Abstract
The Vibrio vulnificus aphB mutant was significantly less virulent than the wild type and was impaired in motility and adherence to host cells. Microarray analysis revealed that AphB of V. vulnificus (AphBVv) influences the expression of over 10% of the V. vulnificus genome. The combined results indicated that AphBVv is a global regulator contributing to the pathogenesis of V. vulnificus.
Microbial pathogenicity is a complex phenomenon that involves the products of many genes, called virulence factors, contributing not only to diseases but also to survival and multiplication on or within the host (20). Most of these virulence factors act cooperatively to obtain maximum effectiveness during pathogenesis, while their expression is coordinately controlled by a common global regulatory system in response to environmental signals (21). This coordinate regulation by a global regulator may facilitate the cooperation of the virulence factors and be crucial for the overall success of the infectious microorganisms during pathogenesis (21). Recently, many regulatory proteins have been implicated as important global regulators controlling the expression of numerous virulence factors in bacterial pathogens (5, 9, 19, 22).
Vibrio cholerae AphB (AphBVc) is a member of the LysR family of transcriptional regulators and plays a central role in the activation of the ToxR virulence cascade (11). AphBVc initiates the expression of the ToxR virulence cascade by directly binding to and activating the promoter of tcpPH in response to yet-unknown environmental signals (11, 12, 13, 14). TcpPH and ToxR function cooperatively in the regulatory cascade, leading to the biogenesis of toxin-coregulated pilus (TCP) and the production of cholera toxin (CT) (4, 7, 11). The TCP is crucial for colonizing the upper intestine (8), and the CT is responsible for the severe diarrhea caused by V. cholerae (10), suggesting that AphBVc is an essential transcriptional regulator modulating the virulence of the organism.
Previously, a homologue of AphBVc from V. vulnificus, a causative agent of food-borne diseases such as gastroenteritis and life-threatening septicemia, was identified (28). The deduced amino acid sequence of AphB from V. vulnificus (AphBVv) was 80% identical to that of AphBVc. However, a search of the genome sequences of both V. vulnificus CMCP6 and YJ016 to find any tcpPH gene homologues with a substantial level of identity was not successful (S. H. Choi, unpublished data). Instead, AphBVv directly activates the transcription of cadC, the product of which facilitates the survival of the pathogen under acid stress (28, 29, 30). This finding indicated that the set of genes regulated by AphB may be broad rather than specific and prompted us to further characterize the functions of the regulatory protein, thus identifying its target genes on a global scale. Accordingly, the present study assessed the functions of AphB by comparing phenotypes of the V. vulnificus aphB mutant with those of the parental wild type in vitro and in mice. Transcriptome analysis using a V. vulnificus whole-genome microarray was also performed and resulted in the identification of at most 489 genes as members of the AphB regulon. Fourteen genes, randomly chosen from the pool of the newly identified genes, were experimentally verified to be regulated by AphB.
Unless noted otherwise, V. vulnificus strains were grown in Luria-Bertani medium supplemented with 2.0% (wt/vol) NaCl at 30°C. For transcriptome analysis, the V. vulnificus whole-genome TwinChip, manufactured and kindly provided by the 21C Frontier Microbial Genomics and Applications Center (Daejeon, South Korea), was used. Total cellular RNA from the V. vulnificus strains grown to an optical density at 600 nm of 0.5 was isolated with an RNeasy midi kit (Qiagen, Valencia, CA), and aminoallyl cDNA was synthesized using an aminoallyl cDNA-labeling kit according to the protocols of the manufacturer (Ambion, Austin, TX). The aminoallyl cDNA from the aphB mutant and that from the wild type were labeled with Cy3 and Cy5 (Ambion), respectively, and equal amounts of the labeled cDNA were combined and used to hybridize the microarray slides at 42°C for 16 h. After hybridization, the arrays were washed, dried, and scanned using GenePix 4000B (Axon Instruments, Foster City, CA). Data from three independent experiments were normalized and then analyzed using the GenePix Pro 3.0 software (Axon Instruments). The open reading frame spots that showed a 2.828-fold or greater difference in expression with a P value of ≤0.05 were considered to represent open reading frames regulated by AphB.
AphB is essential for cytotoxicity toward epithelial cells in vitro.
Previously, we constructed an isogenic aphB mutant of V. vulnificus, JR312, in which the wild-type aphB gene on the chromosome was replaced with the ΔaphB allele (28). To complement the aphB mutation, pHG0602 was constructed by subcloning aphB amplified by PCR using primers APHB033F (5′-AAGAGCTCGATGTGTCAGGAAATATG-3′) and APHB033R (5′-TAGGTACCCTACATCGTTAGTGGATG-3′) into the broad-host-range vector pJH0311 (6).
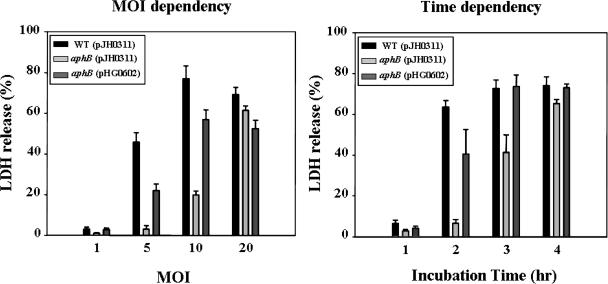
Cytotoxicity is evaluated by the quantification of cytoplasmic lactate dehydrogenase (LDH) activity released by the damage of plasma membranes (31). The preparation of the INT-407 (ATCC CCL-6) human intestinal epithelial cells and infection with the V. vulnificus strains were performed as described previously (27). Monolayers of INT-407 cells infected with the wild type, the aphB mutant, and a complemented strain at different multiplicities of infection (MOI) were incubated for 2 h, and then the LDH activities in the supernatants were determined (Fig. 1). When the MOI was up to 10, cells infected with the aphB mutant JR312 exhibited significantly less LDH activity than those infected with the wild type. The level of LDH activity from the INT-407 cells infected with the aphB mutant was almost 10-fold lower than that from the cells infected with the wild type when the MOI was 5. Also, the LDH activities from INT-407 cells infected at an MOI of 10 were compared at different incubation times, as shown in Fig. 1. When incubated for as long as 4 h, the cells infected with the aphB mutant JR312 exhibited lower levels of LDH activity than the cells infected with the wild type. The lower levels of LDH activity were restored to levels comparable to those obtained from the cells infected with the wild type when the cells were infected with the complemented strain, JR312(pHG0602) (Fig. 1). Therefore, it was confirmed that the attenuated cytotoxic activity of the aphB mutant resulted from the inactivation of functional aphB rather than from any polar effects on genes downstream of aphB. These results suggest that AphB is essential for V. vulnificus to infect and injure host cells.
FIG. 1.
Effect of aphB mutation on virulence of V. vulnificus for INT-407 cells. INT-407 cells were infected with the wild type (WT), the aphB mutant [aphB (pJH0311)], or the complemented strain [aphB (pHG0602)] at various MOI for 2 h (left) or at an MOI of 10 for various incubation times (right). The data are the means plus standard errors of the means (SEM) of results from three independent experiments.
AphB is required for adhesion to epithelial cells and motility in vitro.
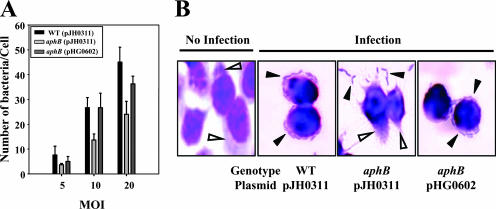
For the adhesion assay, INT-407 cells seeded onto glass coverslips were infected at different MOIs for 1 h and then washed to remove nonadherent bacteria as described previously (16, 27). When INT-407 cells were infected at an MOI of 5, the wild-type strain adhered to the INT-407 cells and reached a concentration of 8 CFU per cell after 1 h of infection (Fig. 2A). The amount of adhered bacteria increased as the infection level (MOI) increased and reached about 46 CFU per cell at an MOI of 20 (Fig. 2A). In contrast, the aphB mutant JR312 was consistently and significantly less adherent than the wild type at all MOI tested. When cells were infected at an MOI of 10, the number of CFU of the aphB mutant per INT-407 cell was about twofold less than that of CFU of the wild type (Fig. 2A). The wild type and JR312(pHG0602) formed small clusters of aggregated bacteria on the INT-407 cell surfaces. With JR312, a much smaller area of the intestinal cell surface was covered with bacteria and no clusters of aggregated bacteria were observed (Fig. 2B). These results clearly revealed that the aphB mutant was significantly impaired in its ability to attach to the epithelial cells.
FIG. 2.
Adhesion of the V. vulnificus strains. (A) INT-407 cells were infected at different MOI, as indicated. After incubation with the bacteria for 1 h, the INT-407 cells were rinsed to remove any nonadhering bacteria. Adherent bacteria were quantified and expressed as the number of bacteria per cell in the coverslip tissue cultures. WT, wild type; [aphB (pJH0311)], aphB mutant; [aphB (pHG0602)], complemented strain. (B) INT-407 cells were infected with the wild type, the aphB mutant, or the complemented strain at an MOI of 10 for 1 h as indicated and morphologically observed using a light microscope (original magnification, ×1,200) after Giemsa staining. The adhered V. vulnificus cells (closed arrowheads) and the cytoplasm of the INT-407 cells (open arrowheads) are indicated.
It was also noteworthy that light micrographs revealed that marked cellular damage such as cytoplasmic loss and nuclear-material condensation appeared in many Giemsa-stained INT-407 cells after infection with the wild type and JR312(pHG0602) (Fig. 2B). Fewer dead cells were observed after infection with the aphB mutant JR312. The cells infected with JR312 exhibited less surface damage and less cytoplasmic loss than those infected with the other strains. The general patterns of the abilities of the V. vulnificus strains to damage INT-407 cells were similar to the patterns of the cytotoxic activities as determined by measuring LDH activity (Fig. 1).
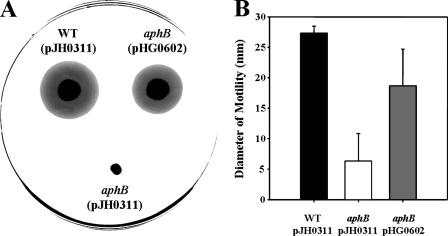
To determine whether or not AphBVv is required for motility, JR312 was tested for its ability to migrate on a semisolid plate surface with 0.3% agar compared to that of the wild type. As shown in Fig. 3, the growth of JR312 away from the inoculation point decreased compared to that of the wild type, and the diameter of the swimming area of the mutant was significantly reduced, to 20% of that of the wild type. The reintroduction of recombinant aphB substantially increased motility (Fig. 3), indicating that AphB is necessary for the optimum motility of V. vulnificus.
FIG. 3.
Motilities of the V. vulnificus strains. (A) The areas of motility of the wild type (WT), the aphB mutant [aphB (pJH0311)], and the complemented strain [aphB (pHG0602)] grown for 18 h on plates with Luria-Bertani medium supplemented with 2.0% (wt/vol) NaCl and 0.3% agar were photographed by using a digital imaging system (UTA-1100; UMAX Technologies, Inc.). (B) The diameters of motility areas are the means plus SEM of results from three independent experiments.
AphB is essential for virulence in mice.
The 50% lethal doses (LD50s) of the wild type and the aphB mutant were compared using ICR mice (specific-pathogen free; Seoul National University), as described elsewhere (27). The LD50s of the V. vulnificus strains for mice infected intraperitoneally were as follows: wild type, 1.3 × 101 CFU, and JR312, 3.9 × 104 CFU. Each inoculation group comprised six iron-overloaded mice, and inocula ranged from 100 to 108 CFU in 10-fold increments. All manipulations of mice were approved by the Institute of Laboratory Animal Resources of Seoul National University. The contrast in the LD50s of the aphB mutant JR312 and the wild type showed the aphB mutant to be significantly less virulent than the wild type. This result indicates that AphBVv is essential for the virulence of V. vulnificus in mice as well as in tissue cultures. Thus, the results of the present study make it reasonable to conclude that the aphB gene is important for the pathogenicity of the bacterium.
Prediction and verification of the AphB regulon.
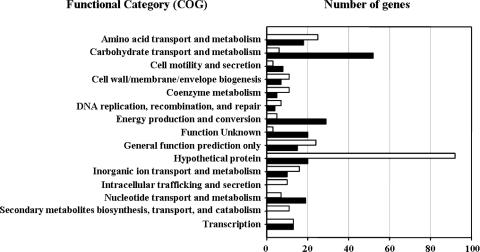
By using a V. vulnificus whole-genome microarray, transcripts from the aphB mutant JR312 were compared to those from the wild type. The microarray screen predicted 489 genes potentially regulated by AphB, 230 of which were up-regulated and 259 of which were down-regulated. The predicted genes of the AphB regulon are distributed throughout the two chromosomes of V. vulnificus. A complete list of names or locus tags of the 489 genes is shown in Table S1 in the supplemental material. Although a substantial portion of the predicted genes are of hypothetical or unknown functions, some belong to functional categories (Fig. 4). Thirteen of these categories, including amino acid transport and metabolism, carbohydrate transport and metabolism, cell motility and secretion, cell wall/membrane/envelope biogenesis, coenzyme metabolism, DNA replication/recombination/repair, energy production and conversion, inorganic-ion transport and metabolism, intracellular trafficking and secretion, nucleotide transport and metabolism, secondary metabolite biosynthesis/transport/catabolism, and transcription, correspond to at least 10 genes (Fig. 4).
FIG. 4.
Number of genes regulated by AphB. Genes with expression ratios of ≥2.828 on the basis of microarray analysis results were considered to be regulated by AphB. Functional categories (COG) corresponding to at least 10 genes are presented and are based on the database for the V. vulnificus CMCP6 genome, which was retrieved from GenBank (accession numbers AE016795 and AE016796). Closed and open bars represent the genes up-regulated and down-regulated by AphB, respectively.
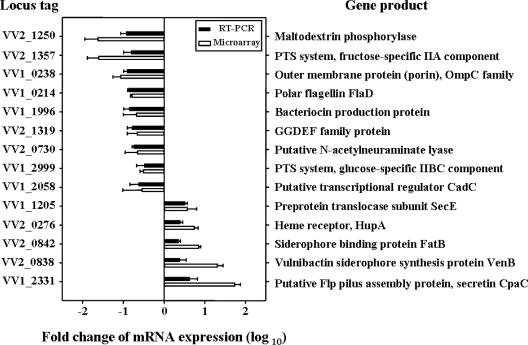
Most of the genes that we predicted to be part of the AphB regulon on the basis of microarray analysis results were not previously reported to be AphB regulated. Therefore, AphB regulation of the newly predicted genes was experimentally verified using a quantitative real-time PCR (RT-PCR). cDNA was synthesized with a SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA), and quantitative RT-PCR amplification of the cDNA was performed with a pair of primers (Table 1) as described previously (2). Sixteen genes, 10 of which were up-regulated and 6 of which were down-regulated by AphB, were randomly selected from the microarray data set, and the regulation of their transcription by AphB was examined. Quantitative RT-PCR revealed that AphB regulates the transcription of 14 of the 16 genes (Fig. 5), suggesting that most of the data set obtained from the microarray analysis is valid. One possible explanation for our inability to demonstrate AphB regulation of the other two genes is that the conditions used for either the microarray analysis or the quantitative RT-PCR were not optimal. Nevertheless, the results suggest that a significant portion of the AphB regulon predicted on the basis of microarray analysis results is indeed regulated by AphB and that AphB is a global regulator in V. vulnificus.
TABLE 1.
Oligonucleotides used for quantitative RT-PCR
| Primer name | Oligonucleotide sequence (5′-3′)
|
Locus taga | |
|---|---|---|---|
| Forward | Reverse | ||
| BACT001 | ACAGGGCTTTCAGGCACAGAC | ACTCTTCCACCACTCCGAACTTG | VV1_1996 |
| CADC001 | ACTGCGTGATGGGCGAGAAG | GCTGGCTCCAACAATTCAACCTC | VV1_2058 |
| CPAC001 | AGTGGTGGTCTGTTGTGGTGTG | GATAACGGAATCAGGCGGCTTTG | VV1_2331 |
| FATB001 | GCTATTGGTGATTGACGGTGATAAG | GACGGTCTCTTTGATGTCGGAAAC | VV2_0842 |
| FLAD001 | TGAGATACCGTCGTTGGCGTTAC | GCAATGACAGCACAGCGTTACC | VV1_0214 |
| FRU001 | TCGGTTTGACGCCCTCTTCG | CTTCAATGGACGGCATCGCTTAC | VV2_1357 |
| GGDEF001 | AGTTCTCGCTCCGCCATCTTG | TGCCGACAACCCATTAGTGACC | VV2_1319 |
| GLU001 | GAACACCACCAGCAACGACAAC | GGCGGTAAAGCAAACATCACAGG | VV1_2999 |
| HUPA001 | GCTCTACTTGATTTGCCGTTCCG | ACTGTAGGTGAAGATGGCGAAGG | VV2_0276 |
| MAL001 | CTACAAGAAGTGCGTGCCAAGTG | GACTGCGTAAGAGCCAATCACAC | VV2_1250 |
| NANA001 | GCAATCGCTTTTCGCTCTTCAAC | GATCGCCGCTCCTCATACACC | VV2_0730 |
| OMPC001 | AGCCCAGAAATCGTCGCCTTC | CGCCAATCTAACACCATCCGTTG | VV1_0238 |
| SECE001 | TGTATGGTGAACTTTCTGTGGTGATTC | GCATAGTTTCTTGGCGAGTAGGC | VV1_1205 |
| VENB001 | TCGGTCTGTCGGATGTTGGTTAG | GCCAGATTCATCGCCAGTTGTTG | VV2_0838 |
FIG. 5.
Verification of the newly predicted genes as part of the AphB regulon. Fourteen genes from the pool of AphB regulon members newly predicted on the basis of microarray analysis results were analyzed by RT-PCR. Each column represents the mRNA expression level in the aphB mutant relative to that in the wild type. Averages and SEM were calculated from the results of at least three independent experiments. Locus tags are based on the database of the V. vulnificus CMCP6 genome as described in the legend to Fig. 4, and the products of the 14 genes are listed on the right. PTS system, phototransferase system.
These results provide the first direct evidence that AphB is essential for the virulence of bacterial pathogens in which TCP and CT are not found. However, transcript profiles of the aphB mutant revealed that virulence factors that directly injure host cells, such as hemolysin, elastolytic protease, and RtxA, are not regulated by AphB (see Table S1 in the supplemental material) (15, 23, 32). Rather, many genes involved primarily in the metabolism and utilization of nutrients were identified as members of the AphB regulon (Fig. 4 and 5; see Table S1 in the supplemental material). Fifty-eight genes, including genes encoding maltodextrin phosphorylase, a fructose-specific IIA component, a glucose-specific IIBC component, and a putative N-acetylneuraminate lyase, are classified into the Clusters of Orthologous Groups (COG) category of carbohydrate transport and metabolism, and 43 genes are classified into the COG category of amino acid transport and metabolism (Fig. 4 and 5; see Table S1 in the supplemental material). Additionally, genes involved in the uptake of iron, such as fatB, hupA, and venB, are also regulated by AphB (Fig. 5) (1, 17, 18). These results suggest that AphB regulates numerous genes involved in the acquisition and metabolism of nutrients and aids growth and adaptation in host environments, potentially contributing to the overall success of V. vulnificus in pathogenesis.
The motility of the pathogenic bacteria is essential for a successful infectious process because it facilitates adhesion to and the colonization of host epithelial cells (24). The aphB mutant was clearly impaired in adherence and motility (Fig. 2 and 3), and many genes known to be potentially important for the motility of bacteria were identified as members of the AphB regulon (Fig. 4 and 5; see Table S1 in the supplemental material). Besides flaD (Fig. 5), 11 genes involved in the biosynthesis, assembly, and transport of flagellum proteins are apparently regulated by AphB (Fig. 4; see Table S1 in the supplemental material). Altering the physiochemical characteristics of the V. vulnificus cell surface has also been postulated to modify the relative adhesive properties of the bacterium (25, 26, 27). Consistent with this possibility, 18 genes including cpaC, encoding the putative Flp pilus assembly protein classified into the COG category of cell wall/membrane/envelope biogenesis, appeared to be regulated by AphB (Fig. 4 and 5; see Table S1 in the supplemental material). Additionally, a protein with a GGDEF domain is regulated by AphB (Fig. 5). Proteins with GGDEF domains are predicted to control the cellular level of a second messenger, the cyclic di-GMP, and thus regulate bacterial cell surface adhesiveness (3). Undoubtedly, adhesion to intestinal epithelial cells is an important step in the disease process of V. vulnificus infection. However, the exact role of AphB in the pathogenicity of V. vulnificus needs additional study.
Microarray data accession number.
All primary microarray data are available from CIBEX at http://cibex.nig.ac.jp/cibex2/index.jsp under accession number CBX42.
Supplementary Material
Acknowledgments
This study was supported by grants to S.H.C. from the Korea Health 21 research and development project, Ministry of Health and Welfare (grant no. A060356); the MarineBio 21 project, Ministry of Maritime Affairs and Fisheries; and the National Research Laboratory, Korea Science and Engineering Foundation (grant no. R0A-2007-000-20039-0), South Korea. Microarray slides, the V. vulnificus whole-genome TwinChips, were kindly provided by the 21C Frontier Microbial Genomics and Applications Center program, Korean Ministry of Science and Technology, South Korea.
Footnotes
Published ahead of print on 14 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1995. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17197-204. [DOI] [PubMed] [Google Scholar]
- 2.Choi, J. J., D. W. Shin, D., and S. R. Ryu. 2007. Implication of quorum sensing in Salmonella enterica serovar Typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 754885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 1502497-2502. [DOI] [PubMed] [Google Scholar]
- 4.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 885403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink, R. C., M. R. Evans, S. Porwollik, A. Vazquez-Torres, J. Jones-Carson, B. Troxell, S. J. Libby, M. McClelland, and H. M. Hassan. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 1892262-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goo, S. Y., H. J. Lee, W. H. Kim, K. L. Han, D. K. Park, H. J. Lee, S. M. Kim, K. S. Kim, K. H. Lee, and S. J. Park. 2006. Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 745586-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häse, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 1681487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 661056-1065. [DOI] [PubMed] [Google Scholar]
- 10.Kaper, J. B., J. R. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 1814250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 1823228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41393-407. [DOI] [PubMed] [Google Scholar]
- 14.Kovacikova, G., and K. Skorupski. 2002. Binding site requirements of the virulence gene regulator AphB: differential affinities for the Vibrio cholerae classical and El Tor tcpPH promoters. Mol. Microbiol. 44533-547. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. H., M. W. Kim, B. S. Kim, S. M. Kim, B. C. Lee, T. S. Kim, and S. H. Choi. 2007. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45146-152. [PubMed] [Google Scholar]
- 16.Lim, M. S., M. H. Lee, J. H. Lee, H. M. Ju, N. Y. Park, H. S. Jeong, J. E. Rhee, and S. H. Choi. 2005. Identification and characterization of the Vibrio vulnificus malPQ operon. J. Microbiol. Biotechnol. 15616-625. [Google Scholar]
- 17.Litwin, C. M., and B. L. Byrne. 1998. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect. Immun. 663134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litwin, C. M., T. W. Rayback, and J. Skinner. 1996. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect. Immun. 642834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4132-137. [DOI] [PubMed] [Google Scholar]
- 20.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 1741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. F., J. J. Mekalanos, and S. Falkow. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243916-922. [DOI] [PubMed] [Google Scholar]
- 22.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55165-199. [DOI] [PubMed] [Google Scholar]
- 23.Oliver, J. D., J. E. Wear, M. B. Thomas, M. Warner, and K. Linder. 1986. Production of extracellular enzymes and cytotoxicity by Vibrio vulnificus. Diagn. Microbiol. Infect. Dis. 599-111. [DOI] [PubMed] [Google Scholar]
- 24.Ottemann, K. M., and J. F. Miller. 1997. Role for motility in bacterial-host interactions. Mol. Microbiol. 241109-1117. [DOI] [PubMed] [Google Scholar]
- 25.Paranjpye, R. N., J. C. Lara, J. C. Pepe, C. M. Pepe, and M. S. Strom. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 665659-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 731411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, N. Y., J. H. Lee, M. W. Kim, H. G. Jeong, B. C. Lee, T. S. Kim, and S. H. Choi. 2006. Identification of Vibrio vulnificus wbpP gene and evaluation of its role in virulence. Infect. Immun. 74721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee, J. E., H. G. Jeong, J. H. Lee, and S. H. Choi. 2006. AphB influences acid tolerance of Vibrio vulnificus by activating expression of the positive regulator CadC. J. Bacteriol. 1886490-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee, J. E., J. H. Rhee, P. Y. Ryu, and S. H. Choi. 2002. Identification of the cadBA operon from Vibrio vulnificus and its influence on survival to acid stress. FEMS Microbiol. Lett. 208245-251. [DOI] [PubMed] [Google Scholar]
- 30.Rhee, J. E., K. S. Kim, and S. H. Choi. 2005. CadC activates pH-dependent expression of the Vibro vulnificus cadBA operon at a distance through direct binding to an upstream region. J. Bacteriol. 1877870-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sepp, A., R. M. Binns, and R. I. Lechler. 1996. Improved protocol for colorimetric detection of complement-mediated cytotoxicity based on the measurement of cytoplasmic lactate dehydrogenase activity. J. Immunol. Methods 196175-180. [DOI] [PubMed] [Google Scholar]
- 32.Wright, A. C., J. G. Morris, Jr., D. R. Maneval, Jr., K. Richardson, and J. B. Kaper. 1985. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect. Immun. 50922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.