Abstract
Under anaerobic growth conditions, an active pyruvate dehydrogenase (PDH) is expected to create a redox imbalance in wild-type Escherichia coli due to increased production of NADH (>2 NADH molecules/glucose molecule) that could lead to growth inhibition. However, the additional NADH produced by PDH can be used for conversion of acetyl coenzyme A into reduced fermentation products, like alcohols, during metabolic engineering of the bacterium. E. coli mutants that produced ethanol as the main fermentation product were recently isolated as derivatives of an ldhA pflB double mutant. In all six mutants tested, the mutation was in the lpd gene encoding dihydrolipoamide dehydrogenase (LPD), a component of PDH. Three of the LPD mutants carried an H322Y mutation (lpd102), while the other mutants carried an E354K mutation (lpd101). Genetic and physiological analysis revealed that the mutation in either allele supported anaerobic growth and homoethanol fermentation in an ldhA pflB double mutant. Enzyme kinetic studies revealed that the LPD(E354K) enzyme was significantly less sensitive to NADH inhibition than the native LPD. This reduced NADH sensitivity of the mutated LPD was translated into lower sensitivity of the appropriate PDH complex to NADH inhibition. The mutated forms of the PDH had a 10-fold-higher Ki for NADH than the native PDH. The lower sensitivity of PDH to NADH inhibition apparently increased PDH activity in anaerobic E. coli cultures and created the new ethanologenic fermentation pathway in this bacterium. Analogous mutations in the LPD of other bacteria may also significantly influence the growth and physiology of the organisms in a similar fashion.
Escherichia coli, a facultative heterotroph, grows under aerobic and anaerobic conditions. During aerobic growth, this bacterium metabolizes glucose through the reactions of glycolysis, pyruvate dehydrogenase (PDH), and the tricarboxylic acid cycle. The NADH generated during these enzyme-catalyzed reactions is oxidized ultimately by oxygen. Under anaerobic conditions and in the absence of external electron acceptors, organic compounds generated from glucose during glycolysis serve as the electron acceptors to maintain the redox balance and continued growth of the bacterium. Due to the differences in electron acceptors between the two growth modes, the reported [NADH]/[NAD+] ratio of an anaerobic cell is severalfold higher (about 0.75) than that of an aerobic cell (about 0.03) (13, 33).
The PDH complex that connects glycolysis and tricarboxylic acid cycle enzymes is composed of multiple subunits of three enzymes, pyruvate decarboxylase (dehydrogenase; enzyme 1 [E1]; EC 1.2.4.1), dihydrolipoamide acetyltransferase (enzyme 2 [E2]; EC 2.3.1.12), and dihydrolipoamide dehydrogenase (LPD) (enzyme 3 [E3]; EC 1.8.1.4) (14). NADH, a product of the PDH reaction, is a competitive inhibitor of the PDH complex (15, 30, 31). The NADH sensitivity of the PDH complex has been demonstrated to reside in LPD, the enzyme that interacts with NAD+ as a substrate (29, 30, 38). Although PDH is critical for aerobic growth of the bacterium, this activity was also detectable in cell extracts of E. coli grown under anaerobic conditions (13, 32, 33). However, based on the product profile, the PDH activity in vivo in anaerobic E. coli cultures is either very low or undetectable (33).
In an anaerobically growing E. coli strain lacking PDH activity, pyruvate is metabolized by an alternative enzyme, pyruvate-formate lyase, to acetyl coenzyme A (acetyl-CoA) with conservation of the reductant as formate (9). Formate is ultimately removed as H2 and CO2 without influencing the [NADH]/[NAD+] ratio of the cell (28). In order to maintain the redox balance, the NADH generated during the oxidation of glyceraldehyde-3-phosphate in the glycolysis pathway is oxidized using acetyl-CoA as the electron acceptor, with the production of ethanol (9). However, reduction of acetyl-CoA to ethanol by alcohol dehydrogenase requires two NADH molecules for each acetyl-CoA molecule, a demand that is not met by fermenting E. coli. Due to this constraint, the fermentation profile of a growing E. coli strain includes equimolar quantities of ethanol and acetate.
We recently isolated and described E. coli mutants that produced ethanol as the main fermentation product (19). The mutation in one of these mutants was mapped in the genes of the pdh locus (pdhR, aceEF, and lpd). Based on the phenotype and genetic analysis, it was inferred that PDH, the enzyme that is normally inactive in an anaerobic E. coli cell, plays a pivotal role in ethanol production by this mutant. Conversion of glucose to two acetyl-CoA molecules by the glycolytic enzymes and PDH would yield four NADH molecules per glucose molecule, and these four NADH molecules can be oxidized using the two acetyl-CoA molecules as the electron acceptors and alcohol dehydrogenase as the catalyst, with production of two equivalents of ethanol. For PDH to be active in an anaerobic cell, the LPD component of the PDH complex is expected to have lost at least part of its sensitivity to NADH inhibition.
Based on the DNA sequence, we localized the mutations in the ethanologenic E. coli mutants to a single change in the LPD amino acid sequence. The results presented in this paper show that the PDH from two such ethanologenic mutants, strains SE2377 and SE2378, are less sensitive to NADH inhibition. The alteration of the LPD and PDH complex to reduced sensitivity to NADH inhibition apparently allowed the enzyme to function in an anaerobic E. coli culture, which changed the fermentation profile of the mutant.
MATERIALS AND METHODS
Materials.
Biochemicals were purchased from Sigma-Aldrich. Organic and inorganic chemicals were purchased from Fisher Scientific and were analytical grade. DNA restriction endonucleases, T4 DNA ligase, DNA polymerases, and other DNA modification enzymes and reagents were obtained from New England Biolabs Inc., Invitrogen, or Clontech Laboratories. The quantitative reverse transcription (RT)-PCR reagent was obtained from Bio-Rad Laboratories. Oligonucleotide primers were synthesized by Invitrogen or Sigma-Genosys.
Bacterial strains, bacteriophages, and plasmids.
The bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1. All E. coli strains are derivatives of strain K-12.
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| W3110 | Wild type | ATCC |
| AH241 | W3110, ΔldhA | 19 |
| AH242 | W3110, ΔldhA Δ(focA-pflB)-Km | 19 |
| BW25113 | lacIqrrnBT14 ΔlacZW116hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | B. Wanner |
| JM109(λDE3) | F′ [traD36 proA+B+ lacIq Δ(lacZ)M15] Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 (λDE3) | Promega |
| SE2377 | AH242, lpd102(H322Y) | This study |
| SE2378 | AH242, lpd101(E354K) | 19 |
| SE2382 | AH242, lpd101(E354K) | This study |
| SE2383 | AH242, lpd102(H322Y) | This study |
| SE2384 | AH242, lpd102(H322Y) | This study |
| SE2385 | AH242, lpd101(E354K) | This study |
| YK1 | SE2378, Kms | This study |
| YK29 | AH242, Kms | This study |
| YK87 | YK1, ΔadhE-Km | This study |
| YK100 | AH242, Δlpd | This study |
| YK110 | YK100, lpd+ | YK100 × P1(W3110) |
| YK111 | YK100, lpd101 | YK100 × P1(SE2378) |
| YK128 | YK100, Plpd+ (pKY32) | This study |
| YK129 | YK100, Plpd101 (pKY33) | This study |
| YK139 | YK100, lpd102 | YK100 × P1(SE2377) |
| YK141 | YK100, lpd101 | YK100 × P1(SE2382) |
| YK175 | AH241, ΔadhE-Km | AH241 × P1(YK87) |
| YK176 | YK141, ΔadhE-Km | YK141 × P1(YK87) |
| YK181 | YK139, ΔadhE-Km | YK139 × P1(YK87) |
| Plasmids | ||
| pKY10 | pUC19, PpdhRSE2378 | This study |
| pKY13 | pUC19, PpdhR+ | This study |
| pKY15 | pTL61t, PpdhSE2378-lacZ | This study |
| pKY17 | pTL61t, PpdhW3110-lacZ | This study |
| pKY32 | pTrc99a, lpd+bla | This study |
| pKY33 | pTrc99a, lpd101 bla | This study |
| pKY36 | pET15b, lpd+bla | This study |
| pKY37 | pET15b, lpd101 bla | This study |
| pKY38 | pET15b, lpd102 bla | This study |
| Phages | ||
| λYK1 | λ(PpdhW3110-lacZ) | This study |
| λYK2 | λ(PpdhSE2378-lacZ) | This study |
Media and growth conditions.
L broth (LB), used as the rich medium, and mineral salts medium were prepared as described previously (21). After the medium was autoclaved, sugars were added at final concentrations of 3 g/liter for aerobic growth and 10 g/liter for anaerobic growth. The media used for propagation of phages P1 and λ, as well as transduction, were prepared as described by Miller (25). Batch fermentation without pH control was carried out in screw-cap tubes (13 by 100 mm) filled to the top with the appropriate medium (19). The inoculum (1%, vol/vol) for the fermentations was grown aerobically for about 16 h. Antibiotics were added, as needed, at initial concentrations of 100 mg/liter of ampicillin and 50 mg/liter of kanamycin.
Genetic methods.
Gene deletions in E. coli were constructed as described by Datsenko and Wanner (11). Appropriate genes were amplified by PCR and cloned into plasmid pCR2.1-TOPO (Invitrogen). After deletion of part of the gene, a DNA cassette containing a kanamycin resistance gene flanked by FRT sites was integrated into the deleted area. The antibiotic resistance gene with the flanking E. coli DNA was PCR amplified, and the PCR product was transformed into E. coli strain BW25113(pKD46) that was pregrown in LB containing arabinose as described previously (11). Transformants with the gene deletion were selected and verified by PCR. The deletion mutation was transduced by phage P1 to other genetic backgrounds before use. All molecular biology experiments were performed as described previously (28).
In vitro mutagenesis of lpd.
The lpd gene in plasmid pKY32 was mutagenized by using either hydroxylamine or error-prone PCR. Hydroxylamine mutagenesis was performed as described by Davis et al. (12). Error-prone PCR was conducted as described previously (24) with the same primer set that was used for cloning the lpd gene into plasmid pET15b. In addition to the Taq polymerase buffer (New England Biolabs), the following ingredients were also added to increase the mutation rate: 0.8 mM dTTP, 0.8 mM dCTP, 4.8 mM MgCl2, and 0.5 mM MnCl2. PCR was performed using the following conditions with a Bio-Rad thermal cycler: 1 min at 95°C, followed by five cycles of 1 min at 95°C, 30 s at 45°C, and 2 min at 72°C, by 30 cycles of 1 min at 95°C, 30 s at 55°C, and 2 min at 72°C, and finally by 15 min at 72°C. The PCR product was purified, cloned into plasmid vector pTrc99a, and transformed into E. coli strain AH242. Transformants that grew anaerobically were selected, and the lpd gene in the plasmid was sequenced to identify the nature of the mutation.
Level of transcription of pdh operon.
The pdh operon promoter DNA was removed from either plasmid pKY13 (W3110) or plasmid pKY10 (SE2378) after hydrolysis with BlpI and AflII. The DNA fragment was treated with the Klenow fragment of DNA polymerase and cloned into the SmaI site of plasmid pTL61t (23) upstream of a promoterless lacZ gene. The plasmid constructs (pKY15 for SE2378 Ppdh-lac and pKY17 for W3110 Ppdh-lac) were selected after transformation of E. coli TOP10 (Invitrogen) as blue colonies on LB containing ampicillin with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml). The cloned pdh promoter DNA was sequenced to confirm the sequence. The Ppdh-lac fusion was transferred to λRZ5 and transduced into E. coli as described previously (28).
Lysogens carrying the λPpdh-lacZ fusion were cultured under aerobic (LB) or anaerobic (LB containing glucose) conditions to the mid- to late exponential phase of growth. The β-galactosidase activity of the cells was determined as described by Miller (25). The specific activity of β-galactosidase was expressed in nmol·min−1·mg cell protein−1.
Quantitative RT-PCR.
For isolation of total RNA, aerobic cultures were grown in 10 ml of LB in 250-ml flasks at 37°C with shaking at 200 rpm. Anaerobic cultures were grown in 9 ml of LB containing glucose in screw-cap tubes (13 by 100 mm) filled to the top. Cells were harvested at the early to mid-exponential phase of growth. Total RNA was extracted by the hot phenol method as described previously (34). Quantitative RT-PCR was performed as described previously (35).
LPD expression plasmids.
Different alleles of the lpd gene were cloned into plasmid pTrc99a and expressed from a lactose-regulated promoter for complementation experiments. For construction of the plasmids, DNA encoding a specific lpd allele was amplified by PCR from appropriate E. coli genomic DNA. The forward primer (GCGACCATGGAGAAGGAGATATACCATGAGTACT) contained an NcoI restriction site at the 5′ end (underlined), and the reverse primer (GCGAAAGCTTTTACTTCTTCTTCGCTTTCG) contained a HindIII restriction site at the 5′ end (underlined). A Shine-Dalgarno sequence (ribosomal binding site) was also located 7 nucleotides upstream of the start codon (ATG) in the forward primer. Both the PCR product and plasmid pTrc99A were hydrolyzed with restriction enzymes NcoI and HindIII and ligated to construct plasmids pKY32 and pKY33 containing lpd+ and the lpd101 allele, respectively.
For purification of LPD, the appropriate lpd allele was cloned into a phage T7-based expression vector. For construction of plasmids pKY36 (lpd+), pKY37 (lpd101), and pKY38 (lpd102), the appropriate lpd gene was amplified by PCR with the following primers: forward primer GAGCCTCGAGATGAGTACTGAAATC and reverse primer GCGTGGATCCTTACTTCTTCTTCG. The forward primer contained an XhoI restriction site at the 5′ end (underlined), and the reverse primer had a BamHI restriction site at the 5′ end (underlined). The PCR products, digested with XhoI and BamHI, were ligated with plasmid pET-15b also digested with XhoI and BamHI. E. coli TOP10 cells were transformed with the ligation product, and the transformants were selected for resistance to ampicillin on LB containing ampicillin. The insert sequences in the plasmids were verified by sequencing the lpd gene.
Purification of LPD.
For purification of LPD, the enzyme was produced in strain JM109(λDE3) transformed with plasmid pKY36, pKY37, or pKY38. A 500-ml culture in LB containing ampicillin in a 2.8-liter Fernbach flask was grown at 37°C with shaking at 250 rpm to an optical density at 420 nm of 0.6 (Beckman DU640 spectrophotometer). Arabinose (1.5%) was added to the culture to induce the T7 RNA polymerase (26). After 4 h of incubation at room temperature with shaking, cells were harvested by centrifugation (10,000 × g, 10 min, 4°C), washed twice with 25 ml of 50 mM potassium phosphate buffer (pH 8.0) (referred to as phosphate buffer below), and resuspended in 5 ml of the same buffer. All operations were conducted at 4°C. Cells were passed through a French pressure cell at 20,000 lb/in2. The crude extract was clarified by centrifugation (30,000 × g, 45 min), and the supernatant was filtered through a 0.22-μm filter. The filtered protein solution was loaded onto a HiTrap chelating column (5 ml; General Electric) that was prewashed with 0.1 M NiCl2 in the same buffer. Unadsorbed and loosely bound proteins were removed from the column by washing with 5 column volumes of phosphate buffer, followed by 5 column volumes of phosphate buffer with 50 mM imidazole. His-tagged LPD protein was eluted with a 50 mM to 0.5 M imidazole gradient in phosphate buffer. All the fractions containing LPD activity were combined. The N-terminal His tag was cleaved off the protein by incubation with thrombin (150 U; General Electric) at 4°C overnight. Thrombin and the small peptide were removed by gel filtration through a Sephacryl S-200 HR column (2.6 by 60 cm; General Electric) that was preequilibrated with phosphate buffer with 0.1 M NaCl. The protein was eluted with phosphate buffer containing 0.1 M NaCl. All the fractions with LPD activity were combined and dialyzed against phosphate buffer. The purity of the protein was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Both the native LPD protein and the mutated forms of LPD were purified by the same method. Since the purified LPD from strain SE2377, encoded by the lpd102 allele, did not have detectable activity, purification of this LPD allele was followed by SDS-PAGE.
Purification of PDH complex.
The PDH complex was purified as described by Bisswanger, with minor modifications (3), from strains YK175 (native protein), YK176 [lpd101 allele; LPD(E354K)], and YK181 [lpd102 allele; LPD(H322Y)]. Cells were cultured in 6 liters of glucose-mineral salts medium (1 liter per 2.8-liter Fernbach flask). When the culture reached an optical density at 420 nm of about 2.0 (Beckman DU640; late exponential phase of growth), cells were harvested by centrifugation (10,000 × g, 10 min, 4°C), washed with 100 ml of phosphate buffer, and resuspended in 20 ml of phosphate buffer. Cells were lysed by passage through a French pressure cell (20,000 lb/in2) in the presence of a protease inhibitor cocktail (5 ml/20 g [wet weight] of cells; Sigma). DNase I and RNase A were each added to the extract in a centrifuge tube at a concentration of 100 μg/ml and incubated at 37°C for 1 h with gentle mixing to reduce the viscosity. All operations after this step were performed at 4°C. The cell extract was centrifuged at 12,000 × g for 30 min to remove cell debris. The supernatant was then centrifuged at 150,000 × g for 4 h to sediment the PDH complex. The supernatant was immediately decanted, and the pellet was dissolved in 6.0 ml of phosphate buffer for 2 h with gentle mixing on a rocker. The protein solution was centrifuged again at 12,000 × g for 15 min to remove particulates that did not dissolve. The supernatant was chromatographed through a hydroxyapatite column (1.5 by 12.0 cm; Bio-Rad) that was equilibrated with phosphate buffer. The protein was eluted from the column with a linear 50 to 500 mM phosphate gradient in phosphate buffer at pH 8.0. Fractions with PDH activity were combined, dialyzed against phosphate buffer, and concentrated. The concentrated protein solution was further purified with a gel filtration column (Sephacryl S-500HR; 2.6 by 35 cm) with phosphate buffer as the eluent. Fractions with activity were pooled and used immediately for the enzyme assay.
Enzyme activity.
LPD activity was assayed as described previously (37). The standard reaction mixture (1.0 ml) for the forward reaction contained 0.1 M KH2PO4 (pH 8.0), 3 mM NAD+, 3 mM dl-dihydrolipoic acid, 1.5 mM EDTA, and the appropriate amount of enzyme. One unit of enzyme activity was defined as the production of 1 μmol NADH·min−1·mg protein−1. The standard reverse reaction mixture (1.0 ml) contained 0.1 M KH2PO4 (pH 8.0), 0.1 mM NAD+, 0.1 mM NADH, 3 mM dl-lipoamide, and 1.5 mM EDTA. Enzyme assays were performed at room temperature, and the rate of NADH oxidation was monitored over time. One unit of enzyme activity was defined as the oxidation of 1 μmol NADH·min−1·mg protein−1.
PDH was assayed both in crude extracts and using purified protein. A standard assay for determination of the activity of the PDH complex in crude extract was based on pyruvate-dependent reduction of NADH at 340 nm (ɛ = 6,220 Μ−1 cm−1) at room temperature, as described by Hinman and Blass (17). Each 1-ml reaction mixture contained thiamine pyrophosphate (0.2 mM), CoA (0.1 mM), MgCl2·6H2O (1 mM), dithiothreitol (0.3 mM), NAD+ (2.5 mM), bovine serum albumin (100 μg/ml), and crude extract or purified protein in 50 mM potassium phosphate buffer (pH 8.0). The reaction was started by addition of pyruvate (5 mM). Enzyme activity was expressed in μmol NADH produced·min−1·mg protein−1. The effect of NADH on enzyme activity was determined using the same reaction mixture with addition of various concentrations of NADH.
PDH activity in the crude extracts was also measured by using a partial reaction catalyzed by the pyruvate decarboxylase/dehydrogenase (E1 activity) in crude extracts (13). One milliliter of reaction mixture contained phosphate buffer (50 mM, pH 7.0), MgCl2·6H2O (12.5 mM), thiamine pyrophosphate (0.18 mM), CoA (0.175 mM), NAD+ (2.0 mM), potassium ferricyanide (1.0 mM), pyruvate (5.0 mM), and crude extract. The reaction was initiated by addition of pyruvate. The rate of reduction of ferricyanide was monitored over time at 430 nm (ɛ = 1,030 Μ−1 cm−1). One unit of enzyme activity was defined as the reduction of 1 μmol ferricyanide·min−1· mg protein−1.
The reported results are data from a typical experiment that was repeated at least three times, and the variation in the experimental results was less than 10%. Kinetic properties of both LPD and PDH were determined as described by Cornish-Bowden (10) using the initial linear rates of the reactions.
Analytical methods.
The NAD+ concentration was determined using an Enzychrom NAD+/NADH assay kit (Bioassay Systems, Hayward, CA). Sugars and fermentation products were analyzed by high-performance liquid chromatography as described previously (19). The nucleotide sequence of DNA was determined by the Interdisciplinary Center for Biotechnology Research DNA sequencing core facility at the University of Florida. Protein concentrations were determined using Coomassie blue G-250 as described by Bradford (5) with bovine serum albumin as the standard. SDS-PAGE was performed with 12.5% polyacrylamide gels as described by Laemmli (20).
RESULTS AND DISCUSSION
An E. coli mutant (strain AH242) that lacks pyruvate-formate lyase (pflB) and fermentative lactate dehydrogenase (ldhA) activities is defective for anaerobic growth (9, 19). We derived a set of mutants (e.g., strain SE2378) of strain AH242 that grew anaerobically and produced ethanol as the fermentation product (19). The mutation in strain SE2378 was mapped in or near the genes that encode the components of the PDH complex (pdh operon; pdhR, aceE, aceF, and lpd). Since the expected [NADH]/[NAD+] ratio in anaerobic cells is higher than that in aerobic E. coli cells (13), operation of this new ethanol production pathway in strain SE2378 that includes glycolysis, PDH, and alcohol dehydrogenase suggests that the activity of the PDH complex is less sensitive or insensitive to inhibition by NADH, in addition to its optimal expression.
Expression of PDH in an anaerobic culture of E. coli.
The results presented in Table 2 show that the relative mRNA level of the aceE gene (the second gene in the pdh operon) encoding the E1 enzyme of the PDH complex is independent of the presence of O2 during growth of E. coli strain W3110. Since the pdhR, aceE, aceF, and lpd genes are transcribed as one transcript (27), it is apparent that all the genes encoding the PDH complex are transcribed under both aerobic and anaerobic growth conditions. Although the aceE mRNA level of strain SE2378 was also not altered by the level of O2 during growth, this mRNA level was slightly lower than that of the wild-type strain W3110. The fact that this mRNA is further translated in cells grown under aerobic and anaerobic conditions is shown by the PdhR-lac fusion-based production of β-galactosidase activity and the presence of PDH activity in extracts (Table 2). In wild-type strain W3110, the level of PDH activity of anaerobic cells was about 50% of the level of activity of aerobic cells, while the levels of transcription in the two growth conditions were about the same. It is possible that the NADH-inhibited protein was subject to proteolysis in the anaerobic cells. Although the level of PDH activity in strain SE2378 grown under aerobic conditions was only about 65% of the level of activity in aerobically cultured strain W3110, this level of activity was not that dissimilar from the level in either strain grown anaerobically. These results show that both the wild-type and mutant E. coli strains produced PDH during anaerobic growth. The inability of strain AH242 (an ldhA pflB derivative of strain W3110) to grow under anaerobic conditions suggests that the PDH activity is negligible during anaerobic growth of strain W3110 due to inhibition of the enzyme complex by NADH. Similar conclusions were also reached by Snoep et al. based on the fermentation profile of E. coli B (33).
TABLE 2.
PDH mRNA, transcription, and protein levels in aerobically and anaerobically grown E. coli wild-type strain W3110 and ethanologenic mutant strain SE2378
| Strain | Relative mRNA levela
|
β-Galactosidase activityb
|
PDH activityc
|
|||
|---|---|---|---|---|---|---|
| With O2 | Without O2 | With O2 | Without O2 | With O2 | Without O2 | |
| W3110 | 1.00 | 0.98 | 600 | 630 | 370 | 185 |
| SE2378 | 0.71 | 0.77 | 570 | 680 | 240 | 200 |
Relative mRNA levels were determined by quantitative RT-PCR. The level of aceE mRNA in wild-type strain W3110 grown under aerobic conditions was defined as 1.0, and the relative levels of aceE mRNA for the other growth conditions and strain SE2378 were determined.
β-Galactosidase activities of Ppdh-lacZ fusions using λYK1 and λYK2 at λatt are expressed in nmol·min−1·mg protein−1.
The PDH activity is the PDH (E1) activity of the PDH complex and is expressed in nmol ferricyanide reduced·min−1·mg protein−1. See Materials and Methods for other details.
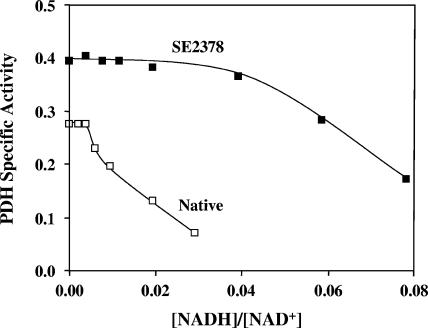
PDH complex is less sensitive to NADH inhibition in strain SE2378.
For the PDH to be active under anaerobic growth conditions, the mutated form of the enzyme in strain SE2378 needs to be significantly less sensitive to NADH inhibition than the native enzyme. When the PDH activity of strain W3110 was assayed in crude extracts, the enzyme was completely inhibited by NADH at a concentration of about 80 μM, corresponding to an [NADH]/[NAD+] ratio of about 0.04. At this ratio of [NADH] to [NAD+], the PDH complex in the crude extract of strain SE2378 was fully active (Fig. 1). Increasing the NADH concentration to obtain a higher ratio did inhibit the enzyme from the mutant, indicating that the mutation did not completely mitigate the NADH inhibition. Taken together, the results presented in Table 2 and Fig. 1 show that the observed anaerobic growth phenotype of strain SE2378 is apparently due to the mutational alteration of the PDH activity.
FIG. 1.
NADH sensitivity of the PDH complex from E. coli wild-type strain W3110 (Native) and an ethanologenic mutant, strain SE2378. Crude extracts were used to assay for PDH complex activity (NADH production) as described in Materials and Methods. The NAD+ concentration in the assay mixture (2 mM for the native enzyme and 1 mM for the enzyme from strain SE2378) was at least five times the Km value for each enzyme. The NADH concentration was varied to obtain the indicated ratios. Specific activity is expressed in μmol min−1 mg protein−1.
Identification of the mutation in lpd.
To localize the mutation that contributed to the reduced sensitivity of PDH to NADH inhibition, the genes comprising the entire pdh operon from six independently isolated ethanologenic E. coli mutants were sequenced. Based on the DNA sequence, a single mutation in lpd was identified in each of the six mutants (Fig. 2). Three mutants, strains SE2377, SE2383, and SE2384, had a base change from C to T at position 964 (A in the ATG codon of the LPD gene was defined as position 1; lpd102), resulting in a change of the histidine at position 322 to tyrosine (Fig. 2). The other three mutants, strains SE2378, SE2382, and SE2385, had a different mutation (G to A at position 1,060; lpd101), resulting in a change of the glutamate at position 354 to lysine (Fig. 2). Only one of the six mutants, strain SE2378, also carried two mutations in the pdhR gene (base change T34C leading to amino acid change S12P; insertion of a TGC sequence between positions 352 and 353, resulting in insertion of leucine between amino acids 117 and 118 in the native protein) besides the E354K change in the LPD protein. In addition, a G-to-A base change upstream of the aceE gene at position −59 from the translational start (A of ATG) was also observed in strain SE2378. These secondary mutations in strain SE2378 may have altered the level of expression of the PDH complex in the cell since the PdhR protein is a known regulator of the pdh operon (16). The mutations in LPD are in agreement with the observed lower sensitivity of the PDH complex in cell extracts to NADH inhibition since the LPD enzyme is the known site of NADH inhibition (29, 30, 38).
FIG. 2.
Amino acid sequence alignment of LPD from E. coli wild-type (wt) strain W3110 and various ethanologenic mutants. A period indicates that the amino acid is same as the amino acid in the wild type. All the other amino acids that are not listed are the same in the LPD from the six mutants and strain W3110.
To confirm that the observed phenotype of anaerobic growth is due only to the lpd mutation and is not associated with a second mutation in an unlinked gene, the following genetic experiment was performed. In the first step, the lpd gene of the ldhA pflB double mutant strain AH242, the parent strain used in construction of the ethanologenic derivatives, was deleted (strain YK100). Strain YK100 is defective for anaerobic growth due to the ldhA and pflB mutations and defective for aerobic growth in glucose-mineral salts medium due to the absence of PDH activity (lpd deletion). Wild-type and lpd101 alleles were either transduced into strain YK100 or introduced via plasmids, and the derivatives with LPD activity were selected for growth under aerobic conditions in glucose-mineral salts medium (Table 3). Only the transductants carrying the lpd101 allele from either strain SE2378 or SE2382 grew anaerobically, while the transductants carrying the wild-type lpd+ gene did not grow anaerobically (Table 3). Strain SE2382 is distinguished from strain SE2378 by its lack of the additional mutations found in the pdh operon of strain SE2378. These results show that the mutation in lpd is responsible for the observed phenotype. Similar results were also obtained when the Δlpd mutation in strain YK100 was complemented with two alleles, lpd+ and lpd101 from a plasmid, expressed from a trc promoter with isopropyl-β-d-thiogalactopyranoside (IPTG). The complementation experiments confirmed that the mutation in the lpd gene alone supports anaerobic growth of an ldhA pflB double mutant. As expected, transductants of YK100 with the lpd102 allele (YK139) also grew anaerobically (data not shown), confirming that the two lpd mutations (lpd101 and lpd102) are interchangeable.
TABLE 3.
Anaerobic growth and fermentation profiles of E. coli strains with different lpd allelesa
| Strain | lpd allele | Anaerobic growth | Concn of fermentation products (mM)
|
||||
|---|---|---|---|---|---|---|---|
| Ethanol | Acetate | Formate | Succinate | Lactate | |||
| W3110 | lpd+ | + | 7.3 | 9.3 | 2.6 | 4.2 | 18.5 |
| SE2378 | lpd101 | + | 107.3 | 4.7 | 0.0 | 4.1 | 0.8 |
| YK110 | lpd+ | − | NG | NG | NG | NG | NG |
| YK111 | lpd101b | + | 125.8 | 2.2 | 0.0 | 5.0 | 0.3 |
| YK141 | lpd101b | + | 114.5 | 3.4 | 0.0 | 6.0 | 0.3 |
| YK128 | Plpd+ | − | NG | NG | NG | NG | NG |
| YK129 | Plpd101 | + | 68.0 | 3.6 | 0.0 | 11.9 | 0.8 |
See Table 1 for a description of strain construction. Anaerobic growth and fermentation were determined using LB containing glucose (1%) in batch fermentations without pH control. Glucose was not completely fermented by strain W3110 due to the accumulation of organic acids that lowered the culture pH to less than 5.0. See text for other details. NG, no growth.
The lpd101 allele in strains YK111 and YK141 was transduced from strains SE2378 and SE2382, respectively.
In order to isolate additional LPD mutations that also would support anaerobic growth of strain AH242 but may map at different parts of LPD, the lpd DNA was specifically mutagenized with hydroxylamine or by error-prone PCR. The PCR mutagenesis did not yield any lpd mutation that supported anaerobic growth of strain AH242 (from a total of about 4,000 transformants). Hydroxylamine treatment of lpd DNA did yield five additional mutants, and the mutation in all five was the same E354K mutation found in strain SE2378.
LPD activity of strain SE2378 is insensitive to NADH inhibition.
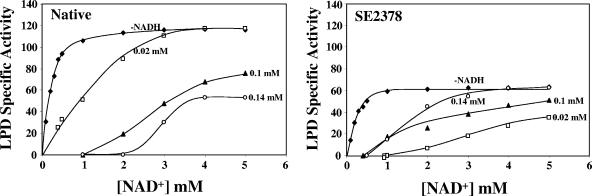
Since, the lpd mutation in all six ethanologenic strains supported anaerobic growth in the ldhA pflB genetic background, the LPD protein was purified from the wild type (native enzyme) and a representative strain SE2378 (mutated enzyme; E354K), and the kinetic properties were determined (Fig. 3 and 4).
FIG. 3.
NADH inhibition of LPD from E. coli wild-type strain W3110 (Native) and mutant strain SE2378. The rate of the forward reaction of LPD as a function of [NAD+] was determined without NADH and with the indicated concentrations of NADH. Specific activity is expressed in μmol min−1 mg protein−1.
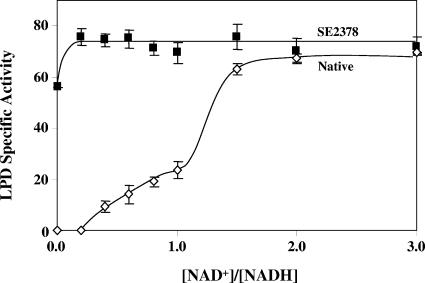
FIG. 4.
Insensitivity of the LPD (reverse reaction) from mutant strain SE2378 to NADH. The reverse reaction of LPD was determined with 0.1 mM NADH and different concentrations of NAD+ to obtain the indicated ratios. Specific activity is expressed in μmol min−1 mg protein−1. Native, wild-type strain.
The native enzyme presented typical Michelis-Menton-type kinetics with respect to NAD+. However, addition of NADH to the reaction mixture progressively inhibited the enzyme activity (Fig. 3). The specific activity of the mutated form of the enzyme was only about 50% of that of the native enzyme, and this altered enzyme was surprisingly more sensitive to inhibition at a lower NADH concentration (0.02 mM) than at higher concentrations of NADH. At an NAD+ concentration of 2 mM, 0.14 mM NADH completely inhibited the activity of the native enzyme, while at the same concentration of NAD+, 0.14 mM NADH inhibited the mutated form of the enzyme only by about 25%. At 4 mM NAD+, the mutated LPD was not inhibited by 0.14 mM NADH, while the native enzyme activity was only about 40% of the control activity without NADH. The Km for NAD+ (0.4 mM) was the same for both forms of the enzyme and is within the range of the reported Km values for the E. coli LPD that varied as a function of pH and NADH concentration (0.09 to 1.0 mM) (29). The Ki for NADH for the native enzyme was 5.2 μM, and this value is close to the value previously reported, 9 μM (30). Due to the complexity of NADH inhibition of the mutated LPD activity (Fig. 3), Ki was not determined for the LPD(E354K) enzyme.
The reverse reaction of native LPD, lipoamide + NADH → dihydrolipoamide + NAD+, depends on NAD+ as an activator (38). Reversal of NADH inhibition of the enzyme activity by NAD+ was biphasic. The first phase showed a gradual increase in activity to about 25 U when the added NAD+ concentration matched that of the substrate, NADH. The activity of the second phase of NAD+-dependent activation of the LPD reverse reaction was significantly higher, reaching the maximum value, about 70 U, when the NAD+ concentration was 1.5 times the concentration of NADH (Fig. 4). The E354K form of the enzyme had about 55 U of activity in the absence of added NAD+, which was about 75% of the maximum activity (about 75 U), which was reached with addition of a very small amount of NAD+ (20 μM). Complete activation of the native enzyme required at least 150 μM NAD+, an amount that was 7.5-fold larger than the amount needed for activation of the E354K form of the enzyme. These results further demonstrate that the sensitivity of LPD to inhibition by NADH is minimal in strain SE2378. An alternative possibility, the possibility that the E354K form of LPD carries a tightly bound NAD+ that overcomes the need for external addition of NAD+ to reverse NADH inhibition, can be ruled out due to the absence of detectable NAD+ in the purified protein (data not shown).
H322Y form of LPD is inactive upon purification.
Three of the mutants isolated as derivatives of strain AH242 that could grow anaerobically carried a mutation that changed the amino acid at position 322 from histidine to tyrosine (H322Y; lpd102) (Fig. 2). The lpd102 DNA was cloned into an expression vector, and the H322Y form of the LPD protein was expressed in recombinant E. coli. Although the cell extract had LPD activity, upon purification the protein lost the activity and associated flavin adenine dinucleotide (FAD) at the gel filtration step. Attempts to reconstitute the enzyme with added FAD as described by Lindsay et al. (22) failed to yield a protein with activity. This form of the enzyme was not studied further.
E. coli PDH complex with LPD mutation is less sensitive to NADH inhibition.
In order to confirm that the lower sensitivity of the LPD from strain SE2378 to NADH inhibition is translated to the entire PDH complex, the PDH complex was purified from the wild type and two mutants, strains SE2377 and SE2378, representing the two alleles. Kinetic properties of the PDH from these strains are presented in Table 4. The native form and the two mutated forms of the enzyme had similar affinities for the substrates, NAD+ and pyruvate. The Km values for pyruvate and NAD+ are within the range of Km values reported for the E. coli PDH complex (0.2 to 0.4 mM for pyruvate and 10 μM for NAD+) (4, 36). However, the native enzyme had a higher Vmax than the two mutated forms of the enzyme. Apparently, the lower specific activity of the LPD than of the native enzyme (Fig. 3) is translated to lower PDH activity. It is interesting that although the LPD with the H322Y mutation (strain SE2377) lost FAD upon purification, the PDH complex with the same mutated form of the LPD was active after purification.
TABLE 4.
Kinetic characteristics of the PDH complex isolated from E. coli wild-type strain W3110 and strains SE2377 (H322Y) and SE2378 (E354K) with a mutation in LPDa
| PDH complex | Km (NAD+) (mM) | Km (pyruvate) (mM) | Ki (NADH) (μM) | Vmax (μmol· min−1·mg protein−1) |
|---|---|---|---|---|
| Native | 0.07 | 0.40 | 1.00 | 46.64 |
| H322Y | 0.11 | 0.48 | 9.60 | 26.70 |
| E354K | 0.08 | 0.30 | 10.00 | 30.28 |
Enzymes were purified from E. coli strains YK175 (native protein), YK181 [LPD(H322Y)], and YK176 [LPD(E354K)]. See Table 1 for the genotypes of the strains.
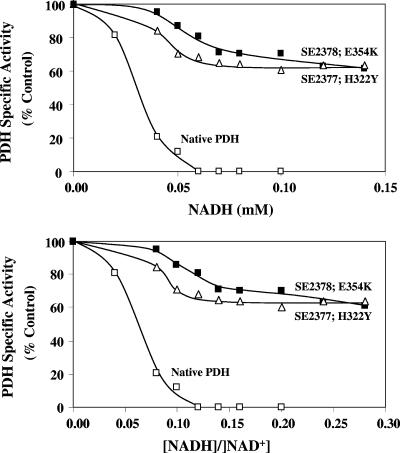
The two mutated forms of the PDH complex had about 10-fold-higher Ki values for NADH, in agreement with the observation that the PDH complex from the two mutants is less sensitive to inhibition by NADH (Table 4 and Fig. 5). The native PDH was completely inhibited at an NADH concentration of 0.06 mM, while the two mutated forms of PDH retained about 70 to 85% of the activity at the same NADH concentration. Even at a higher NADH concentration, the two mutated forms of PDH retained about 70% of the enzyme activity.
FIG. 5.
Differential inhibition of the PDH complex from wild-type E. coli strain W3110 (Native) and the two LPD mutant derivatives by NADH. The NAD+ concentration (0.5 mM) was at least five times the Km value. The specific activity of the native enzyme complex without NADH was 40.16 U. The specific activities of the two PDH complexes with LPD mutations H322Y and E354K without NADH were 17.74 and 31.00 U, respectively.
The intracellular [NADH]/[NAD+] ratio of E. coli grown under aerobic conditions was reported to be about 0.03 to 0.05 (8, 13, 33). At this ratio, the PDH complex retained almost 90% of the maximal activity. The [NADH]/[NAD+] ratio of an anaerobic cell is expected to be higher than that of an aerobic cell (0.7 to 0.8) (13, 33). Even at the lower reported ratio of 0.3, the PDH complex is not active in vitro (Fig. 5), supporting the inactive nature of the PDH complex in anaerobic E. coli. Although it is difficult to extrapolate from in vitro enzyme activity to in vivo levels, it should be noted that the PDH complex, although present, failed to support anaerobic growth of strain AH242 that lacked lactate dehydrogenase and pyruvate- formate lyase (19). The specific mutation, either H322Y or E354K, that made the LPD less sensitive to NADH inhibition also reduced the NADH sensitivity of the PDH complex and allowed the enzyme to support anaerobic growth of E. coli strain SE2378.
Histidine at position 322 was conserved in all LPD sequences tested, including the human LPD sequence, indicating the importance of this amino acid in the activity of LPD and PDH. Although the glutamate at position 354 was conserved in the LPD of all tested bacteria in the family Enterobacteriaceae, it is not conserved across bacterial families. However, a significant level of sequence identity can be found in this region; proline at position 348 is conserved in all the LPD sequences tested, including the human sequence. A glutamate is located at position 356, with few exceptions. The specific role that these amino acids play in the structure or catalysis of LPD is unclear, but these amino acids apparently are important for the normal function of the enzyme.
Although several spontaneous and induced mutations that affect the activity of LPD were reported in several organisms, none of these mutations was at the two positions reported in this study (1, 2, 6, 7, 18). In addition, none of the reported LPD mutations were reported to significantly alter the NADH sensitivity of the PDH complex. The two LPD mutations described in this study are unique in this regard in that they affect the NADH sensitivity of the protein with minimal effects on the overall activity of the protein. The inhibition of PDH (LPD) by NADH is probably caused by overreduction of the redox centers in LPD by NADH, making the enzyme inactive (14). NADH also increased the Km for NAD+, and there was an associated decrease in the LPD activity (29). It is possible that the two mutated forms of LPD described in this study have a higher dissociation constant (Kd) for NADH and/or a reduced rate of FAD reduction by NADH compared to the native LPD. The mechanism of this reduction in NADH sensitivity has not been determined yet.
Conclusion.
The results presented here show that a mutation in LPD led to a PDH complex that is less sensitive to inhibition by NADH. The altered enzyme was active even in the presence of a higher level of NADH in the cell and supported anaerobic growth of strains SE2378 and SE2382 and other similar mutants. The combined functions of glycolysis and the PDH complex in these mutants produced four NADH molecules during conversion of one glucose molecule to two acetyl-CoA molecules. These four NADH molecules were reoxidized by reduction of the two acetyl-CoA molecules to two ethanol molecules by the alcohol/acetaldehyde dehydrogenase (adhE), the observed phenotype of strain SE2378 and other mutants. These results raise an interesting possibility, that a similar alteration in the LPD of other organisms could also lead to PDH complexes that are less sensitive to NADH inhibition and active during anaerobic growth. The presence and functional activity of such an NADH-insensitive PDH may have significant unexplored physiological and biotechnological applications.
Acknowledgments
This study was supported by funds from the U.S. Department of Energy (grant DE-FC36-04-GO14019) and the State of Florida University of Florida Agricultural Experiment Station.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Allison, N., C. H. Williams, Jr., and J. R. Guest. 1988. Overexpression and mutagenesis of the lipoamide dehydrogenase of Escherichia coli. Biochem. J. 256741-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benen, J., W. van Berkel, Z. Zak, T. Visser, C. Veeger, and A. de Kok. 1991. Lipoamide dehydrogenase from Azotobacter vinelandii: site-directed mutagenesis of the His450-Glu455 diad. Spectral properties of wild type and mutated enzymes. Eur. J. Biochem. 202863-872. [DOI] [PubMed] [Google Scholar]
- 3.Bisswanger, H. 1981. Substrate specificity of the pyruvate dehydrogenase complex from Escherichia coli. J. Biol. Chem. 256815-822. [PubMed] [Google Scholar]
- 4.Bisswanger, H., and U. Henning. 1971. Regulatory properties of the pyruvate-dehydrogenase complex from Escherichia coli. Eur. J. Biochem. 24376-384. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brautigam, C. A., J. L. Chuang, D. R. Tomchick, M. Machius, and D. T. Chuang. 2005. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J. Mol. Biol. 350543-552. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, J. M., V. Levandovskiy, N. Mackay, J. Raiman, D. L. Renaud, J. T. Clarke, A. Feigenbaum, O. Elpeleg, and B. H. Robinson. 2006. Novel mutations in dihydrolipoamide dehydrogenase deficiency in two cousins with borderline-normal PDH complex activity. Am. J. Med. Genet. A 1401542-1552. [DOI] [PubMed] [Google Scholar]
- 8.Cassey, B., J. R. Guest, and M. M. Attwood. 1998. Environmental control of pyruvate dehydrogenase complex expression in Escherichia coli. FEMS Microbiol. Lett. 159325-329. [DOI] [PubMed] [Google Scholar]
- 9.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 5223-234. [DOI] [PubMed] [Google Scholar]
- 10.Cornish-Bowden, A. 1995. Fundamentals of enzyme kinetics. Portland Press, London, United Kingdom.
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 13.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixeira de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 1812351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Kok, A., A. F. Hengeveld, A. Martin, and A. H. Westphal. 1998. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385353-366. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, H. G., and U. Henning. 1966. Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Biophys. Acta 122355-358. [DOI] [PubMed] [Google Scholar]
- 16.Haydon, D. J., M. A. Quail, and J. R. Guest. 1993. A mutation causing constitutive synthesis of the pyruvate dehydrogenase complex in Escherichia coli is located within the pdhR gene. FEBS Lett. 33643-47. [DOI] [PubMed] [Google Scholar]
- 17.Hinman, L. M., and J. P. Blass. 1981. An NADH-linked spectrophotometric assay for pyruvate dehydrogenase complex in crude tissue homogenates. J. Biol. Chem. 2566583-6586. [PubMed] [Google Scholar]
- 18.Hopkins, N., and C. H. Williams, Jr. 1995. Lipoamide dehydrogenase from Escherichia coli lacking the redox active disulfide: C44S and C49S. Redox properties of the FAD and interactions with pyridine nucleotides. Biochemistry 3411766-11776. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y., L. O. Ingram, and K. T. Shanmugam. 2007. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl. Environ. Microbiol. 731766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. H., P. Patel, P. Sankar, and K. T. Shanmugam. 1985. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J. Bacteriol. 162344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay, H., E. Beaumont, S. D. Richards, S. M. Kelly, S. J. Sanderson, N. C. Price, and J. G. Lindsay. 2000. FAD insertion is essential for attaining the assembly competence of the dihydrolipoamide dehydrogenase (E3) monomer from Escherichia coli. J. Biol. Chem. 27536665-36670. [DOI] [PubMed] [Google Scholar]
- 23.Linn, T., and R. St. Pierre. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 1721077-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumura, I., and A. D. Ellington. 2001. Methods in molecular biology, 2nd ed. Humana Press Inc., Totowa, NJ.
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor laboratory, Cold Spring Harbor, NY.
- 26.Narayanan, N., M. Y. Hsieh, Y. Xu, and C. P. Chou. 2006. Arabinose-induction of lac-derived promoter systems for penicillin acylase production in Escherichia coli. Biotechnol. Prog. 22617-625. [DOI] [PubMed] [Google Scholar]
- 27.Quail, M. A., D. J. Haydon, and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 1295-104. [DOI] [PubMed] [Google Scholar]
- 28.Rosentel, J. K., F. Healy, J. A. Maupin-Furlow, J. H. Lee, and K. T. Shanmugam. 1995. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol. 1774857-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahlman, L., and C. H. Williams, Jr. 1989. Lipoamide dehydrogenase from Escherichia coli. Steady-state kinetics of the physiological reaction. J. Biol. Chem. 2648039-8045. [PubMed] [Google Scholar]
- 30.Schmincke-Ott, E., and H. Bisswanger. 1981. Dihydrolipoamide dehydrogenase component of the pyruvate dehydrogenase complex from Escherichia coli K12. Comparative characterization of the free and the complex-bound component. Eur. J. Biochem. 114413-420. [DOI] [PubMed] [Google Scholar]
- 31.Shen, L. C., and D. E. Atkinson. 1970. Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J. Biol. Chem. 2455974-5978. [PubMed] [Google Scholar]
- 32.Smith, M. W., and F. C. Neidhardt. 1983. 2-Oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J. Bacteriol. 15681-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snoep, J. L., M. R. de Graef, A. H. Westphal, A. de Kok, M. J. Teixeira de Mattos, and O. M. Neijssel. 1993. Differences in sensitivity to NADH of purified pyruvate dehydrogenase complexes of Enterococcus faecalis, Lactococcus lactis, Azotobacter vinelandii and Escherichia coli: implications for their activity in vivo. FEMS Microbiol. Lett. 114279-283. [DOI] [PubMed] [Google Scholar]
- 34.Tao, H., R. Gonzalez, A. Martinez, M. Rodriguez, L. O. Ingram, J. F. Preston, and K. T. Shanmugam. 2001. Engineering a homo-ethanol pathway in Escherichia coli: increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. J. Bacteriol. 1832979-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, H., A. Hasona, P. M. Do, L. O. Ingram, and K. T. Shanmugam. 2005. Global gene expression analysis revealed an unsuspected deo operon under the control of molybdate sensor, ModE protein, in Escherichia coli. Arch. Microbiol. 184225-233. [DOI] [PubMed] [Google Scholar]
- 36.Visser, J., H. Kester, K. Jeyaseelan, and R. Topp. 1982. Pyruvate dehydrogenase complex from Bacillus. Methods Enzymol. 89399-407. [DOI] [PubMed] [Google Scholar]
- 37.Wei, W., H. Li, N. Nemeria, and F. Jordan. 2003. Expression and purification of the dihydrolipoamide acetyltransferase and dihydrolipoamide dehydrogenase subunits of the Escherichia coli pyruvate dehydrogenase multienzyme complex: a mass spectrometric assay for reductive acetylation of dihydrolipoamide acetyltransferase. Protein Expr. Purif. 28140-150. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, K. D., and C. H. Williams, Jr. 1981. NADH inhibition and NAD activation of Escherichia coli lipoamide dehydrogenase catalyzing the NADH-lipoamide reaction. J. Biol. Chem. 2562307-2314. [PubMed] [Google Scholar]