Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that is one of the most refractory to therapy when it forms biofilms in the airways of cystic fibrosis patients. To date, studies regarding the production of an immunogenic and protective antigen to inhibit biofilm formation by P. aeruginosa have been superficial. The previously uncharacterized outer membrane protein (OMP) Opr86 (PA3648) of P. aeruginosa is a member of the Omp85 family, of which homologs have been found in all gram-negative bacteria. Here we verify the availability of Opr86 as a protective antigen to inhibit biofilm formation by P. aeruginosa PAO1 and several other isolates. A mutant was constructed in which Opr86 expression could be switched on or off through a tac promoter-controlled opr86 gene. The result, consistent with previous Omp85 studies, showed that Opr86 is essential for viability and plays a role in OMP assembly. Depletion of Opr86 resulted in streptococci-like morphological changes and liberation of excess membrane vesicles. A polyclonal antibody against Opr86 which showed reactivity to PAO1 cells was obtained. The antibody inhibited biofilm formation by PAO1 and the other clinical strains tested. Closer examination of early attachment revealed that cells treated with the antibody were unable to attach to the surface. Our data suggest that Opr86 is a critical OMP and a potential candidate as a protective antigen against biofilm formation by P. aeruginosa.
Pseudomonas aeruginosa is a major opportunistic pathogen responsible for many human diseases, such as cystic fibrosis and nosocomial infections such as bacteremia and pneumonia in immunocompromised hosts. Control of infection frequently proves difficult due to high levels of antibiotic resistance (44) and the fact that the organism is known to exist in the body in a biofilm state, which confers even more resistance (6, 26). Biofilms are adherent aggregates of bacterial cells that form matrix-enclosed, complex, and organized communities on biotic and abiotic surfaces (18, 19, 46). Bacteria in biofilms are resistant to antibiotics (8, 27) and less conspicuous to the immune system (6, 14).
Host immunological defenses prevent human pathogens from infecting particular areas of the body, and this is well characterized in pathology, immunology, and bacteriology. In the last decade, outer membrane proteins (OMPs) belonging to the Omp85 family have been identified as antigens in pathogenesis and immunity. For example, several studies of Haemophilus influenzae infection using animal models demonstrated that 85-kDa OMP D15 confers protection against homologous and heterologous strains (25, 52). Similarly, Oma87 of Pasteurella multocida has been shown to elicit protection in an animal model of infection (41). D15 and Oma87, as well as Tp92 of Treponema pallidum, Omp85 of Neisseria meningitidis, and YaeT of Escherichia coli, are members of the Omp85 family, and Omp85 homologs are highly conserved among gram-negative bacteria (5, 43, 49). Omp85/YaeT has recently been well characterized in N. meningitidis and E. coli. The genes encoding Omp85/YaeT are essential for viability and positioned within an operon coding for a range of genes required for outer membrane (OM) biogenesis (7, 11, 49, 50, 51). Moreover, YaeT plays a role in the assembly and insertion of β-barrel proteins into the OM by forming complexes with lipoproteins such as YfgL, YfiO, NlpB, and SmpA (28, 40, 47, 51).
P. aeruginosa OMPs have been studied as candidates for vaccine antigens in the form of purified OM preparations (22, 24), isolated OMPs (38, 48), or protein fusions (1, 21). OMPs are more suitable as antigens than lipopolysaccharides, exopolysaccharides, or isolated flagella are for routine clinical use because of the safety and efficacy of OMPs. As one example, OprF has been well studied as a vaccine target because of the major porin (33), high antigenicity, and high homology among Pseudomonas strains (12, 23, 24). However, an immune response against OMPs involved in biofilm formation by P. aeruginosa has not been reported. Our studies have focused on examining if the OMP antigen immune reaction includes biofilm inhibition and, if so, identifying the individual OMP antigen involved in eliciting such a response.
In this study, we focused on a member of the Omp85 family for its potential as an immunogenic surface antigen due to the presence of extracellular domains as well as conserved regions among closely related species. We investigated whether Opr86, which is an Omp85 homolog of P. aeruginosa and a previously uncharacterized protein, might serve as a new candidate for a protective antigen against biofilm formation by P. aeruginosa. We first characterized Opr86 by constructing a chromosomal opr86 deletion mutant and polyclonal antibodies of recombinant Opr86. We showed that Opr86 of P. aeruginosa is essential for viability and affects the assembly of OMPs. Moreover, we showed that an antibody against Opr86 has a competitive inhibitory effect against biofilm formation by PAO1 as well as several Pseudomonas clinical isolates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. E. coli was grown in LB medium (Lennox; Nacalai, Kyoto, Japan) at 37°C. When necessary, the following antibiotics were used: ampicillin, 100 μg/ml; and gentamicin, 10 μg/ml. P. aeruginosa was grown in LB medium or M63 minimal salts medium (37) supplemented with MgSO4 (1 mM) as the base medium at 37°C. M63 medium was supplemented with the following carbon sources: glucose, 0.2%; Casamino Acids, 0.5%; and arginine, 0.4%. When necessary, the following antibiotics were used: carbenicillin, 200 μg/ml; and gentamicin, 100 μg/ml. For complementation of PS186, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 100 μM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strain | ||
| P. aeruginosa | ||
| PAO1 | Wild type (formally PAO1-Tokai) | 29 |
| PAO1-H | Wild type (formally PAO1-Holloway) | 15 |
| PS186 | PAO1 opr86 mutant carrying pMMB6786 | This study |
| 287 | Clinical isolate | Okayama University |
| 401 | Clinical isolate | Okayama University |
| 428 | Clinical isolate | Okayama University |
| 443 | Clinical isolate | Okayama University |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+ lacIqlacZΔM15] | TaKaRa |
| BL21 | E. coli B, F−dcm ompT hsdS(rB+ mB+) gal | TaKaRa |
| S17-1 | pro thi hsdR recA Tpr Smr; chromosome::RP4-2 Tc::Mu-Km::Tn7 | 45 |
| Plasmid | ||
| pMMB67EH | Broad-host-range vector; Ampr IncQ | 10 |
| pMMB6786 | pMMB67EH derivative carrying the opr86 gene | This study |
| pHSG398 | Cloning vector; Cmr | TaKaRa |
| pG19II | Derivative of pK19 mob sacB; Gmr | 30 |
| pGEX6P1 | Cloning vector; Ampr | GE Healthcare |
| pGEX6p86 | pGEX6P1 derivative carrying p-opr86 | This study |
| pGEX6o86 | pGEX6P1 derivative carrying o-opr86 | This study |
Deletion of the chromosomal opr86 gene.
To construct a mutant lacking the opr86 gene, PCR primers for amplification of the region containing the opr86 gene were synthesized based on the nucleotide sequence from the Pseudomonas genome database (http://www.pseudomonas.com). After amplification of 0.8-kbp DNA fragments upstream and downstream of opr86 from chromosomal DNA with PAO1 as a template, using Fopr861 (5′-CCCAAGCTTGGATGGCTGGCTGAAGGG-3′) and Ropr862 (5′-CGGGATCCGAGAATCTGCCGCAAAGGGC-3′) or Ropr863 (5′-CGGGATCCGTACCGAGTAACGAAACCATGC-3′) and Ropr864 (5′-GGAATTCTGCGACAGGCTGGCATAGGCC-3′), the amplified DNA fragments were digested with HindIII/XbaI or XbaI/EcoRI (underlined) and ligated into the HindIII-EcoRI site in a multicloning site of pG19II to yield pG19-delopr86. A 2.4-kbp opr86 gene fragment amplified with Fopr865 (5′-GGAATTCGTCAGTTGGAAAAAAAGGACTCCATGAAAACG-3′) and Ropr866 (5′-CCCAAGCTTATCAGAAGGTCTGGCCCAGGG-3′) was digested with EcoRI-HindIII (underlined) and ligated into a multicloning site of pMMB67EH to yield pMMB6786. The plasmid pG19-delopr86 was introduced into the mobilizer strain E. coli S17-1 and then was transferred to PAO1 by conjugation as reported previously (30) with some modification. After the first crossover events, the plasmid pMMB6786 was introduced into the conjugant. Then, the transformant was cultured in the presence of 100 μM IPTG, and the second crossover events were performed. The deletion of the chromosomal opr86 gene was confirmed by PCR analysis.
Bacterial viability assay.
The viability of PAO1 and PS186 in planktonic shaking culture was determined by using a BacLight Live/Dead bacterial viability staining kit (Molecular Probes Inc., Eugene, OR). Two stock solutions of stain (SYTO9 and propidium iodide) were each diluted to a concentration of 3 μl/ml in medium. Live SYTO9-stained cells and dead propidium-stained cells were excited with a cool argon laser and a helium neon laser, respectively, and detected with 560-nm and 505-nm long-pass filters, respectively, by using a scanning confocal laser microscope (LSM 5 PASCAL; Carl Zeiss, Oberkochen, Germany). The results are expressed as the means and standard deviations (SD) from five representative images.
SDS-PAGE and Western blot analysis.
Samples were treated with sodium dodecyl sulfate (SDS) loading buffer including 5% 2-mercaptoethanol at 100°C for 5 min and separated by polyacrylamide gel electrophoresis (PAGE) (42). For Western blotting assays, gels were transferred to polyvinylidene difluoride membranes (Atto, Tokyo, Japan). After incubation of the membrane in Tris-buffered saline (TBS) containing 0.5% skim milk for 1 h, the membrane was incubated with p-Opr86 antiserum (1/1,000 dilution) or o-Opr86 antiserum (1/10,000 dilution) at 37°C for 1 h. The membrane was washed with TBS containing 0.05% Tween 20 (TBST) and incubated with goat anti-rabbit alkaline phosphatase-conjugated immunoglobulin G (IgG) secondary antibodies (1/10,000 dilution) (Sigma-Aldrich, St. Louis, MO) for 1 h. The membrane was washed with TBST and exposed to NBT/BCIP (nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate; Roche, Basel, Switzerland).
Analysis of membrane proteins.
OMPs and inner membrane proteins (IMPs) were isolated from lysed total cell proteins by Sarkosyl separation as described previously (31). IMPs purified from 100 μg of total proteins and OMPs purified from 1 mg of total proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. The band intensities of OMPs were evaluated using densitometric scanning (Image Master 1D Elite; GE Healthcare, Buckinghamshire, United Kingdom). Protein bands were excised from gels, and the identification procedure was followed as previously described (20). The protein concentration was determined by the Bradford method (3).
Preparation of antibodies.
After amplification of DNA fragments using chromosomal DNA of PAO1 as a template with Fopr867 (5′-GGAATTCGAGGTTCACGCCGAGTCCTTCACTG-3′) and Ropr866 or Fopr868 (5′-GGAATTCTACGGATCCACCGAGCGCCTGC-3′) and Ropr869 (5′-CCGCTCGAGGTTGTCGGTCTTGATGCCGTCG-3′), the amplified DNA fragments were digested with EcoRI/HindIII or EcoRI/XhoI (underlined) and ligated into pGEX6P1 to yield pGEXp86 and pGEXo86, and these plasmids were introduced into E. coli BL21. The transformants were cultured at 37°C in LB liquid medium containing ampicillin. At an optical density at 600 nm (OD600) of 0.5, the culture medium was rapidly chilled to 20°C, induced with 100 μM IPTG, and incubated for an additional 3 h at 20°C. After harvesting cells by centrifugation, the pellet was resuspended in 50 mM phosphate buffer (pH 7.5) and disrupted ultrasonically. After adding Triton X-100 (final concentration of 1%) and incubating at 4°C for 30 min, unbroken cells were eliminated by centrifugation. The hybrid proteins were purified by Bulk and RediPack glutathione S-transferase (GST) purification modules (GE Healthcare), and the GST was removed with PreScission protease (GE Healthcare). Antisera against p-Opr86 or o-Opr86 were produced by immunizing infant Japanese White rabbits. Doses of antigen (0.3 mg) were emulsified in complete Freund's adjuvant (BD Biosciences, San Jose, CA) and immunized for priming. Two boosts at 2 and 4 weeks with another 0.1-mg dose of protein in incomplete Freund's adjuvant were performed, and then sera were collected at 6 weeks. Antiserum against PAO1 was generated by using the same protocols except that a heat-inactivated bacterial preparation was used as the immunogen (108 CFU per dose). For purification of IgG, proteins in rabbit serum were extracted by ammonium sulfate precipitation in 60% (wt/vol) saturated ammonium sulfate. After desalting with HiTrap (GE Healthcare), IgG was purified from the extracted proteins by HiTrap protein A HP (GE Healthcare). The fractions of protein were detected at 280 nm.
Scanning electron microscopy observation.
For scanning electron microscopy, bacteria grown in LB medium were dropped onto glasses coated with 0.5% poly-l-lysine hydrobromide (Sigma-Aldrich). Then, sample glasses were fixed with a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.2) for 30 min and washed with PBS. After dehydration through an ascending acetone series, the specimens were dried by critical point drying with a liquid CO2 dryer (HCP-2 type; Hitachi, Tokyo, Japan), coated with osmium with a model NL-OPC 80A osmium plasma coater (Nippon Laser and Electronic Laboratory, Nagoya, Japan), and examined with a scanning electron microscope (model S-5000; Hitachi). For immunogold labeling, bacteria were fixed with 4% paraformaldehyde in PBS for 30 min and washed with 0.1% bovine serum albumin (BSA) in PBS. The specimens were incubated with 1% BSA in PBS for 30 min and washed again with 0.1% BSA in PBS. The specimens were then incubated with rabbit serum at a dilution of 1:100 in PBS containing 1% BSA for 60 min. The specimens were washed with 0.1% BSA in PBS and incubated with colloidal gold-labeled anti-rabbit immunoglobulin (diameter of colloidal gold particles, 15 nm) at a dilution of 1:100 in PBS containing 1% BSA for 60 min, and then they were prepared in the same manner as for scanning electron microscopy.
Immunoassay.
Reactivities of antisera were tested by enzyme-linked immunosorbent assay (ELISA). ELISA plates (IWAKI, Tokyo, Japan) were coated with 100 ng/well of purified protein or lysed cell protein in PBS and incubated overnight at 4°C. The wells were blocked with 1% skim milk in PBS containing 0.05% Tween 20 (PBST). After washing with PBST, several diluents of antisera were added to the wells and incubated at 37°C for 1 h. The wells were washed and then treated with alkaline phosphate-conjugated monoclonal anti-rabbit immunogloblins (Sigma-Aldrich) for 1 h. The reactions were developed using 0.3% disodium p-nitrophenyl phosphate as a substrate for 20 min, and the absorbance was measured at OD405. ELISA units were calculated as (OD of sample − OD of negative control)/(OD of positive control − OD of negative control) and are reported as means ± SD from three wells per sample.
Competitive biofilm inhibition assay using IgGs.
P. aeruginosa cells grown in LB medium overnight were washed with PBS three times and diluted to an OD600 of 0.1 in PBS containing IgG at a 10-fold excess over the optimal concentration. The mixtures were incubated at 37°C for 1 h, and these IgG-treated cells were used in planktonic growth assays, biofilm formation assays, or initial surface attachment assays. (i) In the planktonic growth assays, the mixtures were inoculated into LB medium in a test tube to set an OD600 of 0.01. After static incubation for 8 h, test tubes were vortexed and live cells were plated and counted as CFU. (ii) In the biofilm assays, 10 μl of the mixtures were inoculated into 90 μl of LB or M63 medium in microtiter wells made of polyvinylchloride. M63 medium was supplemented with either 0.2% glucose and 0.5% Casamino Acids or with 0.4% arginine. The plates were incubated at 37°C under aerobic conditions for 8 h. Crystal violet staining and quantification of biofilms were performed as previously described (36). (iii) In the initial surface attachment assay, IgG-treated cells were inoculated into LB medium to a final concentration at OD600 of 0.01 (final IgG concentrations were 400 μg/ml) and glass coverslips were placed within the wells and leaned against the side of the well. After incubation for 1 h, 2 h, or 4 h, the glass coverslips were washed to remove the resumed planktonic bacteria and stained with 0.1% crystal violet. The surface-attached bacteria at the air-liquid interface were observed immediately with LSM 5 PASCAL (Carl Zeiss, Oberkochen, Germany).
RNA isolation and reverse transcription (RT)-PCR.
Total RNA was isolated from exponentially growing bacteria (approximately 108 cells) by using the RNeasy mini kit (Qiagen, Valencia, CA), as described by the manufacturer. Genomic DNA was eliminated by RNase-free DNase I (Qiagen) treatment during the isolation procedure. The cDNA synthesis was performed from 1 μg of RNA using RT SuperScript III (Invitrogen, Carlsbad, CA) with gene-specific primers (Romp8610, 5′-GCCCACCACGTTGATGTGGGAAATCGCGG-3′; or RlpxA1, 5′-CGCATGCCTTCGAAGTTCATGCTGCGGGC-3′). PCR amplification for 25 cycles was performed using a 10% volume of synthesized cDNA as the template and the following primers: for opr86, Fopr8611 (5′-GTCCGCCGGTAGCGTGTTCGCCG-3′) and Ropr8612 (5′-CTTCAGCGCGACGCGGTTGCGCG-3′); and for lpxA, FlpxA2 (5′-GTCCGCACGTGGTGCTCAAGGGCC-3′) and RlpxA3 (5′-TTGCCGATGGCGCTGCCCATGCCG-3′).
Congo red assay.
Five microliters of PAO1 cell mixtures treated with 4 mg/ml IgG were applied to an M63 minimal medium agar plate containing Congo red (20 μg/ml) and Coomassie brilliant blue (0.4 μg/ml) and supplemented with arginine as a carbon source. Plates were incubated at 37°C for 24 h followed by 24 h at room temperature.
DNA sequencing.
Sequencing reactions were carried out using ABI PRISM BigDye sequencing kits (Applied Biosystems, Foster City, CA) on an ABI PRISM 310 genetic analyzer (Applied Biosystems). The Ropr866 primer was used for sequencing.
RESULTS
Opr86 is essential for viability in P. aeruginosa.
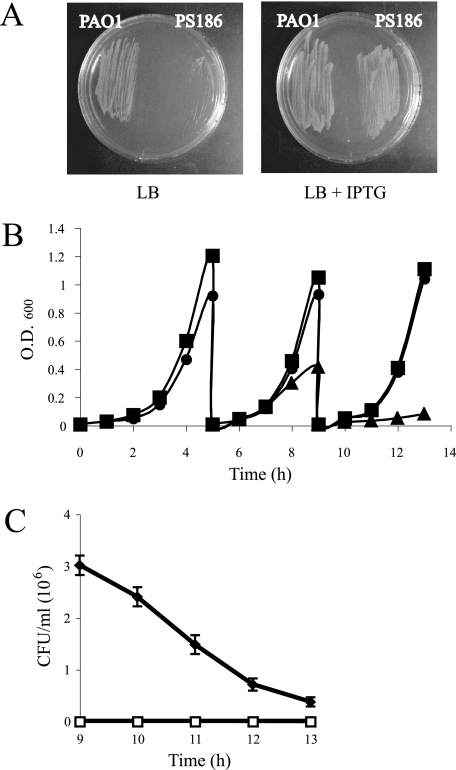
Opr86 (PA3648), an Omp85 homolog of P. aeruginosa, has 32% amino acid identity with neisserial Omp85. The opr86 open reading frame is 2,394 bp in length, encoding a 798-amino-acid protein with a predicted molecular mass of 86 kDa (without probable signal peptide 1-20 predicted by SignalP; www.cbs.dtu.dk/services/SignalP/). To study Opr86 function in P. aeruginosa, we attempted to disrupt the chromosomal copy of the opr86 gene. Based on the data of transcriptional organization around neisserial Omp85 (11), it is predicted that the opr86 gene is located in the middle of a transcriptional unit containing genes involved in lipid A, fatty acid, and phospholipid biogenesis. Therefore, we chose not to replace the opr86 gene with an antibiotic cassette in order to prevent potential transcriptional effects on the downstream genes. Since experiments without a plasmid expressing opr86 in the opr86 deletion background were unsuccessful, it was predicted that the opr86 gene is essential for growth or viability. We constructed a complementation vector pMMB6786 in which opr86 expression is regulated by the tac promoter, and we transformed this plasmid into PAO1 during construction of the chromosomal opr86 deletion to generate a conditional opr86 mutant (termed PS186). pMMB6786 could be extracted from PS186, showing that the plasmid was not integrated into the chromosome. The deletion of the opr86 gene did not affect expression of the downstream lpxA genes in the resulting PS186 strain, as confirmed by RT-PCR (data not shown), suggesting that the opr86 deletion does not have a polar effect on the downstream genes. PS186 could grow on the LB agar only in the presence of IPTG (Fig. 1A). To further examine the growth characteristics, we grew PAO1 in LB broth and PS186 in LB broth supplemented with IPTG to early stationary phase and then in diluted medium with or without IPTG. While PS186 with IPTG grew as well as PAO1, PS186 without IPTG could not grow well when transferred to fresh medium (Fig. 1B), suggesting that Opr86 is essential for growth at least. The CFU experiment of PS186 from the 9-h to 13-h growth point showed that the number of survival cells becomes fewer as time passes (Fig. 1C), suggesting that depletion of Opr86 causes cells to die. To further determine if Opr86 is essential for viability, the cells at the 13-h point shown in Fig. 1B were stained with BacLight Live/Dead stain and examined by confocal microscopy. In this experiment, the viable cell rate was 99% ± 1% for the wild type, 90% ± 3% for PS186, and 16% ± 3% for PS186 without IPTG. These results indicated that Opr86 is essential for not only growth but also viability in P. aeruginosa.
FIG. 1.
Opr86 is essential for viability. (A) PAO1 and a conditional opr86 mutant (PS186) grown on LB agar at 37°C in the absence or the presence of IPTG overnight. (B) Growth curves of PAO1 (squares) and PS186 in the presence (circles) and absence (triangles) of 100 μM IPTG. Initially, PS186 was cultured with IPTG. At the 5-h growth point, cells were diluted to an OD600 of 0.01, and then IPTG was added to one sample of PS186 but not to the other. At the 9-h growth point, cells were diluted to an OD600 of 0.01 in the presence or absence of IPTG again. PAO1 was also cultured and diluted without adding IPTG. The results presented are representative of three independent experiments. (C) The survival rate of PS186 without IPTG. PS186 was cultured without IPTG in the same way described for panel B and plated to an LB plate with (black) or without (white) 100 μM IPTG at hourly intervals after the 9-h growth point. The results are expressed as the mean CFU/ml and SD from three replicates. Similar results were obtained in two independent experiments.
Opr86 plays a role in OMP assembly.
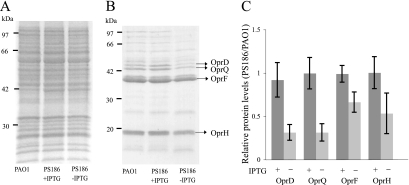
Since previous studies demonstrated that Omp85 of N. meningitidis and YaeT of E. coli play roles in OMP assembly (7, 49, 50), it was expected that Opr86 would also be involved in OMP assembly. To confirm this, OMP levels were examined from cultures of PAO1 and PS186 grown in the presence or absence of IPTG. OMPs and IMPs were purified from cells at the 9-h growth point shown in Fig. 1B, and the protein patterns were analyzed by SDS-PAGE. Although no change was seen in IMP band patterns (Fig. 2A), several OMP bands in PS186 grown without IPTG appeared to be less prominent than the same bands in PAO1 grown without IPTG and PS186 grown with IPTG (Fig. 2B and C). Those bands were isolated and characterized by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The results showed that the main bands include the following proteins: OprF, which is involved in maintaining cell shape and is also important for growth in a low-salt or an anaerobic environment (39, 53); OprH, which replaces OM-stabilizing divalent cations (2); OprD, a component of multidrug resistance efflux pumps (16); and OprQ, a homolog of OprD (35). These are not lipoproteins but OMPs as well as β-barrel proteins in P. aeruginosa. These data suggest that Opr86 has a role in OMP assembly.
FIG. 2.
Opr86 plays a role in OMP assembly. (A, B) Examination of IMP (A) and OMP (B) levels by SDS-PAGE stained with Coomassie brilliant blue. IMPs and OMPs were extracted from homogenized cells at the 9-h growth point prior to the dilution described in the legend for Fig. 1B. WT, PAO1 wild type; PS186 +IPTG, PS186 cultured with IPTG; PS186 −IPTG, PS186 cultured without IPTG. (C) Protein levels from four fractions of OMPs from PS186 cultured with and without IPTG were quantified and expressed as ratios relative to levels of PAO1 OMPs and SD from three independent experiments.
To further examine the OMP assembly function of Opr86, the susceptibilities of PAO1 and PS186 to various small molecules such as detergents and antibiotics were tested. PS186 incubated with IPTG at a concentration of 10 μM, conditions in which opr86 was transcribed at very low levels (as determined by RT-PCR; data not shown), was susceptible to non-ionic detergents such as Triton X-100 and Tween 20 (Table 2). In contrast, PAO1 and PS186 incubated with IPTG at a concentration of 100 μM were not susceptible to the same detergents. In addition, this Opr86-depleted strain exhibited increased susceptibility to all antibiotics tested. These disruptions of the barrier function of the OM also reinforce the role of Opr86 in OMP assembly.
TABLE 2.
Susceptibilities to various detergents and antibiotics
| Compounda | Zone of growth inhibition (mm)b
|
||
|---|---|---|---|
| PAO1 | PS186 (IPTG 100 μM) | PS186 (IPTG 10 μM) | |
| 10% SDS | 0 | 0 | 0 |
| 5% CTAB | 4.0 (±0) | 4.3 (±0.4) | 4.3 (±0.4) |
| 10% Tween 20 | 0 | 0 | 4.7 (±0.5) |
| 10% Triton X-100 | 0 | 0 | 3.7 (±0.5) |
| 0.5 M EDTA | 6.7 (±0.5) | 7.0 (±0.8) | 6.7 (±0.5) |
| 1% Polymyxin B | 11.3 (±0.5) | 11.3 (±0.5) | 12.6 (±0.5) |
| 1% Nalidixic acid | 13.3 (±0.9) | 14.0 (±0.8) | 15.7 (±1.2) |
| 1% Ofloxacin | 18.0 (±0.8) | 17.6 (±0.5) | 26.7 (±1.7) |
| 1% Rifampin | 7.3 (±0.9) | 7.3 (±0.5) | 10.6 (±0.9) |
| 1% Erythromycin | 5.0 (±0) | 5.0 (±0) | 8.0 (±1.4) |
| 1% Chloramphenicol | 13.0 (±0) | 14.0 (±0.8) | 19.0 (±1.6) |
| 1% Tetracycline | 15.0 (±1.6) | 17.3 (±1.7) | 19.3 (±1.7) |
| 1% Gentamicin | 11.0 (±0) | 11.3 (±0.5) | 12.3 (±0.5) |
The compounds were spotted (40 μl) in a cup placed on M63 minimal medium (supplemented with glucose and Casamino Acids as carbon sources) agar containing a lawn of cells plated at 106 cells/ml. Nalidixic acid, ofloxacin, and rifampin were resuspended in 0.02 N NaOH, 0.01 N HCl, and 50% methanol, respectively. Erythromycin, chloramphenicol, and tetracycline were resuspended in 50% ethanol. The zones were measured after 12 h at 37°C. CTAB, cetyltrimethylammonium bromide.
The values are expressed as the averages for triplicate assays. The numbers in parentheses indicate SD.
Opr86 depletion alters cellular morphology.
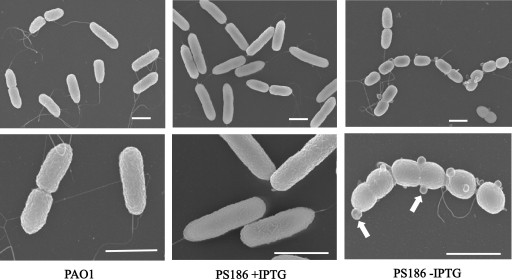
To determine whether the defects in OMP assembly resulting from Opr86 depletion led to observable changes in cellular morphology, the morphologies of PAO1 and PS186 at the 13-h growth point shown in Fig. 1B were examined by scanning electron microscopy (Fig. 3). No difference was observed between PAO1 and PS186 cells when grown with IPTG. In contrast, PS186 cells grown without IPTG were more heterogeneous, some cells showed streptococci-like forms, and the ratio of those forms was approximately 50% when we observed them, suggesting a cell division defect under Opr86 depletion. In addition, many membrane vesicles, for which diameters varied between 50 and 150 nm, were observed on the surface of PS186. Since release of membrane vesicles is an envelope stress response (32), it is possible that overvesiculation under Opr86 depletion would be due to accumulation of many unassembled OMPs in the periplasm.
FIG. 3.
Electron micrographs of PAO1 and PS186 cells at the 13-h growth point from Fig. 1B. Upper panels were taken at low magnification, and lower panels were taken at high magnification. Results are representative images. Arrows indicate membrane vesicles. The bar equals 1 μm in all panels.
Antiserum against Opr86 has reactivity with PAO1.
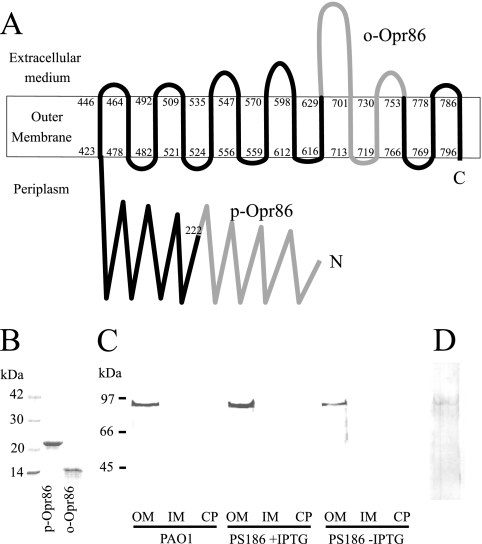
Various computational tools and algorithms (PSORT, PHDhtm, and TMpred) predicted that Opr86 might be an integral OMP. To investigate the localization of Opr86, antibodies against Opr86 were produced. Based on the neisserial Omp85 topology model (49), the structure of YaeT (17), and the topological predictor, PROFtmb, Opr86 is predicted to have a β-barrel structure with 14 transmembrane domains (Fig. 4A). Based on this predicted structure of Opr86, two peptide fragments were selected as antigenic candidates and purified from E. coli expressing GST fusion proteins (Fig. 4B). One fragment is a 23-kDa peptide, p-Opr86 (periplasmic domain of Omp85), composed of amino acid residues 17 to 222, which is the amino terminus and expected soluble fraction. The other fragment is a 14-kDa peptide, o-Opr86 (mostly exposed domain of Opr86), composed of amino acid residues 630 to 752. Western blot analysis of PAO1 cellular fractions confirmed that Opr86 was localized to the OM fraction (Fig. 4C and D).
FIG. 4.
(A) Topology prediction of Opr86 in P. aeruginosa. It is predicted that Opr86 has 14 transmembrane domains. The first and last amino acids of each β-strand are indicated. The domains of p-Omp86 and o-Opr86 are shown in gray. The amino (N) and carboxyl (C) termini of the protein are indicated. (B) SDS-PAGE analysis of p-Opr86 and o-Opr86 purification, performed on a 15% gel. Two micrograms of purified protein was applied per well, and the gel was stained with Coomassie brilliant blue. (C, D) Immunoblot analysis of Opr86 in PAO1. (C) Expression and localization of Opr86 using p-Opr86 antisera. OMPs, IMPs, and cytoplasmic proteins were extracted from homogenized cells at the 9-h growth point prior to the dilution described in the legend for Fig. 1B. Fifty micrograms of protein was separated on a 10% acrylamide SDS gel. WT, PAO1 wild type; PS186 +IPTG, PS186 cultured with IPTG; PS186 −IPTG, PS186 cultured without IPTG. OM, outer menbrane; IM, inner membrane; CP, cytoplasm. (D) Opr86 detection by using o-Opr86 antisera. Fifty micrograms of OMPs extracted from PAO1 were separated on a 10% acrylamide SDS gel.
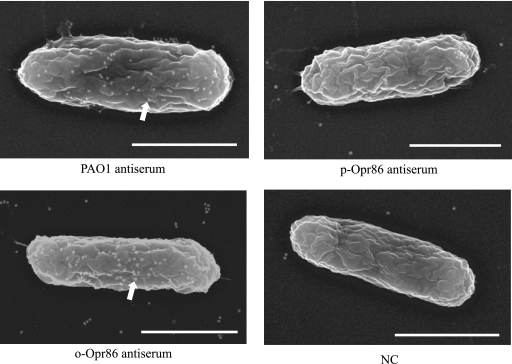
Since the predicted topology of Opr86 implies that the antisera against o-Opr86 will react with intact P. aeruginosa, PAO1 cells were assessed by immunolabeling and scanning electron microscopy in order to verify the reactivity of these antisera with P. aeruginosa. Cells treated with either o-Opr86 antiserum or antiserum against PAO1 showed a high degree of labeling, whereas cells treated with p-Opr86 antiserum were relatively devoid of labeling (Fig. 5). The predicted target of o-Opr86 antiserum is an extracellular portion of Opr86, and the micrograph supports this localization. On the other hand, p-Opr86 antiserum, which is supposed to target the periplasmic portion of Opr86, did not label the surface of PAO1, suggesting that Opr86 has a long N-terminal periplasmic domain as shown in Fig. 4A. Taken together, these data further support the predicted topology of Opr86 as an outer transmembrane protein. Importantly, the antiserum against o-Opr86 has reactivity with PAO1 cells.
FIG. 5.
Immunogold electron micrographs of PAO1. White dots on cells (arrow) indicate gold particles. Exponentially grown cells were probed with the primary antibodies shown below each figure and developed with a secondary gold-conjugated anti-rabbit IgG antibody. NC indicates nonimmunized serum. Bar, 1 μm.
An antibody against Opr86 inhibits biofilm formation.
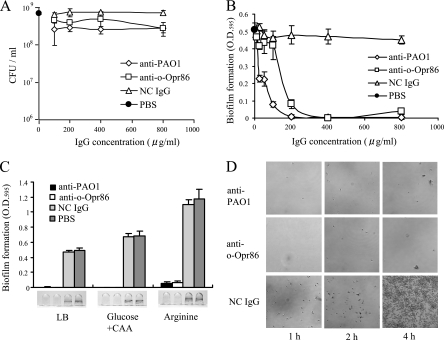
The purpose of this study was to verify the potential of Opr86 as an antigen to inhibit P. aeruginosa biofilms by immunization. It was therefore necessary to examine whether o-Opr86 antiserum is able to inhibit biofilm formation by PAO1. In inhibitory assays, PAO1 antisera and nonimmunized rabbit antiserum were used as positive and negative controls, respectively. To examine the direct reaction of IgG in antisera, IgGs were purified from each antiserum by affinity chromatography. After incubation of several IgGs with PAO1 for 1 h, planktonic growth and static biofilm formation in nutrient rich medium (LB medium) were examined. In the planktonic growth assay, there was very little difference in growth between samples at IgG concentrations ranging from 0 to 800 μg/ml (Fig. 6A), indicating that antibodies against either PAO1 (anti-PAO1) or o-Opr86 (anti-o-Opr86) did not inhibit the growth of PAO1. However, in the static biofilm assay with microtiter plates, biofilm formation by PAO1 cells treated with anti-o-Opr86 was inhibited to a degree comparable with that of anti-PAO1 (Fig. 6B). In contrast, biofilm formation was not inhibited by the p-Opr86 antibody (data not shown). IgG of a nonimmunized rabbit also could not inhibit biofilm formation, indicating that inhibition of biofilm formation by anti-o-Opr86 was not due to crude protein in IgG. These data suggest that the attachment of anti-o-specific IgG to Opr86 could inhibit biofilm formation by PAO1.
FIG. 6.
Immunological influences on planktonic growth or biofilm formation by purified IgG. PAO1 cells were preincubated with anti-PAO1, anti-o-Opr86, nonimmunized IgG (NC IgG), or PBS. (A) Competitive inhibition of planktonic growth. The results are expressed as the mean CFU/ml and SD from three replicates. Similar results were obtained in two independent experiments. Data shown are representative of two independent experiments. (B) Inhibitory biofilm concentrations for each antibody. Biofilms were allowed to form for 8 h. Biofilms attached on wells were stained with crystal violet, solubilized in ethanol, and measured at the OD595. The results are expressed as the mean OD595 and SD from at least four wells per sample. Data shown are representative of three independent experiments. (C) Image and quantification of crystal violet-stained biofilm formation in LB and M63 minimal media supplemented with glucose and Casamino Acids (Glucose +CAA) or arginine. The IgG concentration in the medium was 400 μg/ml. The results are expressed as the means and SD from at least four wells per sample. Data shown are representative of three independent experiments. (D) Visualization of cells attached on glass plates at the air-liquid interface after preincubating with anti-PAO1, anti-o-Opr86, or NC IgG. The magnification is ×400, and the panel size is 105 μm by 105 μm.
To determine whether inhibition of biofilm formation by anti-o-Opr86 was consistent with other media, biofilm formation was examined on minimal medium. It has been previously reported that biofilm formation was robust in a minimal arginine medium and intermediate levels of biofilm were observed in a minimal glucose medium supplemented with Casamino Acids (4). In this work though, biofilm formation was greater on minimal medium supplemented either with glucose plus Casamino Acids or with arginine, compared to levels on LB medium. Inhibition of biofilm after reaction with anti-o-Opr86 was observed on both minimal media (Fig. 6C). Similar results were also obtained at 6 h, 7 h, and 12 h of incubation (data not shown).
Absorption of antibodies inhibits the initial attachment.
The process of biofilm formation by P. aeruginosa begins when planktonic cells initiate surface colonization in a flagellum-dependent manner (36). Cells then progress to a stable surface attachment and develop into a mature biofilm characterized by the synthesis of extracellular matrix components (19). To determine how anti-o-Opr86 inhibits biofilm formation, we examined initial attachment to a glass surface at the air-liquid interface (Fig. 6D). In this experiment, cells treated with either anti-PAO1 or anti-o-Opr86 showed very little surface attachment after 4 h. In contrast, cells treated with nonimmunized IgG showed attachment after only 1 h, and by 4 h, the surface was nearly completely covered with cells forming many clusters. The numbers of viable cells in the bulk fluid phase of these wells did not differ appreciably between these three samples (data not shown). These data suggest that anti-o-Opr86 inhibits biofilm formation at the initial attachment stage.
To further assess the impact of anti-o-Opr86 on biofilm formation, we examined the ability of cells treated with these antibodies to synthesize the polysaccharide component of extracellular matrix using Congo red assays, as Congo red has been shown to bind polysaccharide matrix components (9, 13, 54). When grown on agar plates containing Congo red, colonies of cells treated with either anti-o-Opr86, anti-PAO1, or control IgG were slightly red and there was essentially no difference between colonies (data not shown). These data suggest that inhibition of biofilm formation by absorption with either anti-PAO1 or anti-o-Opr86 was due to a decrease in the ability of cells to attach and did not involve a decrease in extracellular matrix synthesis.
Immunological biofilm inhibition is effective against clinical strains.
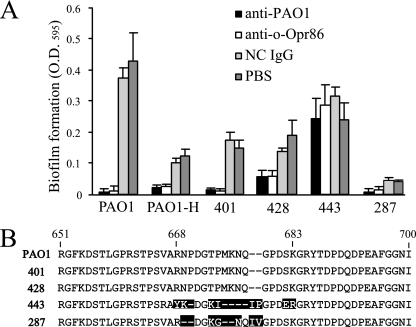
To inhibit biofilm formation in clinical settings, it is important to determine whether these IgGs have effects on clinical strains isolated from patients. Thus, the ability of anti-o-Opr86 to inhibit biofilm formation by another PAO1 strain (termed PAO1-H) and four clinical Pseudomonas strains, 401, 428, 443, and 287, was investigated. In these results, biofilm formations by PAO1-H and clinical strains 401 and 428 were inhibited by anti-o-Opr86 as well as anti-PAO1 (Fig. 7A). Biofilm formation by clinical strain 443 was unaffected by treatment with either antibody. The biofilm formation ability of 287 was potentially defective regardless of the treatment with IgG. These data suggest that anti-o-Opr86 treatment is able to inhibit biofilm formation by several different P. aeruginosa strains. To determine why anti-o-Opr86 could not inhibit biofilm formation by the clinical strain 443, we used PCR to amplify the o-Opr86 region in each clinical strain and found that we could amplify a DNA fragment encoding o-Opr86 from all four of the clinical strains, implying that Opr86 exists in these strains (data not shown). The sequences of the o-Opr86 region in clinical strains 401 and 428 were the same as that of PAO1, but those of 287 and 443 were different from PAO1, most notably in the region from amino acids 668 to 683 of Opr86 (Fig. 7B). This region is a predicted extracellular portion. Indeed, reactivity of anti-o-Opr86 to 443 was remarkably weaker than that to PAO1 (0.091 ± 0.002 and 1.000 ± 0.051 ELISA unit, respectively, at 1/800 dilution of o-Opr86 antiserum) as determined by ELISA. These data suggested that this region would be a target of Opr86 for protective immunity.
FIG. 7.
Immunological biofilm inhibition is effective against clinical strains. (A) Immunological influences on biofilm formation by purified IgG. PAO1 and four different clinical strains were preincubated with anti-PAO1, anti-o-Opr86, non-immunized IgG (NC IgG), or PBS and inoculated into microtiter wells containing 100 μl of LB medium. The IgG concentration in the medium was 400 μg/ml. Biofilms were allowed to form for 8 h. Biofilms attached to the wells were stained with crystal violet, solubilized in ethanol, and measured at OD595. Data shown are representative of three independent experiments. The results are expressed as the means and SD from at least four wells per sample. (B) The amino acid sequences of the o-Opr86 region. The sequence from amino acids 651 to 700 of opr86 in PAO1 and sequences of corresponding regions of clinical strains 401, 428, 443, and 287 are shown. Amino acids different from those of PAO1 are boxed in black.
DISCUSSION
This work focused on elucidating the function of Opr86 by using the opr86 conditional mutant. The chromosomal opr86 mutant PS186 could not grow without IPTG, showing that Opr86 is essential for viability and consistent with previous data in N. meningitidis (11, 49) and E. coli (7, 50, 51). P. aeruginosa is one of the most seriously antibiotic-resistant bacteria because of a low-permeability OM and the expression of a number of broadly specific multidrug efflux systems including MexAB-OprM, MexXY-OprM, MexCD-OprJ, MexEF-OprN, and MexJK-OprM (44). Opr86 is, however, both an OMP and an essential protein for viability. Thus, the targeting of antibiotics directly against Opr86 represents an important approach because it is possible that such antibiotics will be unaffected by efflux pumps.
Regarding the effect of Opr86 on OMP assembly, OMP levels of PS186 without IPTG were less than that of PAO1 (Fig. 2B). The determination of protein identities by matrix-assisted laser desorption ionization-time of flight mass spectrometry revealed that OprD, OprQ, OprF, and OprH were among the OMPs reduced in the PS186 background (Fig. 2C). It was shown in previous work that YaeT affects the assembly and/or insertion of OMPs including TolC, OmpF, OmpC, and OmpA in E. coli (50), and the same effect would be expected in P. aeruginosa. Over the past few years, several studies have focused on OMP assembly in E. coli, and they have shown that lipoproteins such as YfgL, YfiO, NlpB, and SmpA exist in a heterooligomeric complex with YaeT and coordinate the overall OM assembly process (28, 47, 51). Study of these lipoproteins, with the exception of OmlA, has not been reported in P. aeruginosa, yet some of their homologs exist: YfgL is 32% identical to PA3800, YfiO is 41% identical to PA4545 (ComL), and SmpA is 40% identical to OmlA (47). OmlA has already been characterized and plays a nonessential role in OM assembly in P. aeruginosa (34). While a mutant lacking OmlA is more susceptible to only a few antibiotics, PS186 was more susceptible to all antibiotics tested (Table 2), indicating that Opr86 plays a central role in OM assembly with OmlA playing only a supporting role in this process.
The larger goal of this study was to determine whether or not Opr86 was an antigen for a protective vaccine to inhibit biofilm formation by several different P. aeruginosa strains. In order to increase the potential for a broad effect on clinical strains of P. aeruginosa, attention was focused on the region known as o-Opr86, which includes an extracellular domain, for an antigen peptide. The immunolabeling and microscopy studies suggested that the antiserum against o-Opr86 could be applied for suppression of biofilm formation. It was observed that biofilm formation was inhibited completely by treatment with both anti-PAO1 and anti-o-Opr86 while planktonic growth was not affected. Inspection of initial attachment showed that the cells treated with anti-PAO1 or anti-o-Opr86 could not initiate surface association, in contrast to cells treated with the control IgG. It is speculated that the attachment of anti-PAO1 and anti-o-Opr86 to the OMs of cells inhibits the first stage of biofilm development; however, the exact mechanism for this inhibition is unknown.
It was also confirmed that anti-o-Opr86 inhibited biofilm formation by two clinical Pseudomonas isolates, whereas biofilm formation by strain 443 was not affected by treatment with either anti-PAO1 or anti-o-Opr86. One possibility to explain the lack of biofilm inhibition observed for strain 443 is that the amino acid sequence of the extracellular portion of Opr86 in strain 443 differs from those of the other strains and this extracellular portion may be a target for anti-o-Opr86 attachment to Opr86 on the cell surface. Sequencing of the o-Opr86 region of strain 443 did reveal differences in this region relative to the PAO1 reference strain. Furthermore, the possibility that other OMPs were not identical for PAO1 and strain 443 might explain the inability of anti-PAO1 to inhibit biofilm formation by strain 443. Another possibility for the lack of biofilm inhibition by anti-PAO1 and anti-o-Opr86 is that the cell surface of strain 443 is altered in some way, possibly by modification of cell surface proteins such as glycosylation or acetylation of OMPs, and that these alterations prevent biofilm inhibition.
This work has shown that, in P. aeruginosa PAO1, Opr86 is essential for viability and plays a key role in assembly of OMPs. In addition, the antibody against Opr86 can inhibit biofilm formation. In conclusion, Opr86 of P. aeruginosa would be a good target for design of both new antibiotics and vaccine.
Acknowledgments
We thank Sherry L. Kuchma (Dartmouth Medical School) and Philip S. Stewart and Betsey Pitts (Montana State University) for helpful comments on the manuscript and Noriko Saito (National Institute of Infectious Diseases) for the technical work of ultrastructure analysis.
This work was partially supported by a grant to N.N. from Industrial Technology Research Grant Program '04 and '05 of the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Baumann, U., E. Mansouri, and B. von Specht. 2004. Recombinant OprF-OprI as a vaccine against Pseudomonas aeruginosa infections. Vaccine 22840-847. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A., and R. Hancock. 1989. Outer membrane protein H1 of Pseudomonas aeruginosa: purification of the protein and cloning and nucleotide sequence of the gene. J. Bacteriol. 1713211-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Caiazza, N., and G. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 1864476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron, C., S. Lukehart, C. Castro, B. Molini, C. Godornes, and W. Van Voorhis. 2000. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J. Infect. Dis. 1811401-1413. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J., P. Stewart, and E. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Doerrler, W., and C. Raetz. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 28027679-27687. [DOI] [PubMed] [Google Scholar]
- 8.Drenkard, E., and F. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416740-743. [DOI] [PubMed] [Google Scholar]
- 9.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51675-690. [DOI] [PubMed] [Google Scholar]
- 10.Fürste, J., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 11.Genevrois, S., L. Steeghs, P. Roholl, J. Letesson, and P. van der Ley. 2003. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 221780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilleland, H. J., L. Gilleland, and J. Matthews-Greer. 1988. Outer membrane protein F preparation of Pseudomonas aeruginosa as a vaccine against chronic pulmonary infection with heterologous immunotype strains in a rat model. Infect. Immun. 561017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinnebusch, B., R. Perry, and T. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273367-370. [DOI] [PubMed] [Google Scholar]
- 14.Høiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 323-35. [DOI] [PubMed] [Google Scholar]
- 15.Holloway, B., V. Krishnapillai, and A. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 4373-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, H., R. Siehnel, F. Bellido, E. Rawling, and R. Hancock. 1992. Analysis of two gene regions involved in the expression of the imipenem-specific, outer membrane porin protein OprD of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 76267-273. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S., J. Malinverni, P. Sliz, T. Silhavy, S. Harrison, and D. Kahne. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317961-964. [DOI] [PubMed] [Google Scholar]
- 18.Kjelleberg, S., and S. Molin. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5254-258. [DOI] [PubMed] [Google Scholar]
- 19.Klausen, M., M. Gjermansen, J. Kreft, and T. Tolker-Nielsen. 2006. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 2611-11. [DOI] [PubMed] [Google Scholar]
- 20.Kosono, S., K. Asai, Y. Sadaie, and T. Kudo. 2004. Altered gene expression in the transition phase by disruption of a Na+/H+ antiporter gene (shaA) in Bacillus subtilis. FEMS Microbiol. Lett. 23293-99. [DOI] [PubMed] [Google Scholar]
- 21.Larbig, M., E. Mansouri, J. Freihorst, B. Tümmler, G. Köhler, H. Domdey, B. Knapp, K. Hungerer, E. Hundt, J. Gabelsberger, and B. von Specht. 2001. Safety and immunogenicity of an intranasal Pseudomonas aeruginosa hybrid outer membrane protein F-I vaccine in human volunteers. Vaccine 192291-2297. [DOI] [PubMed] [Google Scholar]
- 22.Lee, N., B. Ahn, S. Jung, Y. Kim, H. Kim, and W. Park. 2000. Conformation-dependent antibody response to Pseudomonas aeruginosa outer membrane proteins induced by immunization in humans. FEMS Immunol. Med. Microbiol. 2779-85. [DOI] [PubMed] [Google Scholar]
- 23.Lee, N., B. Ahn, S. Jung, Y. Kim, Y. Lee, H. Kim, and W. Park. 1999. Human anti-Pseudomonas aeruginosa outer membrane proteins IgG cross-protective against infection with heterologous immunotype strains of P. aeruginosa. FEMS Immunol. Med. Microbiol. 25339-347. [DOI] [PubMed] [Google Scholar]
- 24.Lee, N., S. Jung, B. Ahn, Y. Kim, J. Kim, D. Kim, I. Kim, S. Yoon, S. Nam, H. Kim, and W. Park. 2000. Immunization of burn-patients with a Pseudomonas aeruginosa outer membrane protein vaccine elicits antibodies with protective efficacy. Vaccine 181952-1961. [DOI] [PubMed] [Google Scholar]
- 25.Loosmore, S., Y. Yang, D. Coleman, J. Shortreed, D. England, and M. Klein. 1997. Outer membrane protein D15 is conserved among Haemophilus influenzae species and may represent a universal protective antigen against invasive disease. Infect. Immun. 653701-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyczak, J., C. Cannon, and G. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 21051-1060. [DOI] [PubMed] [Google Scholar]
- 27.Mah, T., and G. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 934-39. [DOI] [PubMed] [Google Scholar]
- 28.Malinverni, J., J. Werner, S. Kim, J. Sklar, D. Kahne, R. Misra, and T. Silhavy. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61151-164. [DOI] [PubMed] [Google Scholar]
- 29.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192107-112. [DOI] [PubMed] [Google Scholar]
- 30.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 481320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBroom, A., and M. Kuehn. 2007. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63545-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochsner, U., A. Vasil, Z. Johnson, and M. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 1811099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto, K., N. Gotoh, H. Tsujimoto, H. Yamada, E. Yoshihara, T. Nakae, and T. Nishino. 1999. Molecular cloning and characterization of the oprQ gene coding for outer membrane protein OprE3 of Pseudomonas aeruginosa. Microbiol. Immunol. 43297-301. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 37.Pardee, A., F. Jacob, and F. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1165-178. [Google Scholar]
- 38.Price, B., D. Galloway, N. Baker, L. Gilleland, J. Staczek, and H. J. Gilleland. 2001. Protection against Pseudomonas aeruginosa chronic lung infection in mice by genetic immunization against outer membrane protein F (OprF) of P. aeruginosa. Infect. Immun. 693510-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawling, E., F. Brinkman, and R. Hancock. 1998. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 1803556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robert, V., E. Volokhina, F. Senf, M. Bos, P. Van Gelder, and J. Tommassen. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffolo, C., J. Tennent, W. Michalski, and B. Adler. 1997. Identification, purification, and characterization of the type 4 fimbriae of Pasteurella multocida. Infect. Immun. 65339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 43.Schleiff, E., and J. Soll. 2005. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 61023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer, H. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 248-62. [PubMed] [Google Scholar]
- 45.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118640-659. [DOI] [PubMed] [Google Scholar]
- 46.Singh, P., A. Schaefer, M. Parsek, T. Moninger, M. Welsh, and E. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 47.Sklar, J., T. Wu, L. Gronenberg, J. Malinverni, D. Kahne, and T. Silhavy. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 1046400-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Specht, B., H. Lücking, B. Blum, A. Schmitt, K. Hungerer, and H. Domdey. 1996. Safety and immunogenicity of a Pseudomonas aeruginosa outer membrane protein I vaccine in human volunteers. Vaccine 141111-1117. [DOI] [PubMed] [Google Scholar]
- 49.Voulhoux, R., M. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299262-265. [DOI] [PubMed] [Google Scholar]
- 50.Werner, J., and R. Misra. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 571450-1459. [DOI] [PubMed] [Google Scholar]
- 51.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121235-245. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Y., W. Thomas, P. Chong, S. Loosmore, and M. Klein. 1998. A 20-kilodalton N-terminal fragment of the D15 protein contains a protective epitope(s) against Haemophilus influenzae type a and type b. Infect. Immun. 663349-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon, S., R. Hennigan, G. Hilliard, U. Ochsner, K. Parvatiyar, M. Kamani, H. Allen, T. DeKievit, P. Gardner, U. Schwab, J. Rowe, B. Iglewski, T. McDermott, R. Mason, D. Wozniak, R. Hancock, M. Parsek, T. Noah, R. Boucher, and D. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]
- 54.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]