Abstract
Mycobacterium avium-Mycobacterium intracellulare complex (MAC) is the most common isolate of nontuberculous mycobacteria and causes pulmonary and extrapulmonary diseases. MAC species can be grouped into 31 serotypes by the epitopic oligosaccharide structure of the species-specific glycopeptidolipid (GPL) antigen. The GPL consists of a serotype-common fatty acyl peptide core with 3,4-di-O-methyl-rhamnose at the terminal alaninol and a 6-deoxy-talose at the allo-threonine and serotype-specific oligosaccharides extending from the 6-deoxy-talose. Although the complete structures of 15 serotype-specific GPLs have been defined, the serotype 16-specific GPL structure has not yet been elucidated. In this study, the chemical structure of the serotype 16 GPL derived from M. intracellulare was determined by using chromatography, mass spectrometry, and nuclear magnetic resonance analyses. The result indicates that the terminal carbohydrate epitope of the oligosaccharide is a novel N-acyl-dideoxy-hexose. By the combined linkage analysis, the oligosaccharide structure of serotype 16 GPL was determined to be 3-2′-methyl-3′-hydroxy-4′-methoxy-pentanoyl-amido-3,6-dideoxy-β-hexose-(1→3)-4-O-methyl-α-l-rhamnose-(1→3)-α-l-rhamnose-(1→3)-α-l-rhamnose-(1→2)-6-deoxy-α-l-talose. Next, the 22.9-kb serotype 16-specific gene cluster involved in the glycosylation of oligosaccharide was isolated and sequenced. The cluster contained 17 open reading frames (ORFs). Based on the similarity of the deduced amino acid sequences, it was assumed that the ORF functions include encoding three glycosyltransferases, an acyltransferase, an aminotransferase, and a methyltransferase. An M. avium serotype 1 strain was transformed with cosmid clone no. 253 containing gtfB-drrC of M. intracellulare serotype 16, and the transformant produced serotype 16 GPL. Together, the ORFs of this serotype 16-specific gene cluster are responsible for the biosynthesis of serotype 16 GPL.
Mycobacterial diseases, such as tuberculosis and infection due to nontuberculous mycobacteria (NTM), are still among the most serious infectious diseases in the world. The incidence is increasing because of the spread of drug-resistant mycobacteria and the human immunodeficiency virus (HIV) infection/AIDS epidemic (16, 17, 30). Mycobacterium avium-Mycobacterium intracellulare complex (MAC) is the most common among isolates of NTM and is distributed ubiquitously in the environment. MAC causes pulmonary and extrapulmonary diseases in both immunocompromised and immunocompetent hosts. It affects primarily patients with advanced HIV infection. MAC includes at least two mycobacterial species, M. avium and M. intracellulare, that cannot be differentiated on the basis of traditional physical and biochemical tests (1, 41).
The cell envelope of mycobacteria is a complex and unusual structure. The key feature of this structure is an extraordinarily high lipid concentration (6, 10). To better understand the pathogenesis of MAC infection, it is necessary to elucidate the molecular structure and biochemical features of the lipid components. Among MAC lipids, the glycopeptidolipid (GPL) is of particular importance, because it shows not only serotype-specific antigenicity but also immunomodulatory activities in the host immune responses (2, 9, 23). Structurally, GPLs are composed of two parts, a tetrapeptide-amino alcohol core and a variable oligosaccharide (OSE). C26-C34 fatty acyl-d-phenylalanine-d-allo-threonine-d-alanine-l-alaninol (d-Phe-d-allo-Thr-d-Ala-l-alaninol) is further linked with 6-deoxy talose (6-d-Tal) and 3,4-di-O-methyl rhamnose (3,4-di-O-Me-Rha) at d-allo-Thr and the terminal l-alaninol, respectively. This type of core GPL is found in all subspecies of MAC, shows a common antigenicity, and is further glycosylated at 6-d-Tal to form a serotype-specific OSE.
At present, 31 distinct serotype-specific GPLs have been identified serologically and chromatographically (9). Although the standard technique for differentiation of MAC subspecies has been serotyping based on the OSE residue of its GPL, the complete structures of only 15 GPLs have been defined. In addition to the chemical structures of various GPLs, genes encoding the glycosylation pathways in the biosynthesis of GPL have been identified and characterized (12, 21, 31). Epidemiological studies have shown that MAC serotypes 4 and 8 are the most frequently isolated from patients, and MAC serotype 16 is one of the next most common groups (32, 40). It has been suggested that the serotypes of MAC isolates participate in their virulence (29), and thus, understanding of the structure-pathogenicity relationship of GPLs is necessary. In the present study, we demonstrate the complete OSE structure of the GPL derived from serotype 16 MAC (M. intracellulare), which has a unique terminal-acylated-amido sugar, and we characterized the serotype 16 GPL-specific gene cluster involved in the glycosylation of carbohydrates.
MATERIALS AND METHODS
Bacterial strains and preparation of GPL.
M. intracellulare serotype 16 strain ATCC 13950T (NF 115) was purchased from the American Type Culture Collection (Manassas, VA). Three clinical isolates of M. intracellulare serotype 16 (NF 116 and 117) and M. avium serotype 1 (NF 113) were maintained in The Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association. The preparation of GPL was performed as described previously (18, 24, 26). Briefly, each strain of M. intracellulare serotype 16 was grown in Middlebrook 7H9 broth (Difco Laboratories, Detroit, MI) with 0.5% glycerol and 10% Middlebrook oleic acid-albumin-dextrose-catalase enrichment (Difco) at 37°C for 2 to 3 weeks. The heat-killed bacteria were sonicated, and crude lipids were extracted with chloroform-methanol (2:1, vol/vol). The extracted lipids were dried and hydrolyzed with 0.2 N sodium hydroxide in methanol at 37°C for 2 h. After neutralization with 6 N hydrochloric acid, alkaline-stable lipids were partitioned by a two-layer system of chloroform-methanol (2:1, vol/vol) and water. The organic phase was recovered, evaporated, and precipitated with acetone to remove any acetone-insoluble components containing phospholipids and glycolipids. The supernatant was collected by centrifugation, dried, and then treated with a Sep-Pak silica cartridge (Waters Corporation, Milford, MA) with washing (chloroform-methanol, 95:5, vol/vol) and elution (chloroform-methanol, 1:1, vol/vol) for partial purification. GPL was completely purified by preparative thin-layer chromatography (TLC) of Silica Gel G (20 by 20 cm, 250 μm; Uniplate; Analtech, Inc., Newark, DE). The TLC plate was repeatedly developed with chloroform-methanol-water (65:25:4 and 60:16:2, vol/vol/vol) until a single spot was obtained. After exposure of the TLC plate to iodine vapor, the GPL band was marked, and then, the silica gels were scraped off and the GPL was eluted with chloroform-methanol (2:1, vol/vol).
Preparation of OSE moiety.
β elimination of GPL was performed with alkaline borohydride, and the OSE elongated from d-allo-Thr was released as described previously (18, 24). Briefly, the GPL was dissolved in ethanol, and an equal volume of 10 mg/ml sodium borohydride or borodeuteride in 0.5 N sodium hydroxide was added and then stirred at 60°C for 16 h. The reaction mixture was decationized with Dowex 50W-X8 beads (Dow Chemical Company, Midland, MI), collected, and evaporated under nitrogen to remove boric acid. The dried residue was partitioned in two layers of chloroform-methanol (2:1, vol/vol) and water. The upper aqueous phase was recovered and evaporated. In these processes, the serotype 16-specific OSE was purified as an oligoglycosyl alditol.
MALDI-TOF and MALDI-TOF/TOF MS analyses.
The molecular species of the intact GPL was detected by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with an Ultraflex II (Bruker Daltonics, Billerica, MA). The GPL was dissolved in chloroform-methanol (2:1, vol/vol) at a concentration of 1 mg/ml, and 1 μl was applied directly to the sample plate, and then 1 μl of 10 mg/ml 2,5-dihydroxybenzoic acid in chloroform-methanol (1:1, vol/vol) was added as a matrix. The intact GPL was analyzed in the reflectron mode with an accelerating voltage operating in a positive mode of 20 kV (5). Then the fragment pattern of the OSE was analyzed with MALDI-TOF/TOF MS. The OSE was dissolved in ethanol-water (3:7, vol/vol), and the matrix was 10 mg/ml 2,5-dihydroxybenzoic acid in ethanol-water (3:7, vol/vol). The OSE and the matrix were applied to the sample plate according to the method for intact GPL and analyzed in the lift-lift mode.
GC and GC-MS analyses of carbohydrates and N-acylated short-chain fatty acid.
To determine the glycosyl composition and linkage position, gas chromatography (GC) and GC-MS analyses of partially methylated alditol acetate derivatives were performed. Perdeuteromethylation was conducted by the modified procedure of Hakomori as described previously (18, 20). Briefly, the dried OSE was dissolved with a mixture of dimethyl sulfoxide and sodium hydroxide, and deuteromethyl iodide was added. The reaction mixture was stirred at room temperature for 15 min and then water and chloroform were added. The chloroform-containing perdeuteromethylated OSE layer was collected, washed with water two times, and then completely evaporated. Partially deuteromethylated alditol acetates were prepared from perdeuteromethylated OSE by hydrolysis with 2 N trifluoroacetic acid at 120°C for 2 h, reduction with 10 mg/ml sodium borodeuteride at 25°C for 2 h, and acetylation with acetic anhydride at 100°C for 1 h (8, 18, 25). To identify amino-linked fatty acids, acidic methanolysis of serotype 16 GPL was performed with 1.25 M hydrogen chloride in methanol (Sigma-Aldrich, St. Louis, MO) at 100°C for 90 min, and the fatty acid methyl esters were extracted with n-hexane under the cooled ice. GC was performed using a 5890 series II gas chromatograph (Hewlett Packard, Avondale, PA) equipped with a fused SPB-1 capillary column (30 m, 0.25-mm inner diameter; Supelco Inc., Bellefonte, PA). Helium was used for electron impact (EI)-MS and isobutane for chemical ionization (CI)-MS as a carrier gas. A JMS SX102A double-focusing mass spectrometer (JEOL, Tokyo, Japan) was connected to the gas chromatograph as a mass detector. The molecular separator and the ion source energy were 70 eV for EI and 30 eV for CI, and the accelerating voltage was 8 kV. The d and l configurations of Rha residues were determined by comparative GC-MS analysis of trimethylsilylated (S)-(+)-sec-butyl glycosides and (R)-(−)-sec-butyl glycosides prepared from an authentic standard l-Rha (19).
NMR analysis of GPL.
The GPL was dissolved in chloroform-d (CDCl3)-methanol-d4 (CD3OD) (2:1, vol/vol). To define the anomeric configurations of each glycosyl residue, 1H and 13C nuclear magnetic resonance (NMR) was employed. Both homonuclear correlation spectrometry (COSY) and 1H-detected [1H, 13C] heteronuclear multiple-quantum correlation (HMQC) were recorded with a Bruker Avance-600 (Bruker BioSpin Corp., Billerica, MA), as described previously (9, 18, 24, 34).
Construction of M. intracellulare serotype 16 cosmid library.
A cosmid library of M. intracellulare serotype 16 strain ATCC 13950T was constructed as described previously (18). Bacterial cells were disrupted mechanically, and genomic DNA was extracted with phenol-chloroform and then precipitated with ethanol. Genomic DNA randomly sheared into 30- to 50-kb fragments in the extraction process was fractionated and electroeluted from agarose gels using a Takara Recochip (Takara, Kyoto, Japan). These DNA fragments were rendered blunt ended using T4 DNA polymerase and deoxynucleoside triphosphates and then were ligated to dephosphorylated arms of pYUB412 (XbaI-EcoRV and EcoRV-XbaI), which were the kind gifts of William R. Jacobs, Jr. (Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY). The cosmid vector pYUB412 is an Escherichia coli-Mycobacterium shuttle vector with the int-attP sequence for integration into a mycobacterial chromosome, oriE for replication in E. coli, a hygromycin resistance gene, and an ampicillin resistance gene. After in vitro packaging using Gigapack III Gold extracts (Stratagene, La Jolla, CA), recombinant cosmids were introduced into E. coli STBL2 [F− mcrA Δ(mcrBC-hsdRMS-mrr) recA1 endA1 lon gyrA96 thi supE44 relA1 l Δ(lac-proAB)] and stored at −80°C in 50% glycerol.
Isolation of cosmid clones carrying biosynthesis gene cluster of serotype 16 GPL and sequence analysis.
Isolation of DNA from E. coli transductants was performed as described by Supply et al., with modifications (39). The colonies were picked, transferred to a 1.5-ml tube containing 50 μl of water, and then heated at 98°C for 5 min. After centrifugation at 14,000 rpm for 5 min, the supernatant was used as the PCR template. PCR was used to isolate cosmid clones carrying the rhamnosyltransferase (rtfA) gene with primers rtfA-F (5′-TTTTGGAGCGACGAGTTCATC-3′) and rtfA-R (5′-GTGTAGTTGACCACGCCGAC-3′). rtfA encodes an enzyme responsible for the transfer of Rha to 6-d-Tal in OSE (14, 31). The insert of cosmid clone no. 253 was sequenced using a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 310 gene analyzer (Applied Biosystems). The putative function of each open reading frame (ORF) was identified by similarity searches between the deduced amino acid sequences and known proteins using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and FramePlot (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) with the DNASIS computer program (Hitachi Software Engineering, Yokohama, Japan).
Transformation of M. avium serotype 1 strain with cosmid clone no. 253.
An M. avium serotype 1 strain (NF113) was transformed with pYUB412-cosmid clone no. 253 by electroporation, and hygromycin-resistant colonies were isolated. Alkaline-stable lipids were prepared, and productive GPLs were examined by TLC and MALDI-TOF MS analyses.
Nucleotide sequence accession number. The nucleotide sequence reported here has been deposited in the NCBI GenBank database under accession no. AB355138.
RESULTS
Purification and molecular weight of intact GPL.
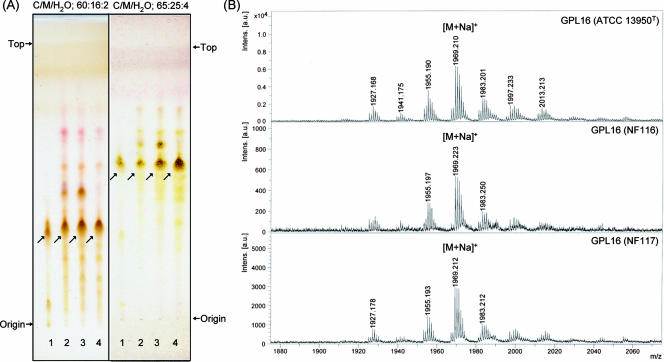
Serotype 16 GPL from M. intracellulare ATCC 13950T (NF 115) was detected as a spot by TLC, and the Rf values were 0.35 and 0.56 when developed with chloroform-methanol-water (60:16:2 and 65:25:4, vol/vol/vol, respectively). Two clinical isolates of M. intracellulare, NF 116 and 117, had serotype 16 GPLs that showed the same Rf values as the serotype 16 GPL derived from strain ATCC 13950T. The serotype 16 GPL of M. intracellulare strain ATCC 13950T was purified repeatedly by TLC and was shown as a single spot by TLC (Fig. 1A). The MALDI-TOF MS spectra of each serotype 16 GPL showed m/z 1969 for [M+Na]+ as the main molecularly related ion in positive mode, with the homologous ions differing by 14 mass units at 1,955 and 1,983 (Fig. 1B). As a result, the main molecular weight of serotype 16 GPL was 1,946, which implied that it has a novel carbohydrate chain elongated from d-allo-Thr.
FIG. 1.
TLC patterns and MALDI-TOF MS spectra of serotype 16 GPL. (A) Serotype 16 GPL purified from M. intracellulare ATCC 13950T (NF 115) and the alkaline-stable lipids derived from ATCC 13950T and two clinical isolates (NF 116 and 117) from left to right were developed on TLC plates with solvent systems of chloroform-methanol-water (65:25:4 and 60:16:2, vol/vol/vol). (B) The MALDI-TOF MS spectra were acquired using 10 mg/ml 2,5-dihydroxybenzoic acid in chloroform-methanol (1:1, vol/vol) as a matrix, and the molecularly related ions were detected as [M+Na]+ in positive mode. Intens., intensity; a.u., absorbance units.
Carbohydrate composition of serotype 16 OSE.
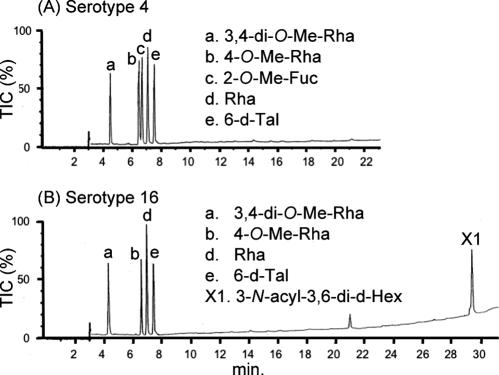
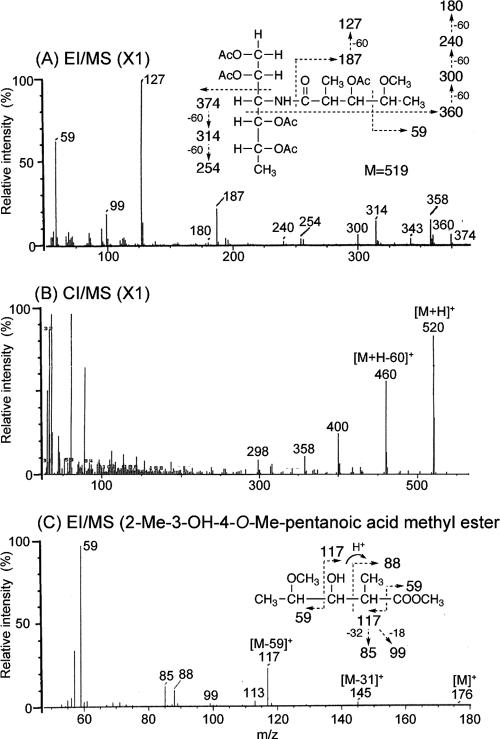
To determine the glycosyl compositions of serotype 16 OSE, alditol acetate derivatives of the serotype 16 GPL were analyzed by GC and GC-MS. The structurally defined serotype 4 GPL was used as a reference standard (9, 35). Comparison of the retention time and GC mass spectra (Fig. 2) with the alditol acetate derivatives of the serotype 16 GPL showed the presence of 3,4-di-O-Me-Rha, 4-O-Me-Rha, Rha, 6-d-Tal, and an unknown sugar residue (X1) in a ratio of approximately 1:1:2:1:1. The alditol acetate of X1 was eluted at a retention time of 29.3 min, greater than that of glucitol acetate on the SPB-1 column. The CI-MS spectrum of X1 was [M+H]+ at m/z 520 as a parent ion and m/z 460 as a loss of 60 (acetate). The fragment ions of X1 sugar showed characteristic patterns in EI-MS. m/z 360 indicated the cleavage of C-3 and C-4, and m/z 300, 240, and 180 were fragmented with a loss of 60 (acetate). Similarly, m/z 374 indicated the cleavage of C-2 and C-3, and m/z 314 and 254 were fragmented with a loss of 60 (Fig. 3A and B). These results indicated that X1 was 3,6-dideoxy hexose (Hex). The odd molecular weight of X1, 519, and m/z 187, 127, and 59 implied the presence of one amido group esterified with a short-chain fatty acid, possibly. After methanolysis of serotype 16 GPL, the resultant fatty acid methyl esters were extracted carefully and analyzed by GC-MS. The EI-MS spectrum of a short-chain fatty acid methyl ester showed mass ions at m/z 176 ([M]+), 145 ([M-31]+), 117 ([M-59]+), 99, 88, 85, and 59 (Fig. 3C) (33, 37). Taking the results together, X1 was structurally determined to be 3-2′-methyl-3′-hydroxy-4′-methoxy-pentanoyl-amido-3,6-dideoxy-Hex.
FIG. 2.
Gas chromatograms of the alditol acetate derivatives from serotype 4 (A) and serotype 16 (B) GPLs. Total ion chromatograms (TIC) are shown. GC was performed on an SPB-1-fused silica column with a temperature program of 160°C for 2 min, followed by an increase of 4°C/min to 220°C, and holding at 220°C for 15 min. Comparison to the GC spectrum of serotype 4 GPL shows that serotype 16 GPL is composed of 3,4-di-O-Me-Rha, 4-O-Me-Rha, Rha, 6-d-Tal, and an unknown X1 sugar residue.
FIG. 3.
EI-MS and CI-MS spectra of the alditol acetate derivative from X1 (A and B) and N-acylated-short-chain fatty acid methyl ester (C). The pattern of prominent fragment ions is illustrated. The CG column and condition were described in the legend for Fig. 2.
Glycosyl linkage and sequence of serotype 16 OSE.
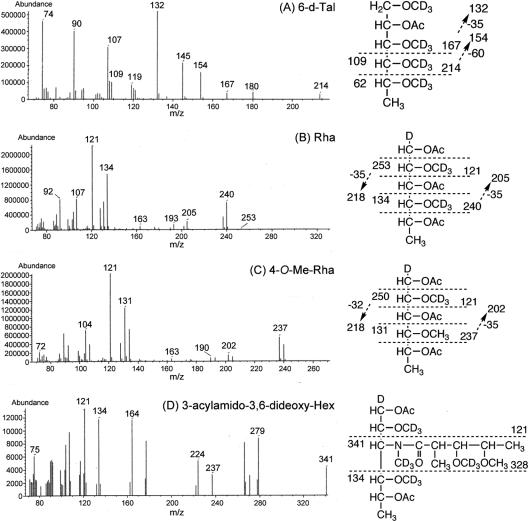
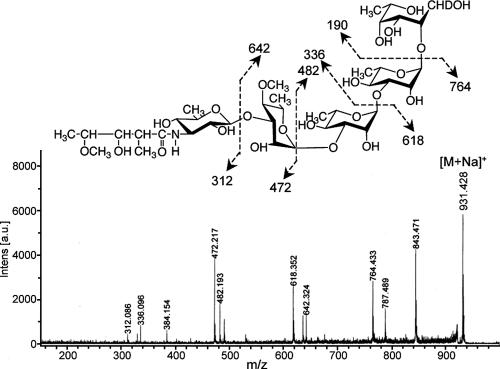
To determine the glycosyl linkage and sequence of the OSE, GC-MS of perdeuteromethylated alditol acetates and MALDI-TOF/TOF MS of the oligoglycosyl alditol from serotype 16 OSE were performed. As shown in Fig. 4, the GC-MS spectra of perdeuteromethylated alditol acetates were assigned four major peaks, 1,3,4,5-tetra-O-deuteromethyl-2-O-acetyl-6-deoxy-talitol (m/z 109, 132, 154, 167, and 214); 2,4-di-O-deuteromethyl-1,3,5-tri-O-acetyl-rhamnitol (m/z 121, 134, 205, 240, and 253); 2-O-deuteromethyl-4-O-methyl-1,3,5-tri-O-acetyl-rhamnitol (m/z 121, 131, 202, and 237); and 2,4-di-O-deuteromethyl-1,5-di-O-acetyl-3-2′-methyl-3′-O-deuteromethyl-4′methoxy- pentanoyl-deuteromethylamido-3,6-dideoxy-hexitol (m/z 121, 134, and 341). These results revealed that the 6-d-Tal residue was linked at C-2; Rha and 4-O-Me-Rha were linked at C-1 and C-3; and the nonreducing terminus, 3-2′-methyl-3′- hydroxy-4′-methoxy-pentanoyl-amido-3,6-dideoxy-Hex, was C-1 linked. The MALDI-TOF/TOF MS spectrum of the oligoglycosyl alditol from serotype 16 OSE afforded the expected molecular ions [M+Na]+ at m/z 931, together with the characteristic mass increments in the series of glycosyloxonium ions formed on fragmentation at m/z 312, 472, 618, and 764 from the terminal sugar N-acyl-Hex to 6-d-Tal and at m/z 336, 482, and 642 from 6-d-Tal to N-acyl-Hex (Fig. 5). Rha residues were determined to be in the l absolute configuration by comparative GC-MS analyses of trimethylsilylated (S)-(+)-sec-butyl glycosides and (R)-(−)-sec-butylglycosides (see Fig. S1 in the supplemental material). Taken together, these results established the sequence and linkage arrangement 3-2′-methyl-3′-hydroxy-4′-methoxy-pentanoyl-amido-3,6-dideoxy-Hex-(1→ 3)-4-O-Me-Rha-(1→3)-l-Rha-(1→3)-l-Rha-(1→2)-6-d-Tal, exclusively.
FIG. 4.
GC-MS spectra of individual perdeuteromethylated alditol acetate derivatives from serotype 16 OSE. The formation of prominent fragment ions is illustrated; fragments were assigned to 1,3,4,5-tetra-O-deuteromethyl-2-O-acetyl-6-deoxy-talitol (A), 2,4-di-O-deuteromethyl-1,3,5-tri-O-acetyl-rhamnitol (B), 2-O-deuteromethyl-4-O-methyl-1,3,5-tri-O-acetyl-rhamnitol (C), and 2,4-di-O-deuteromethyl-1,5-di-O-acetyl-3-2′-methyl-3′-O-deuteromethyl-4′-methoxy-pentanoyl-deuteromethylamido-3,6-dideoxy-hexitol (D).
FIG. 5.
MALDI-TOF/TOF MS spectrum of serotype 16 OSE. The formation of a characteristic increment in fragment ions is illustrated. The matrix was 10 mg/ml 2,5-dihydroxybenzoic acid in ethanol-water (3:7, vol/vol), and it was performed in the lift-lift mode. Intens., intensity; a.u., absorbance units.
NMR analysis of serotype 16 OSE.
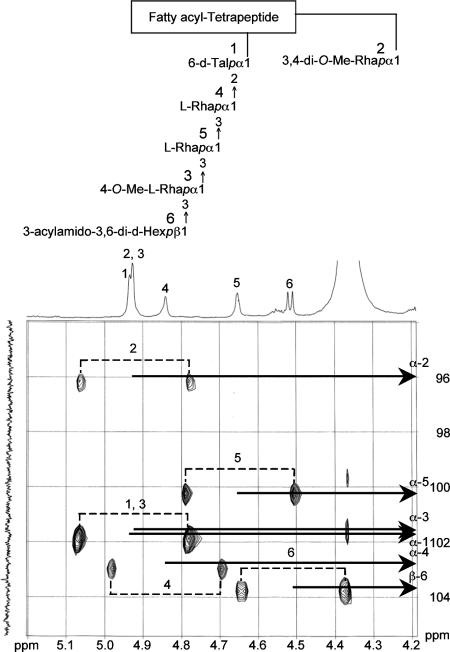
The 1H NMR and 1H-1H COSY analyses of the serotype 16 GPL revealed six distinct anomeric protons with corresponding H1-H2 cross peaks in the low field region at δ4.93, 4.92, 4.92, 4.84, 4.65 (J1-2 = 2 to 3 Hz, indicative of α-anomers) and 4.51 (a doublet, J1-2 = 7.7 Hz, indicative of a β-hexosyl unit). When further analyzed by 1H-detected [1H, 13C] two-dimensional HMQC, the anomeric protons resonating at δ4.93, 4.92, 4.92, 4.84, 4.65, and 4.51 have C-1s resonating at δ101.57, 95.73, 101.40, 102.56, 100.97, and 103.36, respectively (for a summary, see Table S1 in the supplemental material). The JCH values for each of these protons were calculated to be 171, 170, 171, 170, 169, and 161 Hz by measurement of the inverse-detection nondecoupled two-dimensional HMQC (Fig. 6). These results established that the terminal amido-Hex was a β configuration and the others were α-anomers.
FIG. 6.
Nondecoupled 1H-detected [1H, 13C] HMQC spectrum of serotype 16 GPL. Cross-peak labels correspond to those shown on the structure.
Cloning and sequence of serotype 16 GPL biosynthesis cluster.
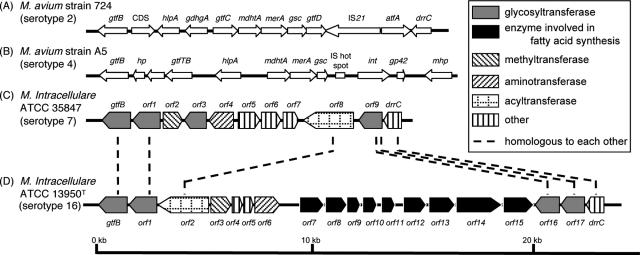
To isolate the serotype 16 GPL biosynthesis cluster, the genomic cosmid library of M. intracellulare serotype 16 strain ATCC 13950T was constructed. Primers were designed to amplify the region corresponding to the rtfA gene. More than 300 cosmid clones were tested using colony PCR with rtfA primers, and the positive clones no. 51 and 253 were isolated from the E. coli transductants. PCR analysis revealed that clone no. 253 contained a drrC gene but that clone no. 51 did not. Thus, we used clone no. 253 for subsequent sequence analysis for the gtfB-drrC region. The 22.9-kb region of M. intracellulare serotype 16 ATCC 13950T was deposited in the NCBI GenBank database (accession no. AB355138). The similarity to protein sequences of each ORF is summarized in Table 1, and the genetic map for the serotype 16 GPL biosynthetic cluster was compared with those of serotype 2, 4, and 7 GPLs (Fig. 7). The gtfB and drrC genes of M. intracellulare serotype 16 ATCC 13950T had 99.8% and 83.7% DNA identities with those of M. intracellulare serotype 7 ATCC 35847, respectively. In the DNA region between gtfB and drrC (20.8 kb), 17 ORFs were observed. Four ORFs (ORF 1, 2, 16, and 17) were homologous to those found in the same region of serotype 7-specific DNA, and the others were unique to the serotype 16 strain. No insertion of insertion elements or transposons was detected in this region. The nucleotide sequences of the ORF 1 and ORF 2 in serotype 16 strain ATCC 13950T were homologous to those of ORF 1 and ORF 8 in serotype 7, respectively, suggesting that these two ORFs have the same function. The similarity of the deduced amino acid sequences suggested the possibility that the functions of ORF 3 and ORF 6 are to encode methyltransferase and aminotransferase, respectively. The deduced amino acid sequences of ORF 4 and ORF 5 showed significant similarities to the WxcM protein, the function of which is not clear. Interestingly, the deduced amino acid sequences of ORF 16 and ORF 17 of serotype 16 were homologous to ORF 9 of serotype 7. ORFs 1, 16, and 17 have considerable homology to glycosyltransferases. Nine ORFs, which are possibly involved in fatty acid synthesis, were detected between ORF 7 and ORF 15. It is notable that ORF 13 had a chimeric structure. The N-terminal half of ORF 13 showed similarity to phosphate butyryl/acetyl transferases, but the C-terminal half showed similarity to short-chain reductase/dehydrogenases. These results suggest that this region of DNA is responsible for the biosynthesis of the serotype 16-specific GPL.
TABLE 1.
Similarity to protein sequences of ORFs in cosmid clone no. 253 derived from M. intracellulare serotype 16 strain ATCC 13950T
| ORF | Predicted molecular mass (kDa) | Predicted pI | Exhibits similarity to: | E value | Amino acid identity (no. matched/total no.) | Accession no. |
|---|---|---|---|---|---|---|
| GtfB | 45.6 | 6.35 | Glycosyltransferase GtfB | 0.0 | 417/418 | BAF45360 |
| Orf 1 | 45.2 | 6.10 | Putative glycosyltransferase | 0.0 | 416/417 | BAF45361 |
| Orf 2 | 78.9 | 8.51 | Putative acyltransferase | 0.0 | 557/728 | BAF45368 |
| Orf 3 | 31.0 | 5.88 | Putative methyltransferase | 2e-89 | 382/421 | NP_218045 |
| Orf 4 | 15.7 | 4.94 | Conserved hypothetical protein | 1e-39 | 73/129 | BAD50406 |
| Orf 5 | 16.0 | 4.69 | Conserved hypothetical protein | 5e-40 | 75/135 | EAX55190 |
| Orf 6 | 41.1 | 5.88 | Aminotransferase/DegT_DnrJ_EryC1 | 6e-119 | 208/357 | ABD68440 |
| Orf 7 | 40.6 | 9.65 | Conserved hypothetical protein | 2e-89 | 178/304 | AAS03547 |
| Orf 8 | 36.7 | 5.32 | Conserved hypothetical protein | 2e-52 | 116/298 | CAE06954 |
| Orf 9 | 22.3 | 9.79 | Putative N-acetyltransferase | 4e-14 | 58/166 | EAU11841 |
| Orf 10 | 25.3 | 7.82 | Short-chain dehydrogenase/reductase | 7e-47 | 101/233 | EAO61220 |
| Orf 11 | 23.8 | 6.05 | Putative hydrolase | 4e-24 | 64/196 | ABG85599 |
| Orf 12 | 37.2 | 6.50 | Ketoacyl-acyl carrier protein synthase III | 3e-55 | 126/331 | EAX48715 |
| Orf 13 | 42.5 | 7.72 | Short-chain dehydrogenase/reductase | 2e-42 | 97/248 | ZP_01289005 |
| Orf 14 | 65.8 | 4.70 | Predicted enzyme involved in methoxymalonyl-acyl carrier protein biosynthesis | 6e-85 | 201/575 | ABB73590 |
| Orf 15 | 50.0 | 6.23 | Acyl coenzyme A synthetases | 2e-128 | 233/445 | EAT27362 |
| Orf 16 | 39.1 | 8.00 | Putative glycosyltransferase | 2e-106 | 196/318 | NP_855197 |
| Orf 17 | 37.7 | 9.46 | Putative glycosyltransferase | 8e-160 | 278/323 | BAF45369 |
| DrrC | 28.6 | 11.47 | Daunorubicin resistance protein C | 6e-132 | 233/261 | BAF45370 |
FIG. 7.
Comparison and overview of genetic maps of GPL biosynthetic cluster. The M. avium strain 724 annotated sequence obtained from GenBank (accession no. AF125999) (A); the M. avium strain A5 annotated sequence obtained from GenBank (accession no. AY130970) (B); the M. intracellulare ATCC 35847 sequenced in our previous study (GenBank accession no. AB274811) (C); the M. intracellulare ATCC 13950T sequenced in this study (GenBank accession no. AB355138) (D). The orientation of each gene is shown by the direction of the arrow. In panels A and B, putative ORFs not showing homology to known proteins sequences are not depicted. The sequences extending upstream in panels A and B and downstream in panel B are not included in the figure.
Expression of cosmid clone no. 253 in M. avium serotype 1 strain.
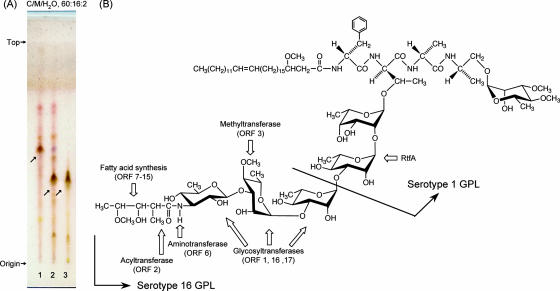
The OSE of serotype 1 GPL was composed of α-l-Rha-(1→2)-6-d-l-Tal (9). The M. avium serotype 1 strain (NF113) was transformed with cosmid clone no. 253 containing a serotype 16-specific gene cluster and produced a new GPL with a different Rf value by TLC compared to serotype 1 GPL (Fig. 8A). The Rf value of the new GPL was identical to that of the serotype 16 GPL. The molecular weight of intact GPL, the fragment pattern of its OSE, and the GC pattern of the alditol acetate derivatives were completely equivalent to those of the serotype 16 GPL (see Fig. S2 in the supplemental material). As a result, the transformant of the serotype 1 strain expressed the cosmid clone no. 253 gene cluster and produced the serotype 16 GPL.
FIG. 8.
TLC pattern of M. avium serotype 1 and its transformant with cosmid clone no. 253 and proposed complete structure of the serotype 16 GPL. (A) The alkaline-stable lipids derived from M. avium serotype 1 (lane 1), its transformant (lane 2), and purified serotype 16 GPL (lane 3) were developed with the solvent system of chloroform-methanol-water (60:16:2, vol/vol/vol). (B) Predicted biosynthesis gene clusters are indicated by arrows.
DISCUSSION
MAC species have serotype-specific GPLs that are characteristic components of the outer layer of the cell wall (6, 9). In addition to their serological differentiation, the chemical structures of 15 serotype-specific GPLs derived from the predominant clinical isolates have been analyzed; however, those of other GPLs remain unclear. The present study demonstrates the chemical structure of the serotype 16 GPL derived from M. intracellulare. We determined the glycosyl composition, linkage positions, and anomeric and ring configurations of the glycosyl residues of the serotype 16 GPL, and its OSE was defined as 3-2′-methyl-3′-hydroxy-4′-methoxy-pentanoyl-amido-3,6-dideoxy-β-Hex-(1→3)-4-O-methyl-α-l-Rha-(1→3)-α- l-Rha-(1→3)-α-l-Rha-(1→2)-6-d-α-l-Tal (Fig. 8B). The serotype 16 GPL should be listed as a group 2 polar GPL in the structural classification of Chatterjee and Khoo (9).
The GPLs of serotypes 7, 12, 17, and 19 have already been classified as group 2 GPLs, which are commonly composed of R→α-l-Rha-(1→3)-α-l-Rha-(1→2)-6-d-l-Tal (R, variable region), possessing a characteristic terminal sugar such as N-acyl-deoxy-Hex. Indeed, the presence of an amido sugar has been reported in only five GPLs, serotypes 7, 12, 14, 17, and 25 (8, 9, 18). It has been determined that the OSE structure of the serotype 17 GPL was 3-2′-methyl-3′-hydroxy-butanoyl-amido-3,6-dideoxy-β-d-Glc-(1→3)-4-O-methyl-α-l-Rha-(1→3)-α-l- Rha-(1→3)-α-l-Rha-(1→2)-6-d-l-Tal (9, 25). Based on the behavior of GPLs in TLC and the GC-MS analysis of alditol acetate derivatives, serotype 16 GPL seems to possess a unique carbohydrate epitope similar to that of serotype 17 GPL. We compared the OSE of serotype 16 GPL to that of serotype 17 GPL. The acylated amido group that was bound to the terminal sugar was different, although the linkage position was identical. Except for the terminal-acylated amido sugar, the other sugar compositions and glycosyl linkage positions were completely identical. An acylated amido group attached to the C-3 position of Hex is very unusual. To our knowledge, 3-amido-Hex is irregular in nature, although 2-amido-Hex is known to be glucosamine or galactosamine, which is frequently isolated as a component of lipopolysaccharides and glycosaminoglycans in prokaryotic and eukaryotic cells (7, 42). Further, existence of short-chain fatty acid 2-methyl-3-hydroxy-4-methoxy-pentanoic acid linked to the amido group of d-Hex is also unique. The characteristic gene cluster is thought to regulate the production of 3-acylated-amido-Hex. It is difficult to determine the species of acylated amido sugars, because no reference standard is available. The terminal sugar of the serotype 17 GPL was reviewed as a gluco-configuration, although firm evidence was not shown (9, 25). The JCH and J1-2 values for the anomeric proton in the terminal sugar were 161 and 7.7 Hz, respectively (Fig. 6; Table S1 in the supplemental material). These results demonstrated unequivocally that the terminal amido-Hex was β configuration and H-2 was in the axial position. The terminal amido-Hex is considered to be derived from glucose or galactose, not Rha.
Next, we explored the genetic mechanism of GPL biosynthesis, because the elongation of carbohydrate chains in serotype-specific GPLs is poorly understood. The ser2 gene cluster of the M. avium serotype 2 strain (31) and a 27.5-kb DNA fragment of the M. avium serotype 4 strain (28) were identified to be responsible for the biosynthesis of each OSE in GPLs. Recently, enzymatic characterizations of glycosyltransferase and methyltransferase of nonpolar GPLs have been reported for Mycobacterium smegmatis (36, 38). In the serotype-specific polar GPL biosynthesis of MAC, only the rtfA gene was functionally clarified to encode the transfer of l-Rha to 6-d-Tal, but which gene cluster transfers the sugars next to l-Rha elongated from 6-d-Tal is unclear.
In this study, we cloned the biosynthetic cluster of the serotype 16 GPL and analyzed its sequence. Seventeen ORFs were detected in the serotype 16 strain, and the sequence homology was analyzed. The transformant of the M. avium serotype 1 strain carrying cosmid clone no. 253 produced serotype 16 GPL. These results strongly implied that this gtfB-drrC region is responsible for the biosynthesis of the serotype 16-specific GPL. From the structural analysis of the serotype 16 GPL and the sequence of cosmid clone no. 253, it is possible to predict the relationship between the biosynthesis of serotype 16 GPL and the function of each ORF.
The genetic map of the serotype 16 GPL biosynthetic cluster was compared to those of serotype 2 GPL from M. avium strain 724, serotype 4 GPL from M. avium strain A5, and serotype 7 GPL from M. intracellulare strain ATCC 35847T (12, 18, 28). Significant differences were found in the neighborhood of the conserved region. The genetic organization of the serotype 16 GPL gene cluster was distinct from that of serotype 7, except for some of the ORFs, and the ORFs in this region of serotype 2 and serotype 4 were completely different from ORFs 1 to 17 in serotype 16 (Fig. 7).
In addition to M. intracellulare serotype 7 (18) and serotype 16 strains, we have analyzed similar gene clusters of M. intracellulare serotype 12 and 17 strains. The sequence homology of the regions of ORF 1 and ORF 17 was highly conserved between only M. intracellulare serotype 16 and 17 strains (unpublished data). ORFs 1, 16, and 17 may lead to transfer of the two additional molecules of l-Rha and terminal amido-Hex. ORF 2 was assigned to acyltransferase and may be responsible for biosynthesis of the 3-2′-methyl-3′-hydroxy-4′-methoxy-pentanoyl-amido group in the terminal Hex. ORF 3 is probably responsible for the transfer of the O-methyl group at the C-4 position in the third l-Rha from 6-d-Tal. ORF 6 is homologous to aminotransferase and possibly associated with the biosynthesis of an amido group in the terminal Hex. The deduced amino acid sequences of ORF 6 in serotype 16 and ORF 4 in serotype 7 have homologies to DegT_DnrJ_EryC1 aminotransferases. However, these two ORFs are dissimilar to each other. Serotype 16 and 7 GPLs have an amido group at the terminal Hex, although the attachment position is different. The serotype 7 GPL has an amido group at the C-4 position in the terminal Hex, but the serotype 16 GPL has it at the C-3 position. Nine ORFs between ORF 7 and ORF 15 are possibly involved in fatty acid synthesis of the acyl chain moiety linked by an amido bond of the terminal Hex. Taken together, this gene cluster may participate in the biosynthetic pathway of the serotype 16-specific GPL, although further study is needed to clarify the function of each ORF.
Recent studies suggest that GPLs play an important role in the phenotype and pathogenicity of MAC. The colony morphology is considered to be influenced by cell wall GPL. MAC colony phenotypes spontaneously occur from smooth to rough type, and this is due to a mutation lacking GPL (3, 13, 22). The deletion of genomic regions encoding GPL biosynthesis may result in the loss of GPL. Danelishvili et al. demonstrated that the uptake by and growth in macrophages of a MAC mutant with the gene belonging to the GPL synthesis pathway inactivated by transposon insertion were decreased (11). Bhatnagar and Schorey have reported that macrophages infected with MAC release exosomes containing GPLs that result in the transfer of the GPLs to uninfected macrophages and induce a proinflammatory response (4). These findings imply that GPL participates in the pathogenicity of MAC. By contrast, our previous studies have demonstrated that anti-GPL antibodies are detected in the sera of most immunocompetent patients with MAC pulmonary disease and that the detection of anti-GPL antibody is useful for the serodiagnosis of MAC disease (15, 26, 27).
To understand the role of GPLs in MAC and its hosts, it is necessary to define the chemical structure and biosynthesis pathways of GPLs. Elucidation of the structure-function relationship of GPL may open a new avenue for controlling MAC disease.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Health Sciences Foundation, and the Ministry of Health, Labor, and Welfare of Japan (Research on Emerging and Reemerging Infectious Diseases).
We are grateful to Sumihiro Hase (Department of Chemistry, Graduate School of Science, Osaka University, Osaka, Japan) and Hiromi Murakami (Osaka Municipal Technical Research Institute, Osaka, Japan) for helpful discussion.
Footnotes
Published ahead of print on 7 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baess, I. 1983. Deoxyribonucleic acid relationships between different serovars of Mycobacterium avium, Mycobacterium intracellulare and Mycobacterium scrofulaceum. Acta Pathol. Microbiol. Immunol. Scand. 91201-203. [DOI] [PubMed] [Google Scholar]
- 2.Barrow, W. W., T. L. Davis, E. L. Wright, V. Labrousse, M. Bachelet, and N. Rastogi. 1995. Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infect. Immun. 63126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 26810517-10523. [PubMed] [Google Scholar]
- 4.Bhatnagar, S., and J. S. Schorey. 2007. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 28225779-25789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt, A., N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, and W. R. Jacobs, Jr. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. USA 1045157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 7.Campo, G. M., S. Campo, A. M. Ferlazzo, R. Vinci, and A. Calatroni. 2001. Improved high-performance liquid chromatographic method to estimate aminosugars and its application to glycosaminoglycan determination in plasma and serum. J. Chromatogr. B 765151-160. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, D., G. O. Aspinall, and P. J. Brennan. 1987. The presence of novel glucuronic acid-containing, type-specific glycolipid antigens within Mycobacterium spp. Revision of earlier structures. J. Biol. Chem. 2623528-3533. [PubMed] [Google Scholar]
- 9.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 582018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39131-203. [DOI] [PubMed] [Google Scholar]
- 11.Danelishvili, L., M. Wu, B. Stang, M. Harriff, S. Cirillo, J. Cirillo, R. Bildfell, B. Arbogast, and L. E. Bermudez. 2007. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc. Natl. Acad. Sci. USA 10411038-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 1492797-2807. [DOI] [PubMed] [Google Scholar]
- 13.Eckstein, T. M., J. M. Inamine, M. L. Lambert, and J. T. Belisle. 2000. A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J. Bacteriol. 1826177-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckstein, T. M., F. S. Silbaq, D. Chatterjee, N. J. Kelly, P. J. Brennan, and J. T. Belisle. 1998. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J. Bacteriol. 1805567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enomoto, K., S. Oka, N. Fujiwara, T. Okamoto, Y. Okuda, R. Maekura, T. Kuroki, and I. Yano. 1998. Rapid serodiagnosis of Mycobacterium avium-intracellulare complex infection by ELISA with cord factor (trehalose 6, 6′-dimycolate), and serotyping using the glycopeptidolipid antigen. Microbiol. Immunol. 42689-696. [DOI] [PubMed] [Google Scholar]
- 16.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field, S. K., D. Fisher, and R. L. Cowie. 2004. Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest 126566-581. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara, N., N. Nakata, S. Maeda, T. Naka, M. Doe, I. Yano, and K. Kobayashi. 2007. Structural characterization of a specific glycopeptidolipid containing a novel N-acyl-deoxy sugar from Mycobacterium intracellulare serotype 7 and genetic analysis of its glycosylation pathway. J. Bacteriol. 1891099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerwig, G. J., J. P. Kamerling, and J. F. G. Vliegenthart. 1978. Determination of the d and l configuration of neutral monosaccharides by high-resolution capillary G.L.C. Carbohydr. Res. 62349-357. [Google Scholar]
- 20.Hakomori, S. 1964. A rapid permethylation of glycolipid, and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J. Biochem. (Tokyo) 55205-208. [PubMed] [Google Scholar]
- 21.Heidelberg, T., and O. R. Martin. 2004. Synthesis of the glycopeptidolipid of Mycobacterium avium serovar 4: first example of a fully synthetic C-mycoside GPL. J. Org. Chem. 692290-2301. [DOI] [PubMed] [Google Scholar]
- 22.Howard, S. T., E. Rhoades, J. Recht, X. Pang, A. Alsup, R. Kolter, C. R. Lyons, and T. F. Byrd. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 1521581-1590. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann, S. H. 2001. How can immunology contribute to the control of tuberculosis? Nat. Rev. Immunol. 120-30. [DOI] [PubMed] [Google Scholar]
- 24.Khoo, K. H., D. Chatterjee, A. Dell, H. R. Morris, P. J. Brennan, and P. Draper. 1996. Novel O-methylated terminal glucuronic acid characterizes the polar glycopeptidolipids of Mycobacterium habana strain TMC 5135. J. Biol. Chem. 27112333-12342. [DOI] [PubMed] [Google Scholar]
- 25.Khoo, K. H., E. Jarboe, A. Barker, J. Torrelles, C. W. Kuo, and D. Chatterjee. 1999. Altered expression profile of the surface glycopeptidolipids in drug-resistant clinical isolates of Mycobacterium avium complex. J. Biol. Chem. 2749778-9785. [DOI] [PubMed] [Google Scholar]
- 26.Kitada, S., R. Maekura, N. Toyoshima, N. Fujiwara, I. Yano, T. Ogura, M. Ito, and K. Kobayashi. 2002. Serodiagnosis of pulmonary disease due to Mycobacterium avium complex with an enzyme immunoassay that uses a mixture of glycopeptidolipid antigens. Clin. Infect. Dis. 351328-1335. [DOI] [PubMed] [Google Scholar]
- 27.Kitada, S., Y. Nishiuchi, T. Hiraga, N. Naka, H. Hashimoto, K. Yoshimura, K. Miki, M. Miki, M. Motone, T. Fujikawa, K. Kobayashi, I. Yano, and R. Maekura. 2007. Serological test and chest computed tomography findings in patients with Mycobacterium avium complex lung disease. Eur. Respir. J. 291217-1223. [DOI] [PubMed] [Google Scholar]
- 28.Krzywinska, E., and J. S. Schorey. 2003. Characterization of genetic differences between Mycobacterium avium subsp. avium strains of diverse virulence with a focus on the glycopeptidolipid biosynthesis cluster. Vet. Microbiol. 91249-264. [DOI] [PubMed] [Google Scholar]
- 29.Maekura, R., Y. Okuda, A. Hirotani, S. Kitada, T. Hiraga, K. Yoshimura, I. Yano, K. Kobayashi, and M. Ito. 2005. Clinical and prognostic importance of serotyping Mycobacterium avium-Mycobacterium intracellulare complex isolates in human immunodeficiency virus-negative patients. J. Clin. Microbiol. 433150-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marras, T. K., and C. L. Daley. 2002. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 23553-567. [DOI] [PubMed] [Google Scholar]
- 31.Maslow, J. N., V. R. Irani, S. H. Lee, T. M. Eckstein, J. M. Inamine, and J. T. Belisle. 2003. Biosynthetic specificity of the rhamnosyltransferase gene of Mycobacterium avium serovar 2 as determined by allelic exchange mutagenesis. Microbiology 1493193-3202. [DOI] [PubMed] [Google Scholar]
- 32.McClatchy, J. K. 1981. The seroagglutination test in the study of nontuberculous mycobacteria. Rev. Infect. Dis. 3867-870. [DOI] [PubMed] [Google Scholar]
- 33.McCloskey, J. A. 1969. Mass spectrometry, p. 402. In J. M. Lowenstein (ed.), Methods in enzymology: lipid, vol. 14. Academic Press, New York, NY. [Google Scholar]
- 34.McNeil, M., H. Gaylord, and P. J. Brennan. 1988. N-formylkansosaminyl-(1-3)-2-O-methyl-d-rhamnopyranose: the type-specific determinant of serovariant 14 of the Mycobacterium avium complex. Carbohydr. Res. 177185-198. [DOI] [PubMed] [Google Scholar]
- 35.McNeil, M., A. Y. Tsang, and P. J. Brennan. 1987. Structure and antigenicity of the specific oligosaccharide hapten from the glycopeptidolipid antigen of Mycobacterium avium serotype 4, the dominant Mycobacterium isolated from patients with acquired immune deficiency syndrome. J. Biol. Chem. 2622630-2635. [PubMed] [Google Scholar]
- 36.Miyamoto, Y., T. Mukai, N. Nakata, Y. Maeda, M. Kai, T. Naka, I. Yano, and M. Makino. 2006. Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis. J. Bacteriol. 18886-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odham, G., and E. Stenhagen. 1972. Fatty acids, p. 211-228. In G. R. Waller (ed.), Biochemical application of mass spectrometry. Wiley-Interscience, New York, NY.
- 38.Patterson, J. H., M. J. McConville, R. E. Haites, R. L. Coppel, and H. Billman-Jacobe. 2000. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 27524900-24906. [DOI] [PubMed] [Google Scholar]
- 39.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36762-771. [DOI] [PubMed] [Google Scholar]
- 40.Tsang, A. Y., J. C. Denner, P. J. Brennan, and J. K. McClatchy. 1992. Clinical and epidemiological importance of typing of Mycobacterium avium complex isolates. J. Clin. Microbiol. 30479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wayne, L. G., and H. A. Sramek. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 51-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods, A., and J. R. Couchman. 2001. Proteoglycan isolation and analysis, p. 10.7.1-10.7.19. In J. S. Bonifacino, M. Dasso, J. B. Harford, J. Lippincott-Schwartz, and K. M. Yamada (ed.), Current protocols in cell biology. Wiley Interscience, Hoboken, NJ. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.