. coli challenged with high osmolarity and may also explain the cross-protection between osmotic and heat stresses, as we discuss below.
There are several lines of evidence in the literature suggesting that both elevated temperature and osmotic stress can lead to oxidative damage. High temperature stimulated the production of free radicals in cells (6, 38, 42), and hyperthermia increased oxidative stress in humans (39). Salinity stress has been shown to enhance the production of ROS in plants (50) and animals (20, 35) through membrane damage resulting in a dysfunction in electron transport (50). Bacteria produce trehalose in response to osmotic stress, and the accumulation of this compound in media of high osmolarity is further stimulated by high temperature (10). However, the levels of trehalose are too low to make a substantial contribution to the cytoplasmic osmolarity, suggesting that it is unlikely to function as an osmoregulatory solute (10). However, it has been suggested that trehalose is an antioxidant (5), and therefore a major role for this disaccharide could be to provide protection against oxidative radicals produced in cells that are challenged with osmotic or heat stress.
Though exposure to high osmolarity enhances the thermotolerance of bacteria, we did not uncover genes with a recognizable thermoprotective function that were induced by high osmolarity at 30 or 43°C. This result could mean that the thermoprotective effect of elevated osmolarity is mediated either by proteins whose thermoprotective function is not obvious or by a collection of weakly induced proteins or that the regulation is exerted posttranscriptionally. However, because the production and chemical reactivity of ROS are enhanced with increasing temperature (6, 38, 42), it is possible that excessive oxidative damage is an important contributing factor in the impairment of growth or viability at elevated temperatures. Thus, by mitigating the deleterious effects of ROS, the osmotic induction of the members of the oxidative-stress regulon may also confer increased resistance to high-temperature stress. Osmotic and/or heat stress can serve as a direct inducer of the oxidative-stress-related genes, or the induction is an indirect consequence of the production of ROS under these conditions. This hypothesis is consistent with the suicide response model of Aldsworth and Dodd (1, 21), which postulates that when actively growing cells are subjected to environmental stress, they produce an excess of free radicals inside cells. The model is supported by two recent studies which demonstrated that a set of common environmental response genes, which were up-regulated under a variety of stressful conditions, included representatives involved in defense against oxidative damage (11, 27).
The second goal of our study was to elucidate the effect of GB on E. coli gene expression under osmotic and heat stresses. GB is one of the most potent osmoprotectants for E. coli, and its presence in the medium not only allows cells to better survive hyperosmotic conditions but it also reduces the accumulation of K+, glutamate, and trehalose (12). Although we found a number of genes that were down-regulated upon the addition of GB to the high-osmolarity medium, we did not see an across-the-board repression of osmotically controlled genes by this osmoprotectant. The model in which K+ is the global regulatory signal of the induction of osmotically controlled gene expression, coupled with the observation that GB decreases the accumulation of K+ (12), predicted that GB should dampen the induction of all of the osmoregulatory responses (22, 56). Our data do not corroborate this theory.
Acknowledgments
Our thanks go to John Lynch for manuscript proofreading and to the members of the Center for Genomics Research at Wright State University for access to their facility. We are also thankful to anonymous reviewers for valuable comments and constructive criticism.
O.P. was supported by Wright Brothers Institute award WBSC9004A and by Wright State University grant 229077, and L.N.C. was supported by USDA NRI Food Safety Program grant 2004-04377.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Aldsworth, T. G., R. L. Sharman, and C. E. Dodd. 1999. Bacterial suicide through stress. Cell. Mol. Life Sci. 56378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, T., and S. N. Timasheff. 1985. The stabilization of proteins by osmolytes. Biophys. J. 47411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsène, F., T. Tomoyasu, and B. Bukau. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 553-9. [DOI] [PubMed] [Google Scholar]
- 4.Balaji, B., K. O'Connor, J. R. Lucas, J. M. Anderson, and L. N. Csonka. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl. Environ. Microbiol. 718273-8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benaroudj, N., D. H. Lee, and A. L. Goldberg. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 27624261-24267. [DOI] [PubMed] [Google Scholar]
- 6.Benov, L., and I. Fridovich. 1995. Superoxide dismutase protects against aerobic heat shock in Escherichia coli. J. Bacteriol. 1773344-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossemeyer, D., A. Borchard, D. C. Dosch, G. C. Helmer, W. Epstein, I. R. Booth, and E. P. Bakker. 1989. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 26416403-16410. [PubMed] [Google Scholar]
- 8.Bradley, T. M., E. Hidalgo, V. Leautaud, H. Ding, and B. Demple. 1997. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucleic Acids Res. 251469-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldas, T., N. Demont-Caulet, A. Ghazi, and G. Richarme. 1999. Thermoprotection by glycine betaine and choline. Microbiology 145(Pt. 9)2543-2548. [DOI] [PubMed] [Google Scholar]
- 10.Cánovas, D., S. A. Fletcher, M. Hayashi, and L. N. Csonka. 2001. Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1833365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cayley, S., B. A. Lewis, and M. T. Record, Jr. 1992. Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol. 1741586-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers, S. T., and C. M. Kunin. 1987. Isolation of glycine betaine and proline betaine from human urine. Assessment of their role as osmoprotective agents for bacteria and the kidney. J. Clin. Investig. 79731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, K. J., V. Badarinarayana, D. W. Selinger, D. Janse, and G. M. Church. 2003. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 13206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbin, R. W., O. Paliy, F. Yang, J. Shabanowitz, M. Platt, C. E. Lyons, Jr., K. Root, J. McAuliffe, M. I. Jordan, S. Kustu, E. Soupene, and D. F. Hunt. 2003. Toward a protein profile of Escherichia coli: comparison to its transcription profile. Proc. Natl. Acad. Sci. USA 1009232-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig, E. A., B. D. Gambill, and R. J. Nelson. 1993. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57402-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 18.Cui, X., and G. A. Churchill. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demple, B. 1999. Radical ideas: genetic responses to oxidative stress. Clin. Exp. Pharmacol. Physiol. 2664-68. [DOI] [PubMed] [Google Scholar]
- 20.Dmitrieva, N. I., M. B. Burg, and J. D. Ferraris. 2005. DNA damage and osmotic regulation in the kidney. Am. J. Physiol. Renal Physiol. 289F2-F7. [DOI] [PubMed] [Google Scholar]
- 21.Dodd, C. E., and T. G. Aldsworth. 2002. The importance of RpoS in the survival of bacteria through food processing. Int. J. Food Microbiol. 74189-194. [DOI] [PubMed] [Google Scholar]
- 22.Epstein, W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 3973-78. [Google Scholar]
- 23.Eshoo, M. W. 1988. lac fusion analysis of the bet genes of Escherichia coli: regulation by osmolarity, temperature, oxygen, choline, and glycine betaine. J. Bacteriol. 1705208-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher, S. A., and L. N. Csonka. 1998. Characterization of the induction of thermotolerance by osmotic stress in Salmonella typhimurium. Food Microbiol. 15307-317. [Google Scholar]
- 25.Fletcher, S. A., D. Rhodes, and L. N. Csonka. 2001. Analysis of the effects of osmoprotectants on the high osmolality-dependent induction of increased thermotolerance in Salmonella typhimurium. Food Microbiol. 18345-354. [Google Scholar]
- 26.Gage, D. J., and F. C. Neidhardt. 1993. Modulation of the heat shock response by one-carbon metabolism in Escherichia coli. J. Bacteriol. 1751961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gort, A. S., D. M. Ferber, and J. A. Imlay. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32179-191. [DOI] [PubMed] [Google Scholar]
- 29.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 30.Gyaneshwar, P., O. Paliy, J. McAuliffe, A. Jones, M. I. Jordan, and S. Kustu. 2005. Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc. Natl. Acad. Sci. USA 1023453-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyaneshwar, P., O. Paliy, J. McAuliffe, D. L. Popham, M. I. Jordan, and S. Kustu. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 1871074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hengge-Aronis, R., R. Lange, N. Henneberg, and D. Fischer. 1993. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 175259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbell, E., W. M. Liu, and R. Mei. 2002. Robust estimators for expression analysis. Bioinformatics 181585-1592. [DOI] [PubMed] [Google Scholar]
- 34.Ignatova, Z., and L. M. Gierasch. 2006. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. USA 10313357-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irarrazabal, C. E., J. C. Liu, M. B. Burg, and J. D. Ferraris. 2004. ATM, a DNA damage-inducible kinase, contributes to activation by high NaCl of the transcription factor TonEBP/OREBP. Proc. Natl. Acad. Sci. USA 1018809-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakubowski, H., and E. Goldman. 1993. Methionine-mediated lethality in yeast cells at elevated temperature. J. Bacteriol. 1755469-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. H., W. S. Yeo, and J. H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 511745-1755. [DOI] [PubMed] [Google Scholar]
- 38.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 807496-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAnulty, S. R., L. McAnulty, D. D. Pascoe, S. S. Gropper, R. E. Keith, J. D. Morrow, and L. B. Gladden. 2005. Hyperthermia increases exercise-induced oxidative stress. Int. J. Sports Med. 26188-192. [DOI] [PubMed] [Google Scholar]
- 40.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 1853696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mujacic, M., M. W. Bader, and F. Baneyx. 2004. Escherichia coli Hsp31 functions as a holding chaperone that cooperates with the DnaK-DnaJ-GrpE system in the management of protein misfolding under severe stress conditions. Mol. Microbiol. 51849-859. [DOI] [PubMed] [Google Scholar]
- 42.Privalle, C. T., and I. Fridovich. 1987. Induction of superoxide dismutase in Escherichia coli by heat shock. Proc. Natl. Acad. Sci. USA 842723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Record, M. T., Jr., E. S. Courtenay, D. S. Cayley, and H. J. Guttman. 1998. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23143-148. [DOI] [PubMed] [Google Scholar]
- 44.Regenberg, B., T. Grotkjaer, O. Winther, A. Fausboll, M. Akesson, C. Bro, L. K. Hansen, S. Brunak, and J. Nielsen. 2006. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 7R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 273821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ron, E. Z., and B. D. Davis. 1971. Growth rate of Escherichia coli at elevated temperatures: limitation by methionine. J. Bacteriol. 107391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherlock, G. 2001. Analysis of large-scale gene expression data. Brief. Bioinform. 2350-362. [DOI] [PubMed] [Google Scholar]
- 48.Sherlock, G. 2000. Analysis of large-scale gene expression data. Curr. Opin. Immunol. 12201-205. [DOI] [PubMed] [Google Scholar]
- 49.Skórko-Glonek, J., D. Zurawa, E. Kuczwara, M. Wozniak, Z. Wypych, and B. Lipinska. 1999. The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol. Gen. Genet. 262342-350. [DOI] [PubMed] [Google Scholar]
- 50.Smirnoff, N. 1995. Antioxidant systems and plant response to the environment, p. 217-243. In N. Smirnoff (ed.), Environment and plant metabolism: flexibility and acclimation. BIOS Scientific Publisher, Oxford, United Kingdom.
- 51.Soupene, E., W. C. van Heeswijk, J. Plumbridge, V. Stewart, D. Bertenthal, H. Lee, G. Prasad, O. Paliy, P. Charernnoppakul, and S. Kustu. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 1855611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steil, L., T. Hoffmann, I. Budde, U. Volker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 1856358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2188-194. [DOI] [PubMed] [Google Scholar]
- 54.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 329348-351. [DOI] [PubMed] [Google Scholar]
- 55.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18207-208. [DOI] [PubMed] [Google Scholar]
- 56.Sutherland, L., J. Cairney, M. J. Elmore, I. R. Booth, and C. F. Higgins. 1986. Osmotic regulation of transcription: induction of the proU betaine transport gene is dependent on accumulation of intracellular potassium. J. Bacteriol. 168805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tesone, S., A. Hughes, and A. Hurst. 1981. Salt extends the upper temperature limit for growth of food-poisoning bacteria. Can. J. Microbiol. 27970-972. [DOI] [PubMed] [Google Scholar]
- 58.Tkachenko, A., L. Nesterova, and M. Pshenichnov. 2001. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch. Microbiol. 176155-157. [DOI] [PubMed] [Google Scholar]
- 59.Tkachenko, A. G., M. R. Pshenichnov, O. Salakhetdinova, and L. Nesterova. 1998. Role of putrescine and potassium transport in regulating the topological state of DNA during adaptation of Escherichia coli to temperature stress. Mikrobiologiya 67601-606. [PubMed] [Google Scholar]
- 60.Tkachenko, A. G., O. Salakhetdinova, and M. R. Pshenichnov. 1997. Exchange of putrescine and potassium between cells and media as a factor in the adaptation of Escherichia coli to hyperosmotic shock. Mikrobiologiya 66329-334. [PubMed] [Google Scholar]
- 61.Tkachenko, A. G., and M. S. Shumkov. 2004. Role of putrescine in regulation of the σS subunit of RNA polymerase in Escherichia coli cells on transition to stationary phase. Biochemistry 69876-882. [DOI] [PubMed] [Google Scholar]
- 62.Vergauwen, B., F. Pauwels, M. Vaneechoutte, and J. J. Van Beeumen. 2003. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J. Bacteriol. 1851572-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber, A., and K. Jung. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 1845502-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber, A., S. A. Kogl, and K. Jung. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 1887165-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wick, L. M., and T. Egli. 2004. Molecular components of physiological stress responses in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 891-45. [DOI] [PubMed] [Google Scholar]
- 66.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 130437-460. [DOI] [PubMed] [Google Scholar]
- 67.Yura, T., M. Kanemori, and T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 68.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 1834562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]