Abstract
NarX-NarL and NarQ-NarP are paralogous two-component regulatory systems that control Escherichia coli gene expression in response to the respiratory oxidants nitrate and nitrite. Nitrate stimulates the autophosphorylation rates of the NarX and NarQ sensors, which then phosphorylate the response regulators NarL and NarP to activate and repress target operon transcription. Here, we investigated both the autophosphorylation and dephosphorylation of soluble sensors in which the maltose binding protein (MBP) has replaced the amino-terminal transmembrane sensory domain. The apparent affinities (Km) for ADP were similar for both proteins, about 2 μM, whereas the affinity of MBP-NarQ for ATP was lower, about 23 μM. At a saturating concentration of ATP, the rate constant of MBP-NarX autophosphorylation (about 0.5 × 10−4 s−1) was lower than that observed for MBP-NarQ (about 2.2 × 10−4 s−1). At a saturating concentration of ADP, the rate constant of dephosphorylation was higher than that of autophosphorylation, about 0.03 s−1 for MBP-NarX and about 0.01 s−1 for MBP-NarQ. For other studied sensors, the published affinities for ADP range from about 16 μM (KinA) to about 40 μM (NtrB). This suggests that only a small proportion of NarX and NarQ remain phosphorylated in the absence of nitrate, resulting in efficient response regulator dephosphorylation by the remaining unphosphorylated sensors.
In Escherichia coli, anaerobic respiratory-gene expression is regulated by the dual-two-component regulatory system, NarX-NarL and NarQ-NarP (28). The paralagous sensors NarX and NarQ are histidine protein kinases (HPKs) that autophosphorylate in response to nitrate and other signals (7, 19, 27, 33, 40). These sensors control the phosphorylation state of the paralogous NarL and NarP response regulators (8, 27, 30, 37). Phospho-NarL and phospho-NarP bind to target operon control regions to regulate transcription (9). During anaerobic growth, this dual-two-component system establishes hierarchal control of gene expression to utilize nitrate in preference to other anaerobic electron acceptors (13, 27).
The NarX and NarQ proteins share three characteristic modules: sensory, central, and transmitter (32). The sensory module includes the periplasmic domain, which is delimited by two transmembrane helices. The periplasmic domain contains the conserved P box element (18 residues), which is involved in sensing nitrate (7, 40). The sensory module also contains a HAMP linker and a signaling helix, both of which are involved in signal transduction (1-3). Little is known about the central module, and its function remains enigmatic.
In general, transmitter modules include both a DHp (dimerization and histidyl phosphotransfer) and a CA (catalytic and ATP-binding) domain (11). The DHp domain consists of the H and X box sequence motifs and contains the conserved autophosphorylated histidyl residue. The CA domain consists of the N, D, F, and G box sequence motifs (also termed N, G1, F, and G2, respectively), which form the nucleotide binding pocket. In conventional transmitters, the CA domain contains all four motifs, where the region between the F and G boxes forms the ATP lid and the conserved glycyl residues of the D and G boxes possibly provide hinges for lid mobility (4, 10, 21, 26, 31, 44). The NarX and NarQ transmitter sequences are in HPK subfamily 7 as defined by Grebe and Stock and Wolanin et al. (14, 42), in which the F box is absent, the distance between the D and G boxes is reduced, and two of the three conserved glycyl residues of the G box are not present.
NarX autophosphorylation has been documented in vitro for native protein, either enriched in membrane vesicles (19, 40) or solubilized by detergent (37). Autophosphorylation in vitro has also been studied for truncated forms of NarX and NarQ reconstituted from inclusion bodies (8, 30). In this investigation, we prepared soluble forms of the proteins by replacing the amino-terminal sensory module with maltose binding protein (MBP) to characterize the kinetics of autophosphorylation and dephosphorylation. The kinetic values obtained indicate that the apparent affinity for ADP is high for phospho-MBP-NarX and phospho-MBP-NarQ in comparison to other HPKs, and the rate constants for dephosphorylation are significantly higher than for autophosphorylation. Therefore, only a low proportion of MBP-NarX and MBP-NarQ remain phosphorylated due to the higher efficiency of ADP-stimulated dephosphorylation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains were cultured at 37°C in tryptone-yeast extract (TY) broth (tryptone, 8 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1) with shaking at 200 rpm. For plasmid selection, TY broth was supplemented with ampicillin (100 μg ml−1). All malE fusion plasmids were introduced into the E. coli host strain JM109 (43). Culture densities were monitored with a Klett-Sumerson photoelectric colorimeter (Klett Manufacturing Co., New York, NY) equipped with a number 66 (red) filter.
DNA manipulation and strain construction.
Fusion proteins are designated according the amino acyl residue of the sensor protein linked to MBP. MBP-NarX185 was constructed from a 2.5-kb PvuII-HindIII fragment of narX+, which contains silent restriction sites introduced at nucleotide positions 550 and 1676 (41), ligated into the StuI-HindIII sites of pMAL-c (39). MBP-NarQ182 was constructed from a 1.5-kb PvuII fragment of narQ+ (40) ligated into the StuI site of pMAL-c (39).
Overexpression and protein enrichment.
Strains containing malE-narX and malE-narQ expression plasmids were inoculated into 200 ml TY plus ampicillin. Expression was induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in the mid-exponential phase (between 30 and 40 Klett units), and incubation continued for 2.5 h. All procedures thereafter were performed at 4°C. Cells were harvested by centrifugation at 10,000 × g for 10 min. The pellets were washed with suspension buffer (20 mM HEPES [pH 7.0], 200 mM KCl, 1 mM dithiothreitol [DTT], and 5% [vol/vol] glycerol) and resuspended in the same buffer supplemented with 1 mM phenylmethanesulfonylfluoride. Cell suspensions were disrupted by passage through a French pressure cell at 20,000 lb/in2. Soluble and particulate fractions were separated by centrifugation (20,000 × g for 30 min). The soluble fraction was subjected to affinity chromatography in a 25-ml column containing 3 ml amylose resin (New England Biolabs, Ipswich, MA) preequilibrated with suspension buffer. The column was washed with 30 ml of suspension buffer and then eluted with 10 ml of suspension buffer supplemented with 1 mM phenylmethanesulfonylfluoride and 10 mM maltose. The protein concentrations of the eluted fractions were determined by the method of Bradford (5). The protein preparations were evaluated by electrophoresis through Laemmli gels (18), which were stained with GelCode Blue Stain Reagent (Pierce, Rockford, IL) and analyzed with a FluorChem imaging system (Alpha Innotech, San Leandro, CA). For this study, protein preparations were stored in 50- to 100-μl aliquots at 4°C, and autophosphorylation of MBP-NarX185 and MBP-NarQ182 was reproducible for 2 to 3 weeks or 4 to 6 weeks, respectively. For the data presented here, a total of five different preparations were used for each protein. Recently, we found that MBP-NarX187 and MBP-NarQ185 retained activity after at least 3 months of storage at −80°C.
Nucleotides.
[γ-32P]ATP (10 mCi ml−1, 3,000 Ci mmol−1; Perkin Elmer, Waltham, MA), ATP, and ADP (Sigma-Aldrich Corp.; catalog no. A-2383 and A-2754, respectively) were ≥98% pure as indicated by the manufacturer.
Autophosphorylation.
All chemicals and reagents were from Sigma-Aldrich Corp. (St. Louis, MO) except as noted. All reactions were performed at 19°C (MiniFridge II; Boekel Scientific, Feasterville, PA). The standard buffer contained 100 mM HEPES (pH 7.0), 50 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 10 mM DTT, and 10% glycerol. Reaction mixtures generally contained 5 μM (final monomer concentration) of enriched MBP-sensor and 30 nM [γ-32P]ATP, with modifications as noted. An ATP-regenerating system (20, 23, 24, 35) consisting of 1 mM phosphoenol pyruvate and 0.01 unit ml−1 pyruvate kinase was included with the standard buffer where indicated. Protein was first incubated with DTT (10 mM final reaction concentration) for 30 min at room temperature (∼25°C), followed by addition of the remaining standard buffer plus 30 nM [γ-32P]ATP to initiate the reaction. Reaction samples were withdrawn at the indicated time intervals, mixed with stop solution, and visualized or quantified by filter binding as described below. The subsequent experiments used these conditions with modifications as noted.
Previously, Nar sensor autophosphorylation has been studied in reactions buffered with either Tris-HCl (pKa = 8.1) at pH 8 (37, 40) or HEPES (pKa = 7.5) at pH 8 (8, 19, 30) containing 5 to 10 mM MgCl2 and 10% glycerol, conducted at or near room temperature. We evaluated these parameters for both MBP-NarX185 and MBP-NarQ182. We found that HEPES was the superior buffer, promoting approximately fivefold-higher autophosphorylation rates. Phospho-MBP-NarQ182 accumulated at an approximately fivefold-higher rate at pH 7 than at pH 8, and the rate was intermediate at pH 7.5. By contrast, phospho-MBP-NarX185 accumulated at similar rates at both pH 7 and 8 (data not shown). Therefore, we used pH 7.0 as the standard condition.
Glycerol stimulated autophosphorylation by two to threefold, as assayed by Laemmli gel electrophoresis (data not shown), so we included 10% glycerol in the standard assay. We used 19°C for all reactions to ensure temperature consistency.
Early experiments suggested that the two sensors exhibited different preferences for divalent cation, so we evaluated this point in a number of experiments using several different protein preparations. Consistently, phospho-MBP-NarQ182 accumulated at higher rates in reactions containing Mn2+ than with Mg2+, but maximum rates required both (data not shown). By contrast, phospho-MBP-NarX185 accumulated at similar rates in the presence of either ion or both. Autophosphorylation was undetectable in the absence of divalent cation, and neither Ca2+, Co2+, nor Zn2+ could substitute for Mg2+ or Mn2+ (data not shown). Accordingly, we included 5 mM each of MgCl2 and MnCl2 in the standard assay buffer.
DTT stimulated accumulation of phospho-MBP-NarX182 by approximately twofold but had little effect on phospho-MBP-NarQ185, as assayed by Laemmli gel electrophoresis (data not shown). Therefore, we included 10 mM DTT as the standard condition. The NarX transmitter module contains four Cys residues, whereas the NarQ transmitter module contains two.
Dephosphorylation.
To prepare phosphorylated sensor proteins, autophosphorylation reactions (150-μl volumes) were performed as described above for 6 min without the addition of an ATP-regenerating system. The reaction mixtures were then filtered four times through Microcon YM-50 filtration devices (Millipore, Bedford, MA) to remove residual [γ-32P]ATP and ADP. The filtration buffer was standard buffer lacking KCl, MgCl2, and MnCl2. Use of buffer without the indicated salts prevented protein precipitation during the filtration steps. The eluted fraction of phosphorylated proteins routinely contained <5% of the initial [γ-32P]ATP concentration (data not shown). The phosphorylated protein was resuspended in standard buffer to a final volume of 300 μl and kept at 4°C until it was ready for use to minimize intrinsic dephosphorylation. Samples were preincubated at 19°C for 3 min, and dephosphorylation reactions were then initiated with addition of different concentrations of ADP as indicated. Samples were withdrawn at the indicated time intervals, mixed with stop solution (see below), and visualized by thin-layer chromatography (TLC) or quantified by filter binding as described below.
Visualization of radiolabeled protein.
For Laemmli gel electrophoresis, 5 μl of reaction samples was added to 5 μl of 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (0.5 M Tris-HCl [pH 6.8], 4.4% [wt/vol] SDS, 20% [vol/vol] glycerol, 2% [vol/vol] β-mercaptoethanol, 0.02% bromophenol blue) after a reaction time of 10 min. Following electrophoresis, the gel was dried for 45 min at 70°C and exposed to a phosphorimager screen overnight.
For TLC, cellulose polyethyleneimine plates (J. T. Baker, Phillipsburg, NJ) were first soaked in deionized water, followed by methanol, and then air dried right before use. Reaction samples (2.5 μl) were added to 2.5 μl stop solution after 5 min and spotted onto the plate. The plates were developed in a solution containing 1 M LiCl and 1 M acetic acid. After being air dried, the TLC plates were exposed to a phosphorimager screen for 1 h. Both Laemmli gel and TLC plate images were analyzed using a Storm Scanner with ImageQuant Software (Molecular Dynamics, Cupertino, CA).
Filter-binding quantification of labeled protein.
The following procedure was adapted from studies of the NtrB-NtrC (NRII-NRI) two-component system (16). Time point samples (5 μl) were taken from the autophosphorylation and dephosphorylation assays described above and added to 200 μl stop solution (20 mM HEPES, pH 7.0, 100 mM EDTA, 0.01% [vol/vol] SDS) with a 1:10,000 dilution of blue dye (food coloring) to mark the slot locations. The blue dye at this dilution did not affect the stability of the labeled proteins (data not shown). Nitrocellulose membranes (Bio-Rad, Hercules, CA) were soaked in 20 mM HEPES [pH 7.0], with 500 mM NaCl and then placed in the Bio-Dot SF (Bio-Rad) apparatus attached to a vacuum. The membranes were vacuum dried, and each slot was equilibrated with 100 μl stop solution. The reaction samples in stop solution were blotted onto the membranes, vacuum dried, and washed with 0.1 M Na2CO3 (200 μl/slot). The membranes were removed from the Bio-Dot SF apparatus and washed three times in 0.1 M Na2CO3 to remove excess [γ-32P]ATP. The membranes were air dried and first visualized by exposure to a phosphorimager screen to ensure slot uniformity (data not shown). The individual slots were then cut and counted by liquid scintillation (Ready Protein; Beckman Coulter, Fullerton, CA). The specific activity of radiolabeled ATP was determined by counting 10 μl of 1:100 and 1:1,000 dilutions of the remaining reaction mixtures directly by liquid scintillation. The levels of phosphosensor were expressed as nmol or pmol of incorporated phosphorus per total amount of protein. All protein concentrations were expressed as monomers.
Kinetics.
For autophosphorylation, the standard buffer contained an ATP-regenerating system to reduce the effects of ADP. Data were collected as three or four time point samples (5 μl) at 30-s intervals from 0 to 60 s or 90 s for the initial rates. For each sample, the measured level of phosphosensor was multiplied by the dilution factor of unlabeled ATP added to 30 nM [γ-32P]ATP. The rates were measured by dividing the slopes of the initial time points by the total amount of protein (per monomer). A Lineweaver-Burk plot was used to estimate the apparent affinity for ATP (KmATP) from the x intercept and the apparent k from the y intercept.
For dephosphorylation, data were collected as two or three time point samples (5 μl) at 30-s intervals from 0 to 30 s or 60 s for MBP-NarX185 and as three or four time point samples (5 μl) at 30-s intervals from 0 to 60 s or 90 s for MBP-NarQ182. The rates were measured by dividing the slopes of the initial time points by the amounts of phosphorylated protein measured at time point zero. The final dephosphorylation rates were then calculated by subtracting the intrinsic dephosphorylation rates (time points 0 to 60 s) from the ADP-stimulated dephosphorylation rates. A Lineweaver-Burk plot was used to estimate the apparent affinity for ADP (KmADP) from the x intercept and the apparent −k from the y intercept.
RESULTS
MBP-NarX and MBP-NarQ proteins.
NarX and NarQ are both intrinsic membrane proteins and therefore must be solubilized by detergent for purification (37). The amino-terminal sensory module contains the transmembrane helices, whereas the central and transmitter modules are cytoplasmic. Previously, amino-terminally truncated forms of NarX and NarQ were produced as histidine-tagged proteins, but they accumulated as inclusion bodies and had to be reconstituted by treatment with 8 M urea (8, 30). Therefore, to obtain soluble forms, we replaced the amino-terminal module with MBP, which increases the solubility of linked protein domains (38). Our constructs, made by S. B. Williams (39), employed a junction similar to that used for the histidine-tagged proteins (30). For NarQ, a native PvuII site spanning codons 181 to 182 within amphiphilic helix AH1 of the HAMP linker was used as the fusion point. For NarX, a PvuII site was introduced at the corresponding position, spanning codons 184 to 185 (41). The resulting fusion proteins, designated MBP-NarX185 and MBP-NarQ182, contain virtually the entire cytoplasmic portions of these sensors, including much of the HAMP linker, the signaling helix, and the central and transmitter modules.
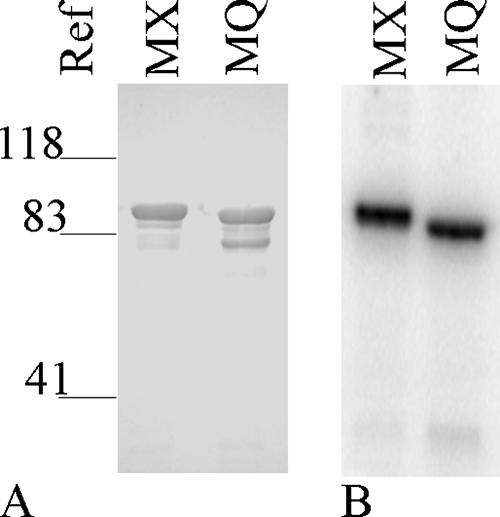
We monitored both soluble and particulate fractions from crude extracts of overexpression cultures and found that the MBP fusion proteins partitioned approximately equally between the two. We made no attempt to recover protein from the particulate fractions. Soluble fractions were subjected to affinity enrichment as described in Materials and Methods, and the protein obtained remained soluble during storage. Results for typical preparations are shown in Fig. 1A. Proteins migrated as expected according to their calculated molecular masses (MBP-NarX185, 87 kDa, and MBP-NarQ182, 84 kDa). In general, MBP-NarX185 preparations were highly enriched for the desired protein with only negligible contaminating species detected by staining (Fig. 1A, lane 1). By contrast, MBP-NarQ182 preparations routinely displayed a higher-mobility contaminating species, which we presume resulted from proteolysis (Fig. 1A, lane 2). This higher-mobility species constituted approximately 20% of the total protein in most preparations, as estimated from gel images (see Materials and Methods). However, it was not observed in assays for autophosphorylation (Fig. 1B, lane 2) and therefore is enzymatically inactive. Accordingly, we normalized the measured levels of phospho-MBP-NarQ182 by 20% to account for this inactive species in calculations of specific activity.
FIG. 1.
Purification and autophosphorylation of MBP fusion proteins MBP-NarX185 (MX) and MBP-NarQ182 (MQ). (A) The purification of MBP fusion proteins was analyzed on a 10% Laemmli gel stained with Coomasie Blue (GelCode Blue Stain Reagent; Pierce) by FluorChem. The relative positions of molecular weight references are indicated. The amount of protein applied for each lane was 1.6 μg. (B) Radiolabeled proteins were prepared and visualized as stated in Materials and Methods.
Autophosphorylation with ATP regeneration.
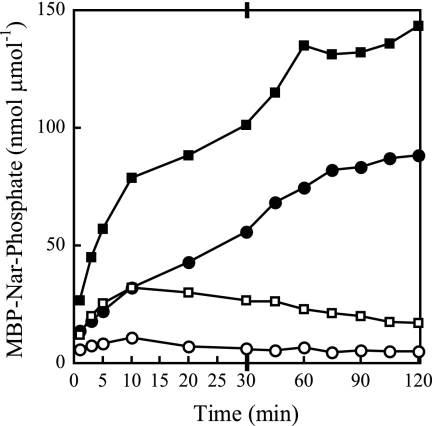
In a previous study of native NarX autophosphorylation in crude vesicles, ATP concentrations as low as 3 μM resulted in inefficient accumulation of phospho-NarX (40). Following on observations with the NtrB (NRII) and CheA HPKs (16, 35), we hypothesized that ADP, both as a contaminant in the ATP used and also generated by autophosphorylation, causes rapid dephosphorylation of phospho-NarX. To examine this, we first analyzed the effects of ATP regeneration (16, 35) on the accumulation of phosphorylated sensors incubated in 30 nM [γ-32P]ATP with 100 μM unlabeled ATP (Fig. 2). These experiments used 5 μM and 4 μM (normalized) concentrations of MBP-NarX185 and MBP-NarQ182 active monomers, respectively. The results are expressed as nmol of incorporated phosphorus per μmol total protein for comparison. In the absence of ATP regeneration, phospho-MBP-NarX185 and phospho-MBP-NarQ182 accumulated for only 10 min (Fig. 2). In the presence of ATP regeneration, phosphosensor accumulation was significantly higher in the first 10 min and continued to increase for at least 60 min. The fractions of phosphorylated monomers were approximately 10% for MBP-NarX185 and 15% for MBP-NarQ182 during the 2-h incubation time.
FIG. 2.
Autophosphorylation of MBP-NarX185 and MBP-NarQ182 with or without an ATP-regenerating system. Labeled proteins were quantified from 5-μl time point samples as described in Materials and Methods. Quantification of phospho-MBP-NarX185 (circles) and phospho-MBP-NarQ182 (squares) with the addition of an ATP-regenerating system (solid symbols) or without (open symbols) in 100 μM [γ-32P]ATP (MBP-NarX185, 2,300 cpm pmol−1, and MBP-NarQ182, 1,800 cpm pmol−1). Specific levels of phosphorylation are expressed as nmol of incorporated phosphate per total amount of protein used, expressed as μmol. Note the change in the x axis scale after 30 min. The data represent one of two independent measurements. Autophosphorylations of MBP-NarX185 and MBP-NarQ182 were conducted as separate experiments.
Autophosphorylation rates for MBP-NarX185 and MBP-NarQ182 in the presence of ATP regeneration increased as a function of the protein concentration over a range of 2 to 8 μM, indicating that ATP (100 μM) was provided in excess (data not shown).
Dephosphorylation.
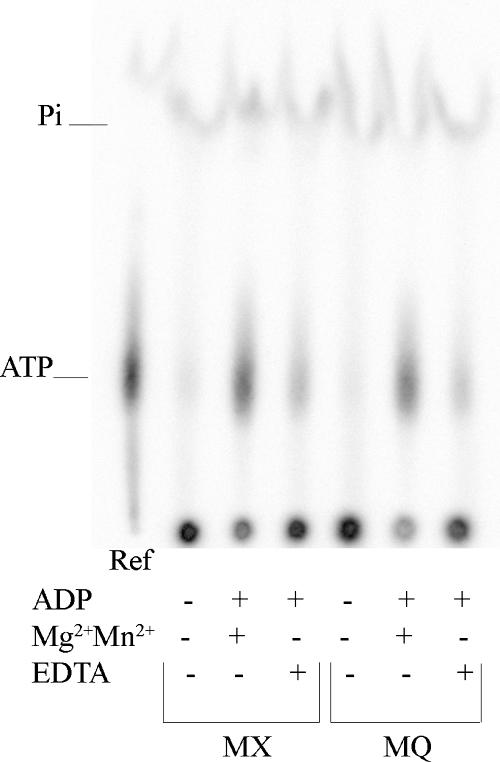
As described above, we observed that dephosphorylation for both phospho-MBP-NarX and phospho-MBP-NarQ proteins was due to ADP. To determine the mechanism of this dephosphorylation, we used TLC to examine the form in which 32P was released from the phosphorylated sensors. The results indicated that, upon addition of 5 μM unlabeled ADP, both sensors dephosphorylated by transferring the phosphoryl group to ADP, forming [32P]ATP (Fig. 3). Dephosphorylation decreased upon replacing divalent cation (Mg2+ plus Mn2+) with 5 mM chelator, EDTA (Fig. 3).
FIG. 3.
Effects of ADP on phosphorylated MBP-NarX185 (MX) and MBP-NarQ182 (MQ) monitored by TLC. Autophosphorylation was first performed for 6 min. The removal of 30 nM [γ-32P]ATP and dephosphorylation reactions were performed as described in Materials and Methods, with the addition of 5 μM ADP. Where indicated, standard reaction buffer was modified by the absence of MgCl2 and MnCl2 and the addition of EDTA (5 mM); 2.5 μl of a 1:104 dilution of 3.3 μM [γ-32P]ATP (3,000 cpm fmol−1) in stop solution was included as a reference (Ref).
Effects of different ATP and ADP concentrations.
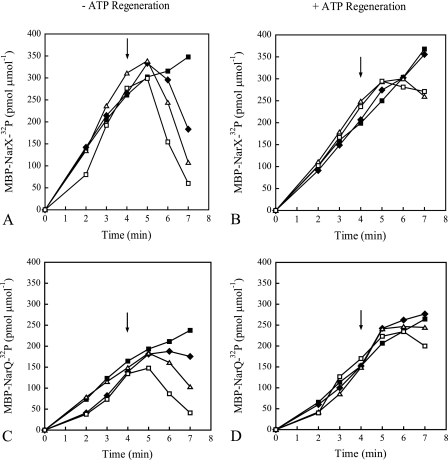
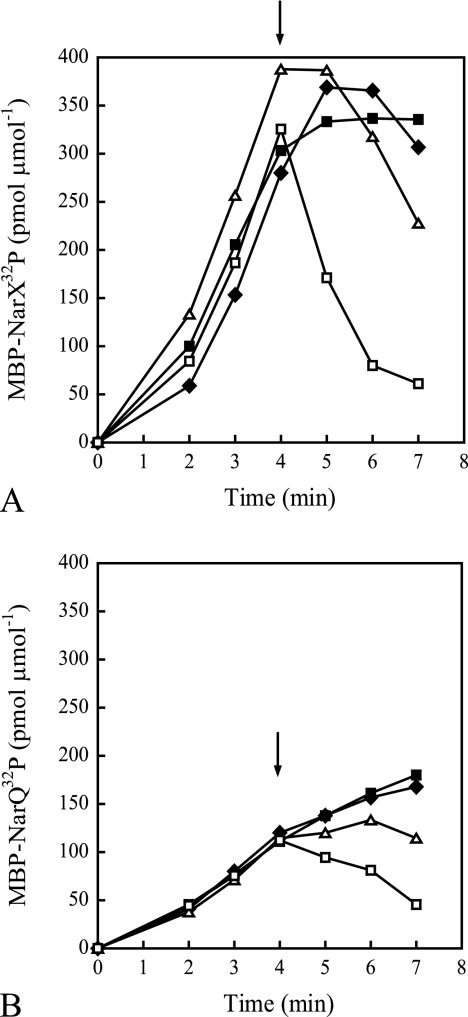
In Fig. 2, the autophosphorylation reactions were performed using a mixture of labeled and unlabeled ATP. Previously, it was shown that addition of unlabeled ATP resulted in the rapid loss of NarX-32P (19). We wanted to determine if this was due to contaminating ADP. Autophosphorylation reactions were performed with the addition of increasing concentrations of unlabeled ATP after 4 min in the presence or absence of ATP regeneration (Fig. 4). An initial [γ-32P]ATP concentration of only 30 nM was included to allow accumulation of labeled protein while minimizing the effects of ADP produced during the time span of the reaction. With phospho-MBP-NarX185, addition of ATP to final concentrations as low as 10 μM resulted in the loss of label (Fig. 4A). With phospho-MBP-NarQ182, loss of label was observed upon addition of 100 μM ATP (Fig. 4C). When ATP regeneration was included in the reactions, no significant loss of label was observed for either protein (Fig. 4B and D). (The levels of labeled protein in the presence of ATP regeneration did not continue to increase after the addition of unlabeled ATP due to dilution of the [γ-32P]ATP.) This indicates that the instability of phosphorylated sensors after the addition of unlabeled ATP is a result of dephosphorylation by contaminating ADP. Since only 30 nM of ATP was used to initiate the reactions in Fig. 4, it is possible that the ATP concentration may be limiting compared to the 100 μM of ATP used in Fig. 2.
FIG. 4.
Effects of unlabeled ATP on the accumulation of radiolabeled MBP-NarX185 and MBP-NarQ182 with the addition of an ATP-regenerating system (B and D) or without (A and C) in 30 nM [γ-32P]ATP (MBP-NarX185, 5,600 cpm fmol−1, and MBP-NarQ182, 5,000 cpm fmol−1). At the time points shown, 5-μl samples of radiolabeled protein were quantified as described in Materials and Methods. Specific levels of phosphorylation are expressed as pmol of incorporated 32P per total amount of protein used, expressed as μmol. Immediately following the removal of the 4-min sample, different concentrations of unlabeled ATP were added to the reaction mixtures (arrows) with the following final concentrations: 1× standard buffer (solid squares), 10 μM ATP (diamonds), 100 μM ATP (triangles), and 1,000 μM ATP (open squares). (A and B) MBP-NarX185. (C and D) MBP-NarQ182. The data represent one of two independent measurements.
To determine the ADP concentration that would cause net dephosphorylation, we performed autophosphorylation reactions as described in the legend to Fig. 4 but instead added different concentrations of unlabeled ADP (Fig. 5). With phospho-MBP-NarX185, addition of ADP to a final concentration as low as 1 μM resulted in the loss of label (Fig. 5A). With phospho-MBP-NarQ182, a significant loss of label was observed with 10 μM ADP (Fig. 5B). This further suggests that the loss of phosphorylated protein observed during autophosphorylation is due to the sensitivity of phospho-MBP sensor to low concentrations of ADP.
FIG. 5.
Effects of ADP on the accumulation of radiolabeled MBP-NarX185 (A) and MBP-NarQ182 (B) in 30 nM [γ-32P]ATP (MBP-NarX185, 3,000 cpm fmol−1, and MBP-NarQ182, 7,000 cpm fmol−1). At the time points shown, 5-μl samples of radiolabeled protein were quantified as described in Materials and Methods. Specific levels of phosphorylation are expressed as pmol of incorporated 32P per total amount of protein used, expressed as μmol. Immediately following the removal of the 4-min sample, different concentrations of ADP were added to the reaction mixture (arrows) with the following final concentrations: 1× standard buffer (solid squares), 0.1 μM ADP (diamonds), 1 μM ADP (triangles), and 10 μM ADP (open squares). The data represent one of two independent measurements.
Autophosphorylation and dephosphorylation kinetics.
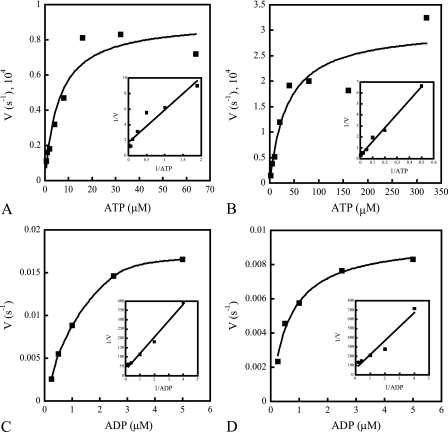
Finally, we analyzed the kinetics for both the forward (autophosphorylation) and reverse (dephosphorylation) reactions. The data were interpreted according to a simplified reaction mechanism where autophosphorylation and dephosphorylation rate constants are designated k and −k, respectively. Initial reaction rates were obtained as described in Materials and Methods. The results from representative single experiments are shown in Fig. 6, and summary data averaged from three independent replicates are presented in Table 1.
FIG. 6.
Kinetics of MBP-NarX185 and MBP-NarQ182 autophosphorylation (A and B) and dephosphorylation (C and D). Quantifications of autophosphorylation and dephosphorylation rates were performed as stated in Materials and Methods. The insets represent Lineweaver-Burk plots of the corresponding reaction velocities as 1/V versus 1/ATP or 1/ADP. Autophosphorylation kinetics were performed with an ATP-regenerating system in 30 nM [γ-32P]ATP (∼7,000 cpm fmol−1) with different concentrations of unlabeled ATP. (A) Autophosphorylation rates of MBP-NarX185 incubated with increasing concentrations of ATP (0.5, 1, 2, 4, 8, 16, 32, and 64 μM). The x intercept (KmATP) was 2.5 μM. The y intercept (k) was 0.59 × 10−4 s−1. (B) Autophosphorylation rates of MBP-NarQ182 incubated with increasing concentrations of ATP (2, 5, 10, 20, 40, 80, 160, and 320 μM). The x intercept (KmATP) was 24.6 μM. The y intercept (k) was 3.0 × 10−4 s−1. (C) Dephosphorylation rates of phospho-MBP-NarX185 (∼3,000 cpm/fmol) incubated with increasing concentrations of ADP (0.25, 0.5, 1, 2.5, and 5 μM). The x intercept (KmADP) was 2.9 μM. The y intercept (−k) was 0.033 s−1. (D) Dephosphorylation rates of phospho-MBP-NarQ182 (∼4,000 cpm/fmol) incubated with increasing concentrations of ADP (0.25, 0.5, 1, 2.5, and 5 μM). The x intercept (KmADP) was 2.2 μM. The y intercept (−k) was 0.015 s−1. The data shown are from one of three independent measurements from various protein purification preparations.
TABLE 1.
Kinetic values of autophosphorylation and dephosphorylation reactions
| Protein | Kinetic valueb
|
Reference | |||
|---|---|---|---|---|---|
| Autophosphorylation
|
Dephosphorylation
|
||||
| KmATP (μM) | k (10−4) (s−1) | KmADP (μM) | −k (s−1) | ||
| MBP-NarX185a | 2.4 ± 0.7 | 0.5 ± 0.2 | 2.3 ± 0.6 | 0.029 ± 0.01 | This study |
| MBP-NarQ182a | 22.8 ± 9.3 | 2.2 ± 1.1 | 1.8 ± 0.7 | 0.013 ± 0.003 | This study |
| CheA | 300 ± 75 | 260 ± 40 | 42 ± 8 | 0.028 ± 0.003 | 35 |
| NRII (NtrB) | 31 ± 1.4 | 118 ± 2.5 | 40 | − | 16, 34 |
| KinA | 74 ± 11 | 19 ± 5 | 16 ± 2.6 | − | 15 |
| EnvZ | 218 | 0.81c | − | − | 17 |
| PhoQ | 20.1 ± 13.1 | −d | − | − | 21 |
Values were determined as described under Materials and Methods and Results expressed per monomer.
± standard errors of the mean. For the Nar sensors, these values were determined from three independent measurements and different protein preparations.
−, not determined or published.
The autophosphorylation rates were biphasic, displaying two distinct linear rates between 0 and 90 s (lower) and 90 and 180 s (higher) (data not shown). The kinetic values displayed in Fig. 6A and B and Table 1 represent the lower rates from the initial time points 0 to 90 s. These initial autophosphorylation rates began to saturate around 16 μM ATP for MBP-NarX185 (Fig. 6A) and around 80 μM ATP for MBP-NarQ182 (Fig. 6B). MBP-NarX185 exhibited an approximately ninefold-greater affinity for ATP than did MBP-NarQ182, but k was approximately fourfold greater for MBP-NarQ182 (Table 1). For the higher autophosphorylation rates observed between 90 and 180 s (data not shown), MBP-NarX185 displayed an average KmATP of 14 μM ATP and an average rate constant of 3.0 × 10−4 s−1. For MBP-NarQ182, the average KmATP was 86 μM, with an average rate constant of 18 × 10−4 s−1. We do not know the cause of the accelerated rates observed between 90 and 180 s.
Intrinsic dephosphorylation rates were measured by adding reaction buffer instead of ADP. Both proteins were dephosphorylated by approximately 20% after 3 min (data not shown). These intrinsic dephosphorylation rates were subtracted from the ADP-stimulated dephosphorylation rates to obtain the final values described below.
For both proteins, rates of dephosphorylation began to saturate around 2.5 μM ADP (Fig. 6C and D), and the calculated values for KmADP were similar (Table 1). However, −k was approximately twofold faster for MBP-NarX185, indicating that it had a slightly higher rate of ADP-stimulated dephosphorylation than MBP-NarQ182. This is consistent with the different responses of the two phosphorylated proteins to added ATP and ADP observed in Fig. 4 and 5. Overall, both proteins exhibited higher rates of dephosphorylation, as well as a higher affinity for ADP observed with MBP-NarQ185. Therefore, MBP-NarX185 and MBP-NarQ182 appear to be more efficient in ADP-stimulated dephosphorylation, resulting in a low accumulation of phosphorylated protein.
DISCUSSION
The NarX and NarQ proteins autophosphorylate inefficiently at millimolar concentrations of ATP (19, 40). Our goal for this study was to determine the reason for this, in order to develop optimized autophosphorylation conditions for future analysis of sensor-response regulator transphosphorylation reactions. The nonlinear accumulation of autophosphorylated MBP-Nar sensor proteins observed with low ATP concentrations was largely due to ADP, since an ATP-regenerating system greatly enhanced autophosphorylation (Fig. 2). Indeed, the rate constants of the reverse reaction (−k) were greater than those of the forward reaction (k) for both proteins, with differences of approximately 600-fold for MBP-NarX185 and 60-fold for MBP-NarQ182 per monomer at saturating nucleotide concentrations. Furthermore, the affinities for ADP were approximately 13-fold higher than for ATP with MBP-NarQ182 (Table 1). Thus, even low ADP concentrations were sufficient to inhibit accumulation of phospho-MBP-Nar sensors.
Autophosphorylation kinetics have previously been examined with several other HPKs (12, 15-17, 21, 25, 31). For these proteins, values for ATP affinity and for phosphoryl transfer rate constants span a broad range (Table 1). The corresponding values for the MBP-Nar sensors are at the lower ends of these ranges. Values for ADP affinity, which are available for three sensors (CheA, NRII, and KinA), range from 16 to 42 μM. The corresponding affinities for the MBP-Nar sensors, about 2 μM, are substantially higher. The rate constant for ADP-stimulated dephosphorylation of CheA is about 0.03 s−1 per monomer (35), roughly equivalent to the rate constants for the MBP-Nar sensors (Table 1). Overall, the kinetic parameters for the MBP-Nar sensors are consistent with relatively efficient dephosphorylation even at low ADP concentrations.
We do not know if MBP-Nar sensor properties in vitro mimic native sensor behavior in vivo. By analogy to other sensors, we presume that Nar sensors adopt either of two conformations in vivo: a ligand-stimulated conformation, which autophosphorylates efficiently in order to catalyze response regulator phosphorylation; and an unstimulated conformation, which dephosphorylates efficiently in order to catalyze response regulator dephosphorylation (27). Our working hypothesis is that the MBP sensors are biased toward the unstimulated conformation. This is based on extrapolated autophosphorylation and dephosphorylation rates of the MBP sensors with ATP and ADP pools of approximately 3,500 μM and 100 μM, respectively, during mid-exponential phase (6). With these nucleotide concentrations, the rate of dephosphorylation for both sensors would be approximately 20-fold faster than the rate of autophosphorylation (34), suggesting that the majority of both proteins would remain unphosphorylated and thereby stimulate phospho-NarL and phospho-NarP dephosphorylation. It should be noted, however, that nucleotide extraction methods may not accurately reflect available nucleotide pools (29). In addition, other HPKs have displayed significant increases in autophosphorylation rates in the presence of their cognate response regulators (15, 16, 35).
The NarX and NarQ transmitters represent a relatively minor sequence subfamily, termed HPK7 (14, 42), which is characterized in part by the absence of an F box motif and an unconventional G box motif in the CA domain. For other transmitters, residues within these sequence motifs form the “ATP lid” element and make other contacts with ATP. For instance, the crystal structure of the PhoQ protein has revealed that residues of the ATP lid interact with the phosphates of the ATP analog, AMPPNP (21). Furthermore, Ala substitutions in the G box motif of the EnvZ protein significantly decrease ATP binding (44). In vitro autophosphorylation of other HPK7 members has been investigated. UhpB exhibited a relatively low affinity for ATP, with a Km of 250 μM (36). In addition, a pulse of unlabeled ATP to a final concentration of approximately 1.7 mM with 32P-labeled DegS resulted in only a modest loss of labeled protein (22). In these studies, however, the affinity of phospho-UhpB or phospho-DegS for ADP was not measured. Therefore, we currently do not know whether the high affinity of MBP-NarX185 and MBP-NarQ182 for ADP results from the unconventional structure of HPK7 transmitters or reflects a more specific property of these Nar proteins.
Acknowledgments
The MBP-Nar expression constructs were fabricated by Stanly Williams, who, along with Gwénola Simon, participated in early studies to characterize Nar protein phosphoryl transfer reactions. We thank Hsia-Yin Lin and Alice Lin for helpful comments and suggestions. We thank the Mitch Singer laboratory for their assistance with TLC and three anonymous reviewers for their useful suggestions.
This study was supported by Public Health Service grant GM036877 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Anantharaman, V., S. Balaji, and L. Aravind. 2006. The signaling helix: a common functional theme in diverse signaling proteins. Biol. Direct. 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleman, J. A., L. L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 1854872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 18589-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilwes, A. M., C. M. Quezada, L. R. Croal, B. R. Crane, and M. I. Simon. 2001. Nucleotide binding by the histidine kinase CheA. Nat. Struct. Biol. 8353-360. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Buckstein, M. H., J. He, and H. Rubin. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavicchioli, R., R. C. Chiang, L. V. Kalman, and R. P. Gunsalus. 1996. Role of the periplasmic domain of the Escherichia coli NarX sensor-transmitter protein in nitrate-dependent signal transduction and gene regulation. Mol. Microbiol. 21901-911. [DOI] [PubMed] [Google Scholar]
- 8.Cavicchioli, R., I. Schröder, M. Constanti, and R. P. Gunsalus. 1995. The NarX and NarQ sensor-transmitter proteins of Escherichia coli each require two conserved histidines for nitrate-dependent signal transduction to NarL. J. Bacteriol. 1772416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin, A. J., K. L. Tyson, S. J. Busby, and V. Stewart. 1997. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol. Microbiol. 25583-595. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2524-28. [DOI] [PubMed] [Google Scholar]
- 11.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34633-640. [DOI] [PubMed] [Google Scholar]
- 12.Foster, J. E., Q. Sheng, J. R. McClain, M. Bures, T. I. Nicas, K. Henry, M. E. Winkler, and R. Gilmour. 2004. Kinetic and mechanistic analyses of new classes of inhibitors of two-component signal transduction systems using a coupled assay containing HpkA-DrrA from Thermotoga maritima. Microbiology 150885-896. [DOI] [PubMed] [Google Scholar]
- 13.Goh, E. B., P. J. Bledsoe, L. L. Chen, P. Gyaneshwar, V. Stewart, and M. M. Igo. 2005. Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J. Bacteriol. 1874890-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41139-227. [DOI] [PubMed] [Google Scholar]
- 15.Grimshaw, C. E., S. Huang, C. G. Hanstein, M. A. Strauch, D. Burbulys, L. Wang, J. A. Hoch, and J. M. Whiteley. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 371365-1375. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, P., J. A. Peliska, and A. J. Ninfa. 2000. Asymmetry in the autophosphorylation of the two-component regulatory system transmitter protein nitrogen regulator II of Escherichia coli. Biochemistry 395057-5065. [DOI] [PubMed] [Google Scholar]
- 17.Kenney, L. J. 1997. Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch. Biochem. Biophys. 346303-311. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lee, A. I., A. Delgado, and R. P. Gunsalus. 1999. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J. Bacteriol. 1815309-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukat, G. S., B. H. Lee, J. M. Mottonen, A. M. Stock, and J. B. Stock. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 2668348-8354. [PubMed] [Google Scholar]
- 21.Marina, A., C. Mott, A. Auyzenberg, W. A. Hendrickson, and C. D. Waldburger. 2001. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J. Biol. Chem. 27641182-41190. [DOI] [PubMed] [Google Scholar]
- 22.Mukai, K., M. Kawata, and T. Tanaka. 1990. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J. Biol. Chem. 26520000-20006. [PubMed] [Google Scholar]
- 23.Ninfa, E. G., A. Stock, S. Mowbray, and J. Stock. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 2669764-9770. [PubMed] [Google Scholar]
- 24.Nørby, J. G. 1988. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156116-119. [DOI] [PubMed] [Google Scholar]
- 25.Nowak, E., S. Panjikar, J. P. Morth, R. Jordanova, D. I. Svergun, and P. A. Tucker. 2006. Structural and functional aspects of the sensor histidine kinase PrrB from Mycobacterium tuberculosis. Structure 14275-285. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 2671-112. [DOI] [PubMed] [Google Scholar]
- 27.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 1753259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabin, R. S., and V. Stewart. 1992. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 898419-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider, D. A., and R. L. Gourse. 2004. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J. Biol. Chem. 2798262-8268. [DOI] [PubMed] [Google Scholar]
- 30.Schröder. I., C. D. Wolin, R. Cavicchioli, and R. P. Gunsalus. 1994. phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J. Bacteriol. 1764985-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart, R. C. 2005. Analysis of ATP binding to CheA containing tryptophan substitutions near the active site. Biochemistry 444375-4385. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 311-10. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, V., L. L. Chen, and H. C. Wu. 2003. Response to culture aeration mediated by the nitrate and nitrite sensor NarQ of Escherichia coli K-12. Mol. Microbiol. 501391-1399. [DOI] [PubMed] [Google Scholar]
- 34.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 35.Tawa, P., and R. C. Stewart. 1994. Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry 337917-7924. [DOI] [PubMed] [Google Scholar]
- 36.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2001. Glucose-6-phosphate-dependent phosphoryl flow through the Uhp two-component regulatory system. Microbiology 1473345-3352. [DOI] [PubMed] [Google Scholar]
- 37.Walker, M. S., and J. A. DeMoss. 1993. phosphorylation and dephosphorylation catalyzed in vitro by purified components of the nitrate sensing system, NarX and NarL. J. Biol. Chem. 2688391-8393. [PubMed] [Google Scholar]
- 38.Waugh, D. S. 2005. Making the most of affinity tags. Trends Biotechnol. 23316-320. [DOI] [PubMed] [Google Scholar]
- 39.Williams, S. B. 1997. Ph.D. thesis. Cornell University, Ithaca, NY.
- 40.Williams, S. B., and V. Stewart. 1997. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12. Mol. Microbiol. 26911-925. [DOI] [PubMed] [Google Scholar]
- 41.Williams, S. B., and V. Stewart. 1997. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J. Bacteriol. 179721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome. Biol. 3REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, Y., and M. Inouye. 2002. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol. Microbiol. 45653-663. [DOI] [PubMed] [Google Scholar]