Abstract
Device-associated infections involving biofilm remain a persistent clinical problem. We recently reported that four methicillin-resistant Staphylococcus aureus (MRSA) strains formed biofilm independently of the icaADBC-encoded exopolysaccharide. Here, we report that MRSA biofilm development was promoted under mildly acidic growth conditions triggered by the addition of glucose to the growth medium. Loss of sortase, which anchors LPXTG-containing proteins to peptidoglycan, reduced the MRSA biofilm phenotype. Furthermore introduction of mutations in fnbA and fnbB, which encode the LPXTG-anchored multifunctional fibrinogen and fibronectin-binding proteins, FnBPA and FnBPB, reduced biofilm formation by several MRSA strains. However, these mutations had no effect on biofilm formation by methicillin-sensitive S. aureus strains. FnBP-promoted biofilm occurred at the level of intercellular accumulation and not primary attachment. Mutation of fnbA or fnbB alone did not substantially affect biofilm, and expression of either gene alone from a complementing plasmid in fnbA fnbB mutants restored biofilm formation. FnBP-promoted biofilm was dependent on the integrity of SarA but not through effects on fnbA or fnbB transcription. Using plasmid constructs lacking regions of FnBPA to complement an fnbAB mutant revealed that the A domain alone and not the domain required for fibronectin binding could promote biofilm. Additionally, an A-domain N304A substitution that abolished fibrinogen binding did not affect biofilm. These data identify a novel S. aureus biofilm phenotype promoted by FnBPA and FnBPB which is apparently independent of the known ligand-binding activities of these multifunctional surface proteins.
Medical device-associated infections caused by pathogens such as Staphylococcus aureus and Staphylococcus epidermidis involve biofilm and are particularly challenging. Accordingly, such infections complicate a wide variety of surgical and medical procedures and seriously drain healthcare resources. The involvement of antibiotic resistant staphylococci, principally methicillin-resistant S. aureus (MRSA), exacerbates the problem. Understanding how staphylococci colonize and persist in the host and evade immune responses (17) is therefore an important area of research. Over the last decade interest in staphylococcal biofilm mechanisms has also intensified, arising initially from the importance of this phenotype as a virulence determinant in S. epidermidis. However, S. aureus is also an adept biofilm former, an attribute which enhances its already considerable virulence capacity.
Comparison of the biofilm mechanisms employed by S. epidermidis and S. aureus reveals interesting differences (48). Production of the icaADBC-encoded exopolysaccharide polysaccharide intercellular adhesin (PIA) (37) or polymeric N-acetyl-glucosamine (PNAG) (39) is now a well-defined mechanism of biofilm development in both S. epidermidis and S. aureus. Carriage of the ica locus is strongly associated with a biofilm-forming capacity in S. epidermidis and is more commonly found in isolates from device-related infections than commensal strains (16, 71). In contrast, the correlation between ica and biofilm formation in S. aureus is more ambiguous even though this locus is maintained and expressed in almost 100% of S. aureus isolates (14, 31, 49). The role of the ica locus in the S. aureus biofilm phenotype is complex, particularly given that icaADBC-independent biofilm development has been described in this organism (3, 15, 49).
Cell surface components such as teichoic acids (22, 60, 61) and surface proteins including microbial surface components recognizing adhesive matrix molecules (18) also contribute to the S. aureus biofilm phenotype. S. aureus can display on its surface up to 21 different LPXTG proteins anchored to the cell wall by sortase (41, 42). Sortase catalyzes cell wall anchoring by transpetidation to peptidoglycan following cleavage at the LPXTG motif which acts as a sorting signal in the C termini of surface proteins. Deletion of srtA in S. aureus interferes with the normal display of LPXTG surface proteins and results in severe virulence defects (41, 42, 46). The LPXTG-containing surface proteins Bap (biofilm-associated protein) (10, 11, 34, 64) and Aap/SasG (S. epidermidis accumulation-associated protein/S. aureus surface protein G) (9, 26, 58, 59) are known mediators of staphylococcal biofilm development. Furthermore, the major cell wall autolysin Atl promotes primary cell attachment to surfaces and is required for biofilm development in S. epidermidis (24) and possibly S. aureus.
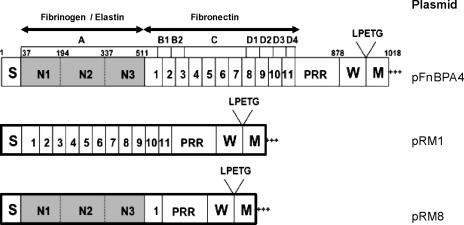
The fibronectin binding proteins A and B (FnBPA and FnBPB) are multifunctional LPXTG-anchored surface proteins that comprise two distinct ligand binding domains (63) (Fig. 1). The N-terminal A domains of FnBPA and FnBPB are 24% identical, and these domains are 24% and 25% identical to the A domain of ClfA, respectively (29, 63, 68). Each binds to the C terminus of the γ chain of fibrinogen by the dock-lock-latch mechanism first reported for the SdrG protein of S. epidermidis (12, 29, 51, 68). The A domains of FnBPA and FnBPB also bind to elastin while the A domain of ClfA does not (13, 29, 56). The A domain of FnBPA is linked to the wall-spanning domain W by 11 tandem repeats of fibronectin binding domains that bind to the N-terminal type I modules of fibronectin by means of the tandem β-zipper mechanism (62).
FIG. 1.
Structural organization of FnBPA from S. aureus 8325-4 and diagrammatic illustration of plasmid constructs lacking regions of FnBPA. Regions B, C, and D (tandem repeats 1 to 11) are required for fibronectin binding. Region A (comprising the subdomains N1, N2, and N3) binds fibrinogen and elastin. The LPXTG motif, required for sortase-mediated anchoring of the protein to the cell wall peptidoglycan, is indicated. The secretory signal sequence (S), wall-spanning (W), and membrane-spanning (M) domains are indicated.
We previously characterized the biofilm phenotypes of 114 MRSA and 98 methicillin-sensitive S. aureus (MSSA) clinical isolates from patients with device-related infections in Beaumont Hospital, Dublin, Ireland. Our studies suggested that glucose-induced biofilms in MRSA isolates are ica independent and involve protein instead of PIA/PNAG (49). In contrast NaCl-induced PIA/PNAG production appears to play a more important role in MSSA biofilm development (49). In this study we have further characterized MRSA biofilm formation by examining the role of cell wall-anchored surface proteins and showed than FnBPs are crucial. The role of SarA in FnBP-mediated biofilm formation and the domain within FnBPA involved in this phenotype were investigated. Our data identify a novel S. aureus protein-dependent biofilm phenotype utilized primarily by MRSA strains that do not produce PIA/PNAG.
MATERIALS AND METHODS
S. aureus strains and plasmids.
The S. aureus strains and the plasmids used in the manipulation of these strains are described in Table 1. The clinical S. aureus isolates used in this study, which have been described previously (49), were collected in Beaumont Hospital, Dublin, Ireland from 1 January 2002 to 30 June 2005.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant detailsa | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| 8325 | NCTC 8325; 11-bp deletion in rsbU | 33 |
| 8325-4 | 8325 derivative cured of prophages; 11-bp deletion in rsbU | 25 |
| SH1000 | Functional rsbU derivative of 8325-4; rsbU+ | 25 |
| RN4220 | Restriction-negative derivative of 8325-4 | 32 |
| DU5972 | sdrCD::Tcr; isogenic mutant of 8325-4 | 47 |
| DU5875 | spa::Tcr; isogenic mutant of 8325-4 | 52 |
| UAMS-240 | sarA::Tcr; isogenic mutant of S. aureus RN6390 | 4 |
| ICA1 | ΔicaADBC::Tcr; isogenic mutant of 8325-4 | 15 |
| DU5883 | fnbA::Tcr, fnbB::Emr; isogenic mutant of 8325-4 | 21 |
| SRTA1 | srtA::Tcr; isogenic mutant of SH1000 | This study |
| CLFA1 | clfA::Tcr; isogenic mutant of RN4220 | This study |
| FNBPB1 | fnbB::Tcr; isogenic mutant of RN4220 | This study |
| DU5944 | clfA::Tn917 clfB::Tcr | 45 |
| SH1000 clfAB fnbAB | clfA::Tn917 clfB::Tcr from DU5944; fnbA::TcrfnbB::Emr from DU5883 | 21, 45 |
| BH1CC | MRSA clinical isolate; biofilm positive; SCCmec type II; MLST type 8; CC8 | 49 |
| BH3(04) | MRSA clinical isolatel; biofilm positive; SCCmec type II; MLST type 8; CC8 | 49 |
| BH4(04) | MRSA clinical isolate; biofilm positive; SCCmec type II; MLST type 8; CC8 | 49 |
| BH10(03) | MRSA clinical isolate; biofilm positive; SCCmec type IV; MLST type 22; CC22 | 49 |
| BH18(04) | MRSA clinical isolate; biofilm positive in BHI glucose medium; SCCmec type IV; MLST type 22; CC22 | 49 |
| BH30(04) | MRSA clinical isolate; biofilm positive; SCCmec type II; MLST type 8; CC8 | 49 |
| BH40(04) | MSSA clinical isolate; biofilm positive; MLST type 1099; CC5 | 49 |
| BH48(04) | MSSA clinical isolate; biofilm positive; MLST type 8; CC8 | 49 |
| BH49(04) | MSSA clinical isolate; biofilm positive; MLST type 6; CC6 | 49 |
| BH51(04) | MSSA clinical isolate; biofilm positive; MLST type 5; CC5 | 49 |
| DAR217 | MRSA reference isolate; biofilm positive; SCCmec type I; MLST type 228; CC5 | 55 |
| DAR22 | MRSA reference isolate; biofilm positive; SCCmec type III; MLST type 5; CC5 | 55 |
| DAR35 | MRSA reference isolate; biofilm positive; SCCmec type II; MLST type 8; CC8 | 55 |
| DAR113 | MRSA reference isolate; biofilm positive; SCCmec type IV; MLST type 22; CC22 | 55 |
| DAR141 | MRSA reference isolate; biofilm positive; SCCmec type II; MLST type 38; CC30 | 55 |
| DAR199 | MRSA reference isolate; biofilm positive; SCCmec II; MLST type 36; CC30 | 55 |
| DAR70 | MRSA reference isolate; biofilm positive; SCCmec type II; MLST type 45; CC45 | 55 |
| E. coli TOPO | recA1 endA1 lac [F′ proAB lac1q Tn10 (Tetr)] | Invitrogen |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kmr Apr | Invitrogen |
| pBluescript | Cloning vector; Apr | Stratagene |
| pT181 | 4.45-kb S. aureus plasmid containing tetA(K) | 30 |
| pBlue-tet | pBluescript containing the tetA gene from pT181 on a 2,352-bp HindIII fragment | This study |
| pBT2 | Temperature-sensitive E. coli-Staphylococcus shuttle vector; Apr (E. coli) Cmr (Staphylococcus) | 5 |
| pLI50 | E. coli-Staphylococcus shuttle vector | 35 |
| pMAD | Shuttle vector for allele replacement; Apr (E. coli) Emr (Staphylococcus); contains bgaB gene encoding a thermostable β-galactosidase | 1 |
| pMAD-Cm | pMAD derivative; Apr (E. coli) Cmr (Staphylococcus) | This study |
| pBT2-tet | 2,352-bp HindIII fragment from pBlue-tet cloned into the HindIII site of pBT2 | This study |
| pBT2-bga | 2,803-bp SmaI-SwaI fragment containing the bgaB gene from pMAD cloned into the SmaI site of pBT2 | This study |
| pSAsrtA1 | 4,570-bp PCR product containing the srtA gene amplified from S. aureus 8325-4 using primers SAsrt1 and SAsrt2 cloned into pCR-Blunt II-TOPO | This study |
| pSAsrtA2 | BamHI-PstI fragment from pSAsrtA1 cloned into the BamHI-PstI sites of pBT2 | This study |
| pSAsrtA3 | 2,361-bp XbaI fragment containing the tetA gene from pBT2-tet cloned into XbaI site in the srtA gene of pSAsrtA2 | This study |
| pSAsrtA4 | 1,080-bp PCR product containing the srtA gene amplified from S. aureus 8325-4 using primers SAsrtAcomp1 and SAsrtAcomp2 cloned into pCR-Blunt II-TOPO | This study |
| pSAsrtA5 | 1,092-bp EcoRI fragment containing the srtA gene from pSAsrtA4 cloned into the EcoRI site of pLI50 | This study |
| pSAclfA1 | 4,420-bp PCR product containing the clfA gene amplified from S. aureus 8325-4 using primers SAclfA1 and SAclfA2 cloned into pCR-Blunt II-TOPO | This study |
| pSAclfA2 | 2,236-bp SwaI-SmaI fragment containing the tetA gene from pBlue-tet cloned into AleI site in the clfA gene of pSAclfA1 | This study |
| pSAclfA3 | 6,656-bp BamHI-XbaI fragment containing the clfA::Tcr allele from pSAclfA2 cloned into the BamHl-Nhel sites of pBT2-bga | This study |
| pSAfnbB1 | 3,100-bp PCR product containing the fnbB gene amplified from S. aureus 8325-4 using primers SAfnbB1 and SAfnbB2 cloned into pCR-Blunt II-TOPO | This study |
| pSAfnbB2 | 2,236-bp SwaI-SmaI fragment containing the tetA gene from pBlue-tet cloned into SwaI site in the fnbB gene of pSAfnbB1 | This study |
| pSAfnbB3 | 5,400-bp BamHI-XbaI fragment containing the fnbB::Tcr allele from pSAfnbB2 cloned into the BamHl-Xbal site of pBT2 | This study |
| pSAfnbB4 | 2,803-bp SmaI-SwaI fragment containing the bgaB gene from pMAD cloned into the SmaI site of pSAfnbB3 | This study |
| pFnBPA4 | Shuttle plasmid derived from pGEM-7Zf (Ampr) and pSK265 (Cmr) carrying the full length fnbA gene | 21 |
| pFnBPB4 | Shuttle plasmid derived from pBluescript (Ampr) and pSK265 (Cmr) carrying the full length fnbB gene | 21 |
| pRM8 | pFnBPA4 expressing FnBPA minus the B, C, and D domains | 40 |
| pRM1 | pFnBPA4 expressing FnBPA minus the A domain | 40 |
MLST, multilocus sequence typing.
Media and growth conditions.
S. aureus strains were grown at 30°C or 37°C on brain-heart infusion (BHI) (Oxoid) medium supplemented, when required, with chloramphenicol (10 to 20 μg/ml), erythromycin, and tetracycline (2.5 μg/ml). BHI broth was supplemented where indicated with 1% glucose, 4% NaCl, or exogenous V8 protease (5 units/ml) (Sigma). Bacteria were grown on Congo red agar to characterize colony morphology as described previously (7). Escherichia coli strains were grown at 37°C on LB medium supplemented, when required, with ampicillin (100 μg/ml) or kanamycin (50 μg/ml).
Genetic techniques.
Genomic and plasmid DNA was prepared using genomic and plasmid DNA purification kits (Sigma). Prior to DNA extraction, cells were pretreated with 5 to 10 μl of a 1-mg/ml concentration of lysostaphin (Ambi Products, New York) in 100 μl of 50 mM EDTA to facilitate subsequent lysis. Restriction and DNA modifying enzymes (Roche, United Kingdom, and New England Biolabs, MA) were used according to the manufacturers ’ instructions. Transformations of plasmid DNA into E. coli and Staphylococcus strains were performed as described previously (7). MWG Biotech, Germany, supplied all oligonucleotide primers used for PCR and reverse transcription-PCR (RT-PCR) (Table 2).
TABLE 2.
Oligonucleotide primers used in this study
| Target gene | Primer name | Primers sequence (5′-3′) |
|---|---|---|
| srtA | SAsrtA1 | CCGTCCATATCTACCGCAAT |
| SAsrtA2 | ATGAAGTCAATGCTGCGATG | |
| srtA | SAsrtAcomp1 | CGGTATACATTGCCGCTTCT |
| SAsrtA4comp2 | ATGCGACACGTTCGTCTATG | |
| clfA | SAclfA1 | ATTCGGCAACAGAAGAGCAT |
| SAclfA2 | TTTCGCCAAGCCAAATTAT | |
| fnbB | SAfnbB1 | AACATGCCGTTGTTTGTTGA |
| SAfnbB2 | AATGTCGGCTTGAAATACGG | |
| fnbB | SAfnbBFOR | CGCGGATCCGTGGAAGTAGAAGAAGGTAG |
| SAfnbBREV | GACGCGTCGACTTAATTAGGTTCCTTTAGTTTATC | |
| fnbA | SAfnbAFOR | CGCGGATCCGGTACAGATGTAACAAGTAAAG |
| SAfnbAREV | GACGCGTCGACTTAATTCGGACCATTTTTCTCATT | |
| gyrB | SAgyrBFOR | TTATGGTGCTGGGCAAATACA |
| SAgyrBREV | CACCATGTAAACCACCAGATA | |
| fnbA | N304AFWD | CGGCTGAACTAGAAATTGCTTTATTTATTGATCC |
| N304AREV | GGATCAATAAATAAAGCAATTTCTAGTTCAGCCG |
Construction of fnbAB::Tcr, sdrCDE::Tcr, spa::Tcr, and sarA::Tcr mutations in clinical isolates.
An sdrCD::Tcr mutation was transduced from S. aureus DU5972 (47) into clinical isolates using phage 80α or 11. Because the sdrCD::Tcr mutation in DU5972 was constructed in 8325-4, a strain that lacks the sdrE gene, using bacteriophage to replace the wild-type sdrCD genes with the sdrCD::Tcr allele from DU5972 cotransduces an sdrE deletion to generate an sdrCDE triple mutation in transductants. The fnbAB::Tcr, spa::Tcr, and sarA::Tcr mutations were transduced into clinical isolates from S. aureus DU5883 (21), DU5875 (52), and UAMS-240 (4), respectively, using phage 80α/501 or 11. Transduction of a ΔicaADBC::Tcr allele into clinical isolates has been described previously (49). Transductants harboring the mutant alleles were selected on BHI agar containing 500 mg/liter sodium citrate and 2.5 μg/ml tetracycline, and the presence of the mutant alleles was confirmed by PCR analysis using the appropriate primers listed in Table 2 (data not shown).
Construction of a srtA::Tcr mutation.
The srtA mutant strain SRTA1 was constructed using the following procedure. A 4,570-bp fragment containing the srtA gene was amplified by PCR with Phusion high-fidelity DNA polymerase (NEB) from S. aureus 8325-4 using the primers SAsrtA1 and SAsrtA2 and cloned into the pCR-Blunt II-TOPO plasmid (Invitrogen) to create pSAsrtA1. The Tcr gene from plasmid pT181 was digested on a 2,352-bp HindIII fragment and subcloned into a temperature-sensitive E. coli-Staphylococcus shuttle vector, pBT2, that had also been digested with HindIII to create pBT2-tet. A BamHI-PstI fragment from pSAsrtA1 was subcloned into pBT2 that had also been digested with BamHI-PstI to create pSAsrtA2. A 2,361-bp XbaI fragment from pBT2-tet containing the Tcr gene was ligated into the XbaI site in the srtA gene of pSAsrtA2 to create pSAsrtA3. The pSAsrtA3 plasmid was first transformed by CaCl2 and heat shock into E. coli, then by electroporation into S. aureus RN4220, and finally into SH1000. Allele replacement of the temperature-sensitive pSAsrtA3 in SH1000 was achieved following repeated growth (three subcultures) at 42°C for 24 h without antibiotic selection and then selection of tetracycline (10 μg/ml)-resistant colonies on BHI agar plates. The colonies were then screened for sensitivity to chloramphenicol to confirm plasmid loss, and PCR analysis was used to verify the presence of the srtA::Tcr allele on the chromosome (data not shown). Bacteriophage 80α was used to transduce the mutation into clinical isolates.
In order to complement the SRTA1 mutant, a 1,080-bp PCR product containing the srtA gene was amplified with Phusion high-fidelity DNA polymerase from S. aureus 8325-4 using the primers SAsrtAcomp1 and SAsrtAcomp2 and cloned initially into pCR-Blunt II-TOPO (pSAsrtA4) and subsequently on an EcoRI fragment into pLI50 to create pSRsrtA5. The pSAsrtA5 plasmid was first transformed by CaCl2 and heat shock into E. coli, then by electroporation into S. aureus RN4220, and finally into SRTA1.
Construction of a clfA::Tcr mutation.
The clfA mutant strain CLFA1 was constructed using the following procedure. A 4,420-bp fragment containing the clfA gene was amplified by PCR with Phusion high-fidelity DNA polymerase (NEB) from 8325-4 using the primers SAclfA1 and SAclfA2 and cloned into the pCR-Blunt II-TOPO plasmid (Invitrogen) to create pSAclfA1. The Tcr gene from plasmid pT181 was digested on a 2,352-bp HindIII fragment and subcloned into pBluescript (Stratagene) to create pBlue-tet. The Tcr gene was subsequently subcloned from pBlue-tet on a 2,236-bp SwaI-SmaI fragment into the AleI site in the clfA gene of pSAclfA1 to create pSAclfA2. A 6,700-bp BamHI-XbaI fragment containing the clfA::Tcr allele from pSAclfA2 was ligated into pBT2-bga digested with BamHI-NheI to create pSAclfA3. The plasmid pBT2-bga contains the bgaB gene from pMAD (1) cloned on a 2,803-bp SmaI-SwaI fragment into the SmaI site of pBT2. The bgaB gene encodes a thermostable β-galactosidase that can be exploited in colorometric blue-white selection during allele replacement mutagenesis (1). The pSAclfA3 plasmid was transformed by CaCl2 and heat shock into E. coli and then by electroporation into S. aureus RN4220.
Allele replacement of the temperature-sensitive pSAclfA3 in RN4220 was achieved following repeated growth (three subcultures) at 42°C for 24 h without antibiotic selection and then selection of tetracycline (10 μg/ml)-resistant colonies on BHI agar plates. Bluo-Gal (50 μg/ml) (Invitrogen) was also included in the medium to facilitate colorimetric (blue-white) screening. White, Tcr colonies were then screened for sensitivity to chloramphenicol to confirm plasmid loss, and PCR analysis was used to verify the presence of the clfA::Tcr allele on the chromosome (data not shown). Bacteriophage 80α was used to transduce the mutation into clinical isolates.
Construction of an fnbB Tcr mutation.
The fnbB mutant strain FNBPB1 was constructed using the following procedure. A 3,100-bp fragment containing the fnbB gene from SH1000 was amplified by PCR with Phusion high-fidelity DNA polymerase (NEB) using the primers SAfnbB1 and SAfnbB2 and cloned into the pCR-Blunt II-TOPO plasmid (Invitrogen) to create pSAfnbB1. The Tcr gene from pBlue-tet was subcloned on a 2,236-bp SwaI-SmaI fragment into the SwaI site in the fnbB gene of pSAfnbB1 to create pSAfnbB2. A 5,400-bp BamHI-XbaI fragment containing the fnbB::Tcr allele from pSAfnbB2 was then ligated into pBT2 digested with BamHI-NheI to create pSAfnbB3. Finally a 2,803-bp SmaI-SwaI fragment containing the bgaB gene from pMAD was ligated into the SmaI site of pSAfnbB3 to create pSAfnbB4. The pSAfnbB4 plasmid was transformed by CaCl2 and heat shock into E. coli and then by electroporation into RN4220.
Allele replacement of the temperature-sensitive pSAfnbB4 in RN4220 was achieved following repeated growth (three subcultures) at 42°C for 24 h without antibiotic selection and then selection of tetracycline (10 μg/ml)-resistant colonies on BHI agar plates. Bluo-Gal (50 μg/ml) (Invitrogen) was also included in the medium to facilitate colorimetric (blue-white) screening. White, Tcr colonies were then screened for sensitivity to chloramphenicol to confirm plasmid loss, and PCR analysis was used to verify the presence of the fnbB::Tcr allele on the chromosome (data not shown). Bacteriophage 80α was used to transduce the mutation into clinical isolates.
Biofilm assays.
Semiquantitative measurements of biofilm formation were determined using Nunclon tissue culture-treated (delta surface) 96-well polystyrene plates (Nunc, Denmark), based on the methods of Christensen et al. (6) and Ziebuhr et al. (71) as described previously (7, 49). Bacteria were grown in individual wells of 96-well plates at 37°C in BHI medium or BHI medium supplemented with 4% NaCl or 1% glucose. After 24 h of growth the plates were washed vigorously. This involved three rounds of plunging the plates into a large volume of distilled water and decanting to remove unattached bacteria. The plates were subsequently dried for 1 h at 60°C prior to staining with a 0.4% crystal violet solution. The absorbance of the adhered, stained biofilms was measured at 492 nm using a microtiter plate reader. Each strain was tested at least three times, and average results are presented. A biofilm-positive phenotype was defined as an A492 of ≥0.17.
The laboratory strain S. aureus RN4220 and its isogenic ΔicaADBC mutant were periodically included in biofilm assays as positive and negative controls, respectively. In our assays, and as reported previously, S. aureus RN4220 is a reliably strong biofilm former (A492 of ≈2 to 4) in BHI, BHI NaCl, and BHI glucose media (7, 49).
Biofilm stability against proteinase K (Sigma) or sodium-metaperiodate (Sigma) treatment was tested as described previously (38, 59).
Biofilm formation was also examined under flow conditions. Bacteria were grown in Biosurface Technologies FC93 flow cells (Bozeman, MT) containing BHI medium supplemented with 4% NaCl at a flow rate of 500 μl/min. Overnight cultures were adjusted to an A600 of 1.0 prior to inoculation; the bacteria were allowed to adhere to the flow cells for 120 min before the flow was turned on, and the biofilms were allowed to mature for 24 h.
Primary attachment assay on polystyrene.
Primary attachment assays were performed based on the method of Lim et al. (36). Briefly, bacteria from an overnight incubation at 37°C in BHI medium supplemented with 1% glucose were diluted in the same medium to give approximately 300 CFU/100 μl. Bacterial concentrations were determined by bacterial plate counts run in parallel to the attachment assay. A total of 100 μl of the adjusted suspension was spread on Nunclon tissue culture-treated (delta surface) petri dishes (Nunc, Denmark). After incubation at 37°C for 30 min, the petri dishes were rinsed gently with 5 ml of sterile phosphate-buffered saline (PBS; pH 7.5) three times and covered with 15 ml of molten 0.8% trichostatin A cooled to 48°C. Primary attachment was expressed as a percentage of CFU remaining on the petri dishes after washing. Each experiment was repeated three times.
RNA purification and analysis.
RNA purification, RT-PCR, and analysis of RT-PCR data were performed as described previously (7, 8, 15, 49). The constitutively expressed gyrB gene was used as an internal standard in these experiments using the primers described by Goerke et al. (20). The primers used to amplify the fnbA, fnbB, and gyrB transcripts are described in Table 2. Reverse transcription was performed at 50°C for 30 min, terminated at 94°C for 15 min, and followed by 20 cycles of PCR for gyrB and up to 30 cycles of PCR for fnbA and fnbB amplification.
PIA/PNAG assays.
PIA/PNAG assays were performed as described elsewhere (27). Briefly 5-ml overnight cultures (approximately 5 × 109 bacteria) were collected by centrifugation, resuspended in 200 to 500 μl of 0.5 M EDTA, and boiled for 5 min. The cell debris was again centrifuged, and the supernatant was treated with 20 μg of proteinase K at 65°C for 1 h. The proteinase K was inactivated by boiling for 5 min, and the samples were diluted as appropriate before being applied onto nitrocellulose (prewetted in Tris-buffered saline [TBS]) using a vacuum blotter. The blots were dried, rewet in TBS, and blocked for 1 h in 1% bovine serum albumin (BSA). The primary antibody (1:5,000 dilution of rabbit anti-PIA/PNAG [a kind gift from Tomas Maira Litran and Gerald Pier] in TBS-Tween and 0.1% BSA) was then applied to the membrane for 1 h. Horseradish peroxidase-linked anti-rabbit immunoglobulin G secondary antibody (1:5,000 dilution in TBS-Tween plus 0.1% skim milk) was then incubated with the membrane for 1 h. A chemiluminescence kit (Amersham) was used to generate light via the horseradish peroxidase-catalyzed breakdown of lumina, which was detected using a Bio-Rad Fluor-S Max charge-coupled-device camera system.
Fibronectin and fibrinogen binding assays.
Fibronectin and fibrinogen binding assays were performed as described previously (23, 50) with the following modifications. Doubling dilutions of human fibronectin (Calbiochem) (0 to 20 μg/ml) or fibrinogen (Enzyme Research Laboratories) (0 to 10 μg/ml) were added in 100-μl volumes to 96-well flat-bottomed polystyrene microtiter plates (Sarstedt) and incubated overnight at 4°C. The microtiter plates were then washed three times in sterile PBS and blocked for 2 h at 37°C with 5% BSA (Sigma). Bacterial cultures were grown at 37°C in BHI broth supplemented with 1% glucose to an A600 of ≈0.5 before being washed and resuspended in sterile PBS, and the cell density was adjusted to an A600 of 1.0. The individual wells of the fibronectin or fibrinogen-coated microtiter plates were inoculated with 100-μl volumes of the bacterial cell suspensions and incubated at 37°C for 2 h. The wells were then washed three times with sterile PBS and fixed with 25% formaldehyde. Wells containing PBS without bacteria were used as a negative control in all assays. Finally, wells were washed once with sterile PBS, stained with 0.4% crystal violet, and quantified by measuring the A570 of crystal violet, which had been solubilized in 5% acetic acid. The binding of each isolate to fibronectin or fibrinogen was measured at least three times.
Antibodies.
To obtain antibodies reactive against the A domain of FnBPA, specific-pathogen-free rabbits were immunized with the A domain of recombinant FnBPA containing residues 37 to 544 (FnBPA37-544), and immunoglobulin was purified as described previously (56).
Site directed mutagenesis.
Oligonucleotide-directed mutagenesis (Quikchange, Stratagene) was employed to mutate the fnbA gene. The plasmid pRM8 (Table 1) carrying the 5′ region of the fnbA gene encoding the A domain was used as a template for the reactions. The complementary oligonucleotides used to construct the N304A mutation, which has previously been implicated in elastin and fibrinogen binding (29), were N304AFwd and N304Arev (Table 2). The mutagenesis resulted in the loss of an AseI restriction site which, together with DNA sequencing, was used to confirm the presence of the desired mutation (data not shown) before the fragment was transformed by electroporation first into S. aureus RN4220 and subsequently into clinical isolates.
Statistical analyses.
Two-tailed, two-sample equal variance Student's t tests (Microsoft Excel 2007) were used to determine statistically significant differences in biofilm-forming capacity and binding to immobilized fibronectin by S. aureus strains.
RESULTS
Environmental regulation of biofilm development in MRSA.
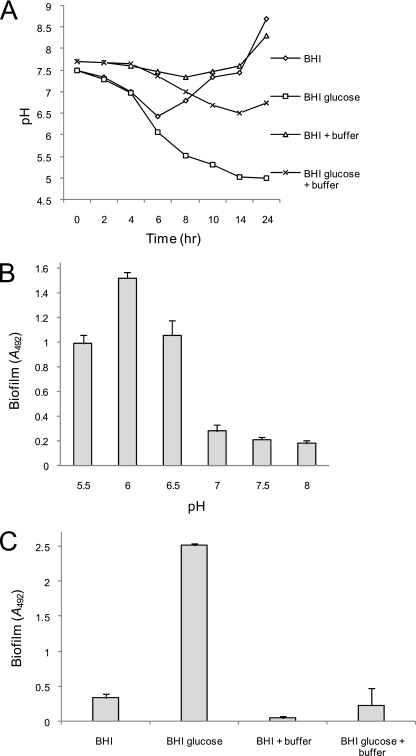
We recently reported evidence for ica-independent biofilm development by MRSA strains (49). Here, we extended our analysis of this phenomenon using a previously described MRSA isolate associated with a device-related infection, BH1CC (type II staphylococcal cassette chromosome mec [SCCmec]; clonal complex 8 [CC8]), (15, 49). Unlike ica-dependent biofilm development, which is triggered by osmotic stress, MRSA biofilm formation is induced primarily in broth supplemented with glucose. Glucose concentrations above 0.3% significantly induced biofilm formation in 96-well plate assays (P < 0.0001) compared to lower concentrations (data not shown). To investigate the effect of glucose on growth, we supplemented BHI broth with 1% glucose. No differences were measured in the rate of growth of BH1CC in exponential phase and the final cell densities of the cultures grown in BHI and BHI glucose media (data not shown). Consistent with the findings of Rice et al. (54), who recently reported that metabolism of glucose and other carbon sources leads to the accumulation of acetic acid, the final pH of BH1CC grown in BHI broth with glucose was less than 5 compared to ca 8.5 in BHI broth without glucose (Fig. 2A). To test the hypothesis that low pH triggers biofilm formation, we reduced the starting pH of BH1CC cultures using acetic acid. Biofilm development by BH1CC was significantly enhanced with a starting pH of 5.5, 6, or 6.5 compared to 7.0, 7.5, or 8.5 (P < 0.0001) (Fig. 2B) but was not affected by the addition of sodium acetate (data not shown). Using 200 mM disodium phosphate to buffer the medium (Fig. 2A), biofilm development by BH1CC in BHI-glucose medium was dramatically inhibited (P < 0.0001) (Fig. 2C). In addition, weak biofilm development by BH1CC in BHI medium alone, which correlates with a drop in pH during exponential growth (Fig. 2A), was inhibited by the addition of buffer (P < 0.0001) (Fig. 2C). Buffering of BHI-glucose medium was also associated with a significant inhibition of biofilm development by the MRSA isolates BH18(04) (type IV SCCmec; CC22), BH10(03) (type IV SCCmec; CC22), BH4(04) (type II SCCmec; CC8), and BH30(04) (type II SCCmec; CC8) (49) (data not shown) (for convenience, these strains are referred to as BH18, BH10, BH4, and BH30, respectively, throughout the text). These data strongly suggest that MRSA biofilm development is activated by low pH in at least two distinct lineages of MRSA.
FIG. 2.
Low pH-induced MRSA biofilm formation is triggered by supplementing the growth medium with 1% glucose. (A) Measurement of culture pH (over 24 h) of BH1CC grown in BHI and BHI-glucose media, BHI medium supplemented with 200 mM disodium phosphate (buffer), and BHI-glucose medium supplemented with 200 mM disodium phosphate (buffer). (B) Activation of BH1CC biofilm development under mildly acidic growth conditions. The starting pH of BHI medium was altered using acetic acid. (C) Biofilm development by BH1CC grown in BHI and BHI-glucose media, BHI medium supplemented with 200 mM disodium phosphate (buffer), and BHI-glucose medium supplemented with 200 mM disodium phosphate (buffer). Biofilm formation after growth for 24 h in tissue culture-treated 96-well plates was measured three times, and standard deviations are indicated.
Contribution of sortase to the biofilm phenotype of MRSA and MSSA strains.
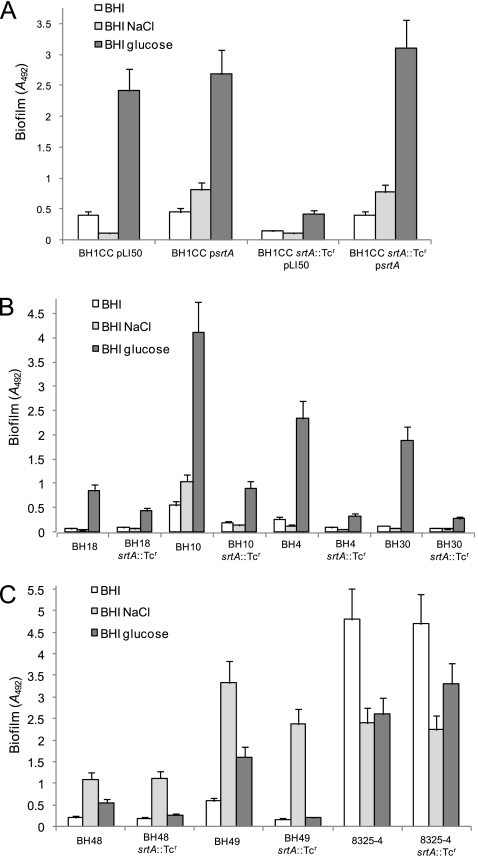
We recently reported that glucose/low pH-induced MRSA biofilm development was independent of the ica operon and that proteinase K dispersed mature MRSA biofilms (49), suggesting that cell surface proteins may play an important role. To investigate the involvement of surface proteins, we first examined the role of sortase. A srtA::Tcr mutation was constructed by allele replacement in the laboratory strain SH1000 and transduced into BH1CC (Fig. 3). The srtA mutant exhibited an approximately sixfold reduction in biofilm density in BHI medium supplemented with 1% glucose compared to wild-type (P < 0.0001) (Fig. 3A). The biofilm defect of the BH1CC srtA::Tcr mutant was complemented by the wild-type srtA gene carried on plasmid pLI50 (P < 0.0001) (Fig. 3A). Transduction of srtA::Tcr into strains BH18, BH10, BH4, and BH30 revealed that sortase was also required for biofilm development in these strains (Fig. 3B). We previously demonstrated that deletion of the icaADBC locus had no significant effect on biofilm in BH1CC, BH18, BH10, BH4, and BH30 (49).
FIG. 3.
Contribution of the srtA gene to biofilm regulation in S. aureus. (A) Impact of an srtA::Tcr mutation and carriage of multicopy psrtA (srtA) and pLI50 (control) plasmids on biofilm development in the MRSA isolate BH1CC. (B and C) Impact of an srtA::Tcr mutation on biofilm regulation in the MRSA isolates BH18, BH10, BH4, and BH38 (B) and the MSSA isolates BH48, BH49, and 8325-4 (C). The strains were grown in BHI medium or in BHI medium supplemented with 4% NaCl or 1% glucose at 37°C for 24 h. Biofilm formation in tissue culture-treated 96-well plates was measured three times, and standard deviations, which were less than 15%, are indicated.
Our previous studies revealed that biofilm development in the laboratory strains RN4220 and 8325-4 and the MSSA clinical isolates BH48(04) and BH49(04) (both from CC8 and referred to as BH48 and BH49 for convenience) was ica dependent (49). Therefore, to investigate the role of sortase in PIA/PNAG-mediated S. aureus biofilm development, the srtA::Tcr mutation was transduced into strains BH48, BH49, and 8325-4. In all strains except BH49, NaCl-induced, PIA/PNAG-dependent biofilm was largely unaffected by the srtA mutation (Fig. 3C). Biofilm formation by BH49 grown in BHI-glucose medium may involve a surface protein adhesin because it was reduced by the srtA mutation (Fig. 3C). However, the srtA::Tcr mutation had no impact on biofilm development by 8325-4 grown in BHI-glucose medium (Fig. 3C), suggesting involvement of a non-LPXTG-anchored surface protein.
In addition, the srtA::Tcr mutation did not affect primary attachment of BH1CC to polystyrene (data not shown), suggesting that this mutation interferes with the cell-to-cell accumulation phase of biofilm development and not initial adhesion of bacterial cells and the plastic surface.
Taken together, these data suggest that sortase-anchored LPXTG surface proteins play an important role in the accumulation phase of biofilm formation by several MRSA strains, a finding consistent with our previous data that the MRSA biofilm matrix is composed of protein(s) (49). In contrast, NaCl-induced PIA/PNAG-mediated biofilm development in MSSA isolates is srtA independent.
Contribution of surface proteins to the MRSA biofilm phenotype.
The negative impact of a srtA mutation on MRSA biofilm prompted us to investigate which cell wall-anchored proteins are involved. A total of 21 S. aureus surface proteins are predicted to contain LPXTG motifs (19, 57). Previously constructed sdrCDE::Tcr (47), spa::Tcr (52), and fnbAB::Tcr (21) mutations, together with a clfA::Tcr mutation constructed in this study, were transduced into selected MRSA isolates that were amenable to genetic manipulation. Transduction of the sdrCDE::Tcr and clfA::Tcr mutations into the MRSA isolates BH1CC, BH10, BH4, and BH30 did not impair biofilm formation in BHI, BHI-NaCl, or BHI-glucose medium (data not shown). We were unable to transduce the spa::Tcr mutation into BH1CC and BH10, but its introduction into BH30 and BH4 had no impact on biofilm under any of the growth conditions (data not shown).
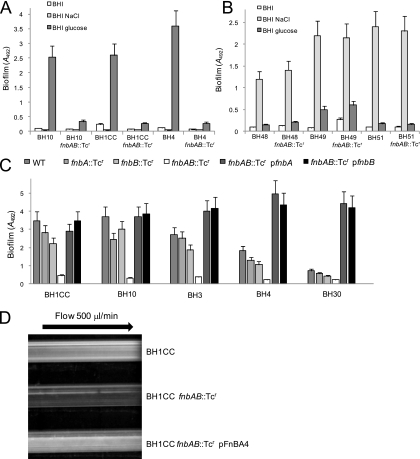
In contrast, glucose-induced biofilm in the MRSA isolates BH10, BH1CC, and BH4 was dramatically reduced following introduction of the fnbA fnbB double mutation (Fig. 4A), whereas the same mutation had no significant impact on NaCl-promoted biofilm in the MSSA isolates BH48, BH49, and BH51(04) (referred to as BH51 for convenience) (Fig. 4B). In order to determine if one or both of the fnbA and fnbB genes are required for glucose-induced MRSA biofilm, we examined the effect of fnbA::Tcr and fnbB::Tcr single mutations in isolates BH1CC, BH10, BH3, BH4, and BH30 (Fig. 4C). Mutation of either the fnbA or fnbB gene alone did not substantially impair glucose-induced biofilm formation compared to the impact of an fnbAB double mutation (Fig. 4C). The biofilm defect of the fnbAB double mutation in each of the MRSA strains was complemented by introducing plasmids carrying either the wild-type fnbA or fnbB gene alone (Fig. 4C). These data strongly suggest that both the FnBPA and FnBPB proteins mediate glucose-induced biofilm development in MRSA. Consistent with our analysis of the srtA::Tcr mutant, primary attachment assays revealed no significant differences between BH1CC and its isogenic fnbAB mutant (data not shown), suggesting that FnBPA and FnBPB are involved in intercellular accumulation and not initial interactions with surfaces. In order to investigate FnBP-promoted biofilm under flow conditions, a Biosurface Technologies FC93 (Bozeman, MT) flow cell system was also used to examine biofilm development by BH1CC in BHI-glucose medium. Consistent with the static biofilm assays, the fnbAB double mutation reduced biofilm formation after growth for 24 h in the flow cells, and the defect was complemented by plasmid pFnBPA4 carrying the wild-type fnbA gene (Fig. 4D).
FIG. 4.
Contribution of the fnbA and fnbB genes to biofilm regulation in S. aureus. (A and B) Impact of a fnbAB::Tcr mutation on biofilm regulation in the MRSA isolates BH10, BH1CC, and BH4 (A) and the MSSA isolates BH48, BH49, and BH51 (B). The strains were grown in BHI, BHI-4% NaCl, and BHI-1% glucose media at 37°C for 24 h. (C) Impact of fnbA and fnbB single and double mutations and carriage of multicopy fnbA (pFnBPA4) and fnbB (pFnBPB4) plasmids on biofilm development in the MRSA isolates BH1CC, BH10, BH3, BH4, and B30. The strains were grown in BHI medium supplemented with 1% glucose at 37°C for 24 h. Biofilm formation in tissue culture-treated 96-well plates was measured three times, and standard deviations, which were less than 20%, are indicated where appropriate. (D) Biofilm development under flow conditions by the MRSA strains BH1CC (vector control), BH1CC fnbAB::Tcr, and BH1CC fnbAB::Tcr (pFnBPA4) using a Biosurface Technologies flow system. Strains were grown for 24 h at 37°C in BHI broth supplemented with 1% glucose, and the flow cells were photographed after 24 h.
Analysis of FnBPA- and FnBPB-induced biofilms.
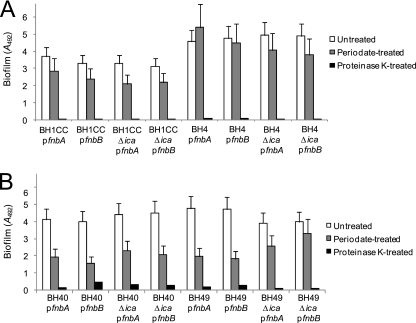
Similar to MRSA, expression of the fnbA or fnbB gene from multicopy plasmids can trigger glucose-induced biofilm development in the MSSA strains BH40(04) (referred to as BH40 for convenience) and BH49 (data not shown). To investigate the extracellular matrix of FnBP-induced biofilms, we examined the ability of sodium metaperiodate and proteinase K to disperse biofilms formed by the MRSA strains BH1CC and BH4 and the MSSA strains BH40 and BH49 carrying fnbA and fnbB plasmids grown in BHI-glucose medium. Sodium metaperiodate had little effect on the MRSA biofilms, whereas they were dispersed by treatment with proteinase K (Fig. 5A). Similar results were observed following treatment of the MSSA biofilms, which were also substantially dispersed with proteinase K and only partially dispersed with sodium metaperiodate. These data are in contrast to our previous findings that NaCl-induced biofilms in MSSA isolates were dispersed with sodium metaperiodate and not proteinase K (49). These data further suggest that the FnBPA and FnBPB surface proteins can promote biofilm formation by S. aureus strains under mildly acidic growth conditions.
FIG. 5.
Susceptibility of fnbA- and fnbB-induced MRSA (A) and MSSA (B) biofilms to treatment with sodium metaperiodate and proteinase K. All biofilms of strains carrying multicopy fnbA (pFnBPA4) and fnbB (pFnBPA4) plasmids were grown in BHI medium supplemented with 1% glucose at 37°C for 24 h prior to treatment.
To investigate if the FnBPA and FnBPB proteins cooperate with PIA/PNAG to mediate biofilm formation (particularly in MSSA isolates), we examined the impact of carriage of the fnbA or fnbB gene on multicopy plasmids in icaADBC deletion mutants of the MRSA isolates BH1CC and BH4 and the MSSA isolates BH40 and BH49. Biofilm formation associated with carriage of the fnbA or fnbB gene on multicopy plasmids was independent of the ica locus (Fig. 5A and B). Furthermore, FnBPA- and FnBPB-induced biofilms in ica mutants of all four isolates were substantially dispersed with proteinase K and to a much lesser extent by sodium metaperiodate (Fig. 5A and B). Apparently, both FnBPA and FnBPB are capable of mediating biofilm formation in MRSA and MSSA strains independently of PIA/PNAG. Furthermore, these findings suggest that although glucose- or acid-induced FnBP-mediated biofilm development is more commonly associated with MRSA isolates, FnBPA and FnBPB may also contribute to biofilm formation by MSSA under appropriate conditions.
Contribution of SarA to FnBP-mediated biofilm in S. aureus.
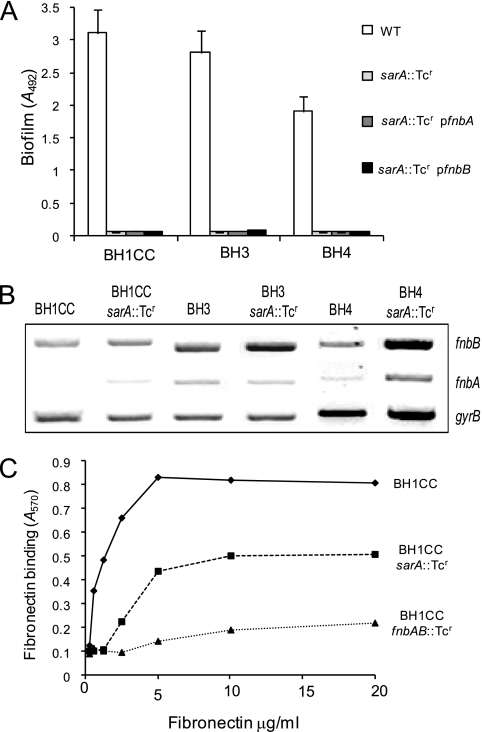
Mutation of the sarA locus resulted in a biofilm-negative phenotype in both MRSA and MSSA (2, 49, 65). Consistent with our earlier studies, sarA mutants of the MRSA strains BH1CC, BH3, and BH4 did not form low pH-induced biofilm in BHI-glucose medium (Fig. 6A). Overexpression of the fnbA or fnbB gene was insufficient to restore biofilm (Fig. 6A). Transcription of the fnbA and fnbB genes was not diminished by the sarA::Tcr mutation as measured by RT-PCR (Fig. 6B). The transcripts were either unchanged (BH1CC and BH3) or were higher (BH4) in the sarA mutant (Fig. 6B). To investigate if lack of sarA might impair the function of FnBPA and FnBPB, we measured the ability of BH1CC and its isogenic sarA and fnbAB mutants to adhere to immobilized fibronectin. Adhesion was reduced by approximately 50% in the sarA mutant (Fig. 6C) compared to the fnbAB mutant, which was completely defective (Fig. 6C).
FIG. 6.
Contribution of sarA to MRSA biofilm development, fnbA and fnbB transcription, and binding to immobilized human fibronectin. (A) Biofilm development by the MRSA isolates BH1CC, BH3, and BH4 and derivatives lacking the sarA gene and the sarA mutants with the multicopy fnbA (pFnBPA4) or fnbB (pFnBPB4) plasmid grown at 37°C for 24 h in BHI medium supplemented with 1% glucose. Biofilm formation was measured at least three times, and standard deviations, which were less than 20%, are indicated. (B) Comparative measurement of fnbA, fnbB, and gyrB (control) transcription in the MRSA isolates BH1CC, BH3, and BH4 and their isogenic ΔsarA mutants. RT-PCR analysis was performed on total RNA extracted from cultures grown at 37°C or 30°C to an A600 of 2.0 in BHI medium supplemented with 1% glucose. These experiments were performed separately for each strain and its isogenic mutant, and representative results have been assembled. (C) Binding of the MRSA isolate BH1CC and BH1CC derivatives harboring mutations in the sarA and fnbAB loci to immobilized human fibronectin. Values represent the mean of triplicate wells.
Inhibition of MRSA biofilm by V8 protease.
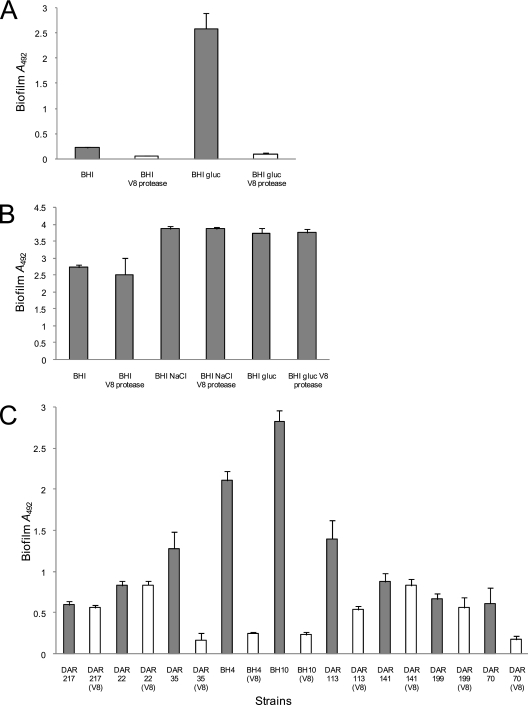
The cell wall-anchored FnBPs are extremely sensitive to degradation by the sspA-encoded V8 serine protease (43). Growth of MRSA strains in the presence of excess V8 protease might inhibit low pH-induced biofilm development, whereas NaCl-induced MSSA biofilm development most likely would be unaffected. To test this hypothesis, we measured the capacity of BH1CC and the laboratory strain RN4220 to form biofilm in the presence and absence of V8 protease. Addition of V8 protease to BH1CC cultures significantly reduced biofilm development in BHI (P < 0.0001) and BHI-glucose (P < 0.0001) (Fig. 7A) media and binding to immobilized fibronectin (P < 0.001) (data not shown), which is consistent with a role for the FnBPs in MRSA biofilm. In contrast, biofilm formation by RN4220 in BHI (P = 0.26), BHI-NaCl (P = 0.97), and BHI-glucose (P = 0.76) media was unaffected by V8 protease (Fig. 7B), whereas binding to immobilized fibronectin was strongly reduced (P < 0.001) (data not shown). We also examined the impact of V8 protease on biofilm development by the MRSA isolates BH18, BH10, BH4, and BH30 and the MSSA isolates BH40, BH48, BH49, and BH51. Like BH1CC, low pH-induced biofilm formation by the MRSA isolates was inhibited by V8 protease whereas NaCl-induced biofilm formation by MSSA isolates was unaffected (data not shown).
FIG. 7.
Susceptibility of S. aureus biofilm development to treatment with exogenous V8 protease. (A) Biofilm development by the MRSA isolate BH1CC grown in BHI and BHI-glucose media with or without exogenous V8 protease (5 units/ml). (B) Biofilm development by the laboratory MSSA strain RN4220 grown in BHI, BHI-NaCl, and BHI-glucose media with or without exogenous V8 protease (5 units/ml). (C) Biofilm development by MRSA isolates DAR217 (CC5), DAR22 (CC5), DAR35 (CC8), BH4 (CC8), BH10 (CC22), DAR113 (CC22), DAR141 (CC30), DAR199 (CC30), and DAR70 (CC45) grown in BHI-glucose medium with or without exogenous V8 protease (5 units/ml). Biofilm formation was measured at least three times, and standard deviations, which were less than 20%, are indicated.
The MRSA strains in our local collection are from a restricted number of lineages (CC8 and CC22), and the evidence for FnBP-mediated biofilm in MRSA strains is limited to a number of clinical isolates that are amenable to genetic manipulation (49). Therefore, we also tested the sensitivity of biofilm formation to V8 protease in biofilm-positive (A600 of >0.5) isolates from genetically diverse MRSA and MSSA strains (55). Consistent with our data from RN4220, biofilm development in BHI-NaCl medium by two MSSA isolates from CC5, five isolates from CC8, two isolates from CC22, and three isolates from CC45 was unaffected by V8 protease (data not shown). Similar to BH1CC, biofilm formation by MRSA isolates DAR35 and BH4 from CC8 (P < 0.0001), BH10 and DAR113 from CC22 (P < 0.0001), and DAR70 from CC45 (P < 0.0001) was significantly reduced by supplementation with V8 protease (Fig. 7C). However, V8 protease did not inhibit biofilm development by MRSA isolates DAR217 and DAR22 from CC5 and isolates DAR141 and DAR199 from CC30 (Fig. 7C). Binding by all of the above isolates to immobilized fibronectin was significantly reduced by V8 protease (P < 0.001) (data not shown). These data are consistent with the idea that FnBP-mediated biofilm development is a common MRSA trait from CC8, CC22, and CC45 lineages whereas biofilm formation by CC5 and CC30 might not be FnBP mediated. However, studies with larger numbers of MRSA isolates are required, and a detailed analysis of the role of FnBPs is required. Also the mechanisms of CC5 and CC30 MRSA biofilm formation require further investigation.
Domains of FnBPs involved in biofilm formation by MRSA strains.
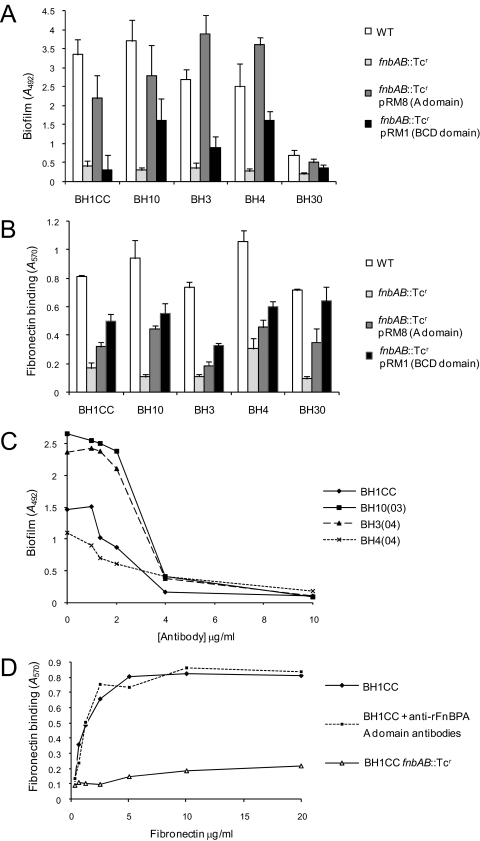
FnBPA is a multifunctional protein with an A domain that binds to fibrinogen and elastin (29, 68) and with BCD domains (tandem repeats 1 to 11) that bind to fibronectin (62). To identify the domain(s) of FnBPA required for biofilm development, we tested the ability of plasmids pRM8 (expressing the FnBPA A domain and one fibronectin binding domain) and pRM1 (expressing only the FnBPA BCD domains) (Fig. 1) to promote biofilm formation in fnbAB::Tcr mutants of the MRSA strains BH1CC, BH10, BH3, BH4, and BH30. Expression of the A domain of FnBPA promoted biofilm formation close to or greater than wild-type levels in all five strains compared to expression of the BCD domains, which did so poorly (Fig. 8A). To determine if the truncated protein lacking the A domain but carrying the BCD domains expressed by pRM1 was functional in the clinical isolates, we tested the ability of strains to adhere to fibronectin. Binding occurred in all strains but at a lower level than wild type (Fig. 8B). Plasmid pRM8 promoted a lower level of adhesion to fibronectin than pRM1, presumably due to the presence of only a single fibronectin binding domain (Fig. 8B).
FIG. 8.
Role of FnBPA structural domains in MRSA biofilm development and binding to immobilized human fibronectin. (A) Complementation of biofilm development in MRSA fnbAB::Tcr mutants by the A (pRM8) or BCD (pRM1) domains of the FnBPA protein. MRSA isolates BH1CC, BH10, BH3, BH4, and BH30 were grown in BHI medium supplemented with 1% glucose at 37°C for 24 h. (B) Complementation of fibronectin binding activity in MRSA fnbAB::Tcr mutants by the A (pRM8) or BCD (pRM1) domains of the FnBPA protein. Cells of BH1CC, BH10, BH3, BH4, and BH30 were tested for their ability to bind to immobilized human fibronectin (5 μg/ml). (C) Inhibition of MRSA biofilm development by anti-FnBPA37-544 A-domain antibodies. MRSA isolates BH1CC, BH10, BH4, and BH3 were grown in BHI medium supplemented with 1% glucose and various concentrations of polyclonal anti-FnBPA A-domain antibodies at 37°C for 24 h. Values represent the mean of triplicate wells. (D) Binding of the MRSA isolate BH1CC to immobilized human fibronectin in the presence and absence of polyclonal anti-FnBPA A-domain antibodies. BH1CC fnbAB::Tcr was used as a negative control. Values represent the mean of triplicate wells.
To further investigate the role of the A domain of FnBPA in biofilm formation, we measured the ability of anti-FnBPA37-544 A-domain antibodies (56) to inhibit biofilm development by BH1CC, BH10, BH3, and BH4 grown in BHI-glucose medium. The Fab fragment (56) was used in order to reduce the steric effects of the large immunoglobulin G molecule's binding to the cell surface-expressed FnBPA A domain and also to eliminate nonimmune binding to protein A, which might interfere. The antibodies inhibited biofilm in all four isolates in a concentration-dependent manner (Fig. 8C) whereas they had no effect on fibronectin binding by BH1CC (Fig. 8D). The extent of biofilm inhibition by the antibodies was somewhat unexpected, given that they do not cross-react with FnBPB (unpublished data). This suggests that FnBPB may also be sterically blocked by the FnBPA antibodies.
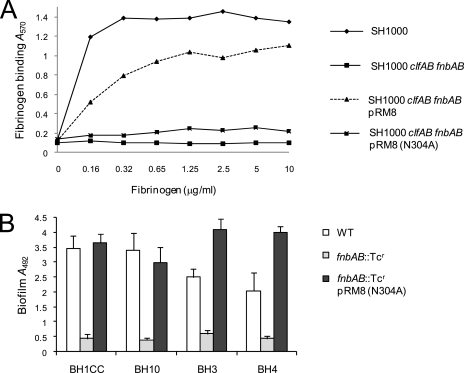
FnBPA promotes bacterial adherence to immobilized fibrinogen and elastin via interactions reliant on the N304 and F306 residues of the A-domain N2 subdomain (Fig. 1) (29, 56, 68). A pRM8 derivative carrying an N304A substitution, which reduced fibrinogen binding activity (Fig. 9A), retained the capacity to complement the biofilm defect in four MRSA fnbAB mutants (Fig. 9B). These data indicate that FnBPA- and FnBPB-mediated biofilm development in S. aureus is a novel function of the A domain of these cell wall-anchored proteins. Interestingly, the amino terminal 500 amino acids of ClfA (including the A domain) shares approximately 25% identity and 42% similarity with the corresponding region of FnBPA. However, as described above, mutation of the clfA gene was not accompanied by a reduction in biofilm (data not shown), suggesting that specific and perhaps unique characteristics of the A domains of FnBPA and FnBPB are required.
FIG. 9.
Binding to human immobilized fibrinogen (A) and complementation of biofilm development (B) in MRSA fnbAB::Tcr mutants by pRM8 (FnBPA A domain) carrying the N304A amino acid substitution. MRSA isolates BH1CC, BH10, BH3, and BH4 were grown in BHI medium supplemented with 1% glucose at 37°C for 24 h. Standard deviations are indicated.
DISCUSSION
Infections involving S. aureus remain a serious threat to hospitalized patients following trauma or surgery or with underlying medical conditions. A significant portion of these infections are caused by MRSA, leading to further complications in treatment. S. aureus expresses an array of enzymes and toxins that subvert the host immune response and enhance its virulence. It also displays a striking capacity to colonize implanted medical devices and form biofilm, a trait more commonly associated with its less pathogenic relative S. epidermidis. Indeed, together coagulase-negative staphylococci and S. aureus are responsible for the majority of device-related infections.
In recent years, we have begun to characterize the biofilm phenotype in clinical isolates of MRSA and MSSA isolated from device-related infections (14, 15, 49). These studies revealed that MSSA isolates are more likely to produce an icaADBC-dependent, PIA/PNAG-mediated biofilm activated by osmotic stress, for example, in medium supplemented with NaCl (15, 49). In contrast, addition of glucose to the growth medium triggered the formation of biofilm by some MRSA strains by an icaADBC-independent mechanism involving protein(s) (15, 49).
The addition of glucose to BHI medium results in a lowering of the pH during growth, which seems to promote biofilm development. Reducing the pH prior to growth allowed biofilm formation without the addition of glucose. Given that many host niches colonized by S. aureus are mildly acidic (e.g., skin, vagina, urinary tract, and mouth) (69), low pH-induced MRSA biofilm development is likely to be of physiological importance. Interestingly, glucose and a lowered pH have been shown to decrease expression of the accessory gene regulator (agr) locus (53), which is known to repress biofilm development (66, 67). Enhanced biofilm formation by agr mutants has been attributed to decreased levels of the RNAIII-encoded δ-toxin, which has surfactant properties (66, 67). However, we have previously reported that mutation of the agr locus in 13 MRSA and 8 MSSA clinical isolates increased biofilm in only 5 of 21 (23%) isolates and had no significant impact on biofilm in the remaining 16 isolates (49). These data make it difficult to correlate modulation of agr locus activity alone with enhanced biofilm development in the presence of glucose.
Mutation of sortase, which anchors LPXTG surface proteins to the cell wall (19, 57), reduced biofilm formation in MRSA but not in MSSA. Mutations in a number of loci encoding LPXTG proteins were introduced into MRSA strains, allowing the involvement of SdrC SdrD SdrE, ClfA, and Spa in the MRSA biofilm phenotype to be ruled out and revealing the requirement for FnBPA and FnBPB. A mutation of either fnbA or fnbB alone was insufficient to impair biofilm development, and overexpression of either gene alone could complement fnbAB double mutants of MRSA clinical isolates, showing that both FnBPA and FnBPB promote biofilm.
Although the fnbA and fnbB genes were not required for NaCl-induced, icaADBC-dependent biofilm development in MSSA isolates, overexpression of either gene potently increased MSSA biofilm development in medium supplemented with glucose, suggesting that the fibronectin binding proteins are capable of promoting biofilm in both MSSA and MRSA isolates under appropriate conditions.
By exploiting the sensitivity of the FnBPs to V8 protease (43), we produced data that suggest that biofilm development by a number of genetically diverse MRSA clinical isolates is likely to be FnBP dependent. However, biofilm formation by MRSA isolates from CC5 and CC30 appeared to be resistant to V8 protease inhibition. FnBPs from these strains may be poorly expressed or resistant to degradation by V8 protease, or different surface proteins may be involved in biofilm formation by these strains. We are currently examining the role of PIA/PNAG- and FnBP-mediated biofilm in these strains.
The global regulator of transcription, SarA, was required for FnBP-mediated biofilm development in MRSA. However, in contrast to a previous study in S. aureus Newman (70), our data do not support a role for SarA as a transcriptional activator of fnbA and fnbB expression in MRSA. Instead, increased expression of proteases (4, 28), including V8 serine protease, may explain reduced biofilm and reduced fibronectin binding in MRSA sarA mutants. McGavin et al. (43) reported that the degradation of cell surface FnBPs as cultures entered stationary phase was due to V8 protease activity. However, unlike biofilm development, fibronectin binding was reduced but not completely abolished in a sarA mutant. Remnants of cleaved FnBPs exposed on the surface, which contain one or more fibronectin binding domains, may retain the capacity to promote low adherence to fibronectin but lack the ability to mediate biofilm cell-cell interactions.
FnBPA and FnBPB are large, multidomain proteins in which fibronectin binding is mediated by tandem repeats located between the N-terminal A domain and C-terminal wall-spanning and LPXTG-anchoring domains (44, 62). Our data indicate that FnBP-mediated biofilm formation is determined primarily by the N-terminal A domain. Altering the FnBPA N304 residue of the N2 subdomain eliminated fibrinogen and elastin binding (29, 56, 68) but did not affect biofilm formation. Experiments are now under way to identify residues in the A domain that are required for biofilm development.
How FnBPA and FnBPB promote biofilm development remains to be determined. Possibly, intercellular accumulation is mediated by direct interactions between the cell wall-anchored FnBPs on different cells, or the FnBP A domains bind to another cell surface molecule. In either case it will be important to investigate further why FnBP-mediated biofilm is associated primarily with MRSA and not MSSA isolates. Presumably, the redundancy of the icaADBC-encoded PIA/PNAG in the MRSA but not MSSA biofilm phenotype (49) partially answers this question, and it will be important to determine why MRSA isolates do not appear to make PIA/PNAG-dependent biofilm even though the ica locus is maintained and transcribed in these strains. Alternatively, FnBPA and FnBPB may be more highly expressed, or their integrity may be more resistant to protease degradation during stationary phase and biofilm growth in MRSA than in MSSA strains. If the FnBPs mediate biofilm through interactions with another cell wall-associated ligand, it is also possible that this receptor is more highly expressed in MRSA than MSSA isolates.
In conclusion, this study reveals that in addition to binding extracellular matrix proteins, FnBPA and FnBPB can also promote intercellular accumulation and biofilm development by S. aureus. Given that implanted surgical and medical biomaterials are rapidly coated by a conditioning film composed primarily of extracellular matrix proteins including fibronectin, our findings suggest that these cell wall-anchored proteins are likely to be important virulence factors in device-related S. aureus infections, particularly those caused by MRSA strains, and may represent attractive therapeutic targets.
Acknowledgments
This study was funded by a Clinical Research Training Fellowship from the Health Research Board (Ireland) to E. O'Neill, grants from the Hospital Infection Society (United Kingdom) and the Health Research Board to J. P. O'Gara, grants from Science Foundation Ireland and the Health Research Board to T. J. Foster, and a grant from the American Heart Association to D. A. Robinson.
We thank M. Smeltzer for strain UAMS-240 carrying a sarA::Tcr allele, S. J. Foster for SH1000, C. Y. Lee for plasmid pLI50, K. K. Jefferson for plasmid pMAD, and T. Maira Litran and G. B. Pier for rabbit anti-PIA/PNAG serum. We acknowledge the helpful comments and experimental advice of L. Holland, F. Keane, and C. D. Murphy during this study.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 714206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1511-8. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 1844400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216173-179. [DOI] [PubMed] [Google Scholar]
- 9.Corrigan, R. M., D. Rigby, P. Handley, and T. J. Foster. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 1532435-2446. [DOI] [PubMed] [Google Scholar]
- 10.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 1832888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella, C., M. A. Tormo, C. Ubeda, M. P. Trotonda, M. Monzon, C. Peris, B. Amorena, I. Lasa, and J. R. Penades. 2004. Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 722177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deivanayagam, C. C., E. R. Wann, W. Chen, M. Carson, K. R. Rajashankar, M. Hook, and S. V. Narayana. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 216660-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downer, R., F. Roche, P. W. Park, R. P. Mecham, and T. J. Foster. 2002. The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277243-250. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2006. Environmental regulation of biofilm development in methicillin-resistant and methicillin-susceptible Staphylococcus aureus clinical isolates. J. Hosp. Infect. 62120-122. [DOI] [PubMed] [Google Scholar]
- 15.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 431973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick, F., H. Humphreys, E. G. Smyth, C. A. Kennedy, and J. P. O'Gara. 2002. Environmental regulation of biofilm formation in intensive care unit isolates of Staphylococcus epidermidis. J. Hosp. Infect. 52212-218. [DOI] [PubMed] [Google Scholar]
- 17.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3948-958. [DOI] [PubMed] [Google Scholar]
- 18.Foster, T. J., and M. Hook.1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6484-488. [DOI] [PubMed] [Google Scholar]
- 19.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 1872426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 681304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, C., D. McDevitt, P. Francois, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 171143-1152. [DOI] [PubMed] [Google Scholar]
- 22.Gross, M., S. E. Cramton, F. Gotz, and A. Peschel. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 693423-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartford, O., P. Francois, P. Vaudaux, and T. J. Foster. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 251065-1076. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 241013-1024. [DOI] [PubMed] [Google Scholar]
- 25.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB Modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 1845457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 1862449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 694742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane, F. M., A. Loughman, V. Valtulina, M. Brennan, P. Speziale, and T. J. Foster. 2007. Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63711-723. [DOI] [PubMed] [Google Scholar]
- 30.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10251-259. [DOI] [PubMed] [Google Scholar]
- 31.Knobloch, J. K., M. A. Horstkotte, H. Rohde, and D. Mack. 2002. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 191101-106. [DOI] [PubMed] [Google Scholar]
- 32.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 33.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 1804814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lasa, I., and J. R. Penades. 2006. Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 15799-107. [DOI] [PubMed] [Google Scholar]
- 35.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 36.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 602048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 704433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massey, R. C., M. N. Kantzanou, T. Fowler, N. P. Day, K. Schofield, E. R. Wann, A. R. Berendt, M. Hook, and S. J. Peacock. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 3839-851. [DOI] [PubMed] [Google Scholar]
- 41.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 975510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 992293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGavin, M. J., C. Zahradka, K. Rice, and J. E. Scott. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 652621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meenan, N. A., L. Visai, V. Valtulina, U. Schwarz-Linek, N. C. Norris, S. Gurusidappa, M. Hook, P. Speziale, and J. R. Potts. 2007. The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. [DOI] [PubMed]
- 45.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30245-257. [DOI] [PubMed] [Google Scholar]
- 46.Novick, R. P. 2000. Sortase: the surface protein anchoring transpeptidase and the LPXTG motif. Trends Microbiol. 8148-151. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien, L., S. W. Kerrigan, G. Kaw, M. Hogan, J. Penades, D. Litt, D. J. Fitzgerald, T. J. Foster, and D. Cox. 2002. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol. Microbiol. 441033-1044. [DOI] [PubMed] [Google Scholar]
- 48.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270179-188. [DOI] [PubMed] [Google Scholar]
- 49.O'Neill, E., C. Pozzi, P. Houston, D. Smyth, H. Humphreys, D. A. Robinson, and J. P. O'Gara. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 451379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peacock, S. J., N. P. Day, M. G. Thomas, A. R. Berendt, and T. J. Foster. 2000. Clinical isolates of Staphylococcus aureus exhibit diversity in fnb genes and adhesion to human fibronectin. J. Infect. 4123-31. [DOI] [PubMed] [Google Scholar]
- 51.Ponnuraj, K., M. G. Bowden, S. Davis, S. Gurusiddappa, D. Moore, D. Choe, Y. Xu, M. Hook, and S. V. Narayana. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115217-228. [DOI] [PubMed] [Google Scholar]
- 52.Poston, S. M., G. R. Glancey, J. E. Wyatt, T. Hogan, and T. J. Foster. 1993. Co-elimination of mec and spa genes in Staphylococcus aureus and the effect of agr and protein A production on bacterial adherence to cell monolayers. J. Med. Microbiol. 39422-428. [DOI] [PubMed] [Google Scholar]
- 53.Regassa, L. B., R. P. Novick, and M. J. Betley. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 603381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice, K. C., J. B. Nelson, T. G. Patton, S. J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 473926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roche, F. M., R. Downer, F. Keane, P. Speziale, P. W. Park, and T. J. Foster. 2004. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 27938433-38440. [DOI] [PubMed] [Google Scholar]
- 57.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 1492759-2767. [DOI] [PubMed] [Google Scholar]
- 58.Rohde, H., E. C. Burandt, N. Siemssen, L. Frommelt, C. Burdelski, S. Wurster, S. Scherpe, A. P. Davies, L. G. Harris, M. A. Horstkotte, J. K. Knobloch, C. Ragunath, J. B. Kaplan, and D. Mack. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 281711-1720. [DOI] [PubMed] [Google Scholar]
- 59.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 551883-1895. [DOI] [PubMed] [Google Scholar]
- 60.Sadovskaya, I., E. Vinogradov, S. Flahaut, G. Kogan, and S. Jabbouri. 2005. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 733007-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sadovskaya, I., E. Vinogradov, J. Li, and S. Jabbouri. 2004. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr. Res. 3391467-1473. [DOI] [PubMed] [Google Scholar]
- 62.Schwarz-Linek, U., J. M. Werner, A. R. Pickford, S. Gurusiddappa, J. H. Kim, E. S. Pilka, J. A. Briggs, T. S. Gough, M. Hook, I. D. Campbell, and J. R. Potts. 2003. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423177-181. [DOI] [PubMed] [Google Scholar]
- 63.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trotonda, M. P., A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penades. 2005. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 1875790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 481075-1087. [DOI] [PubMed] [Google Scholar]
- 66.Vuong, C., S. Kocianova, Y. Yao, A. B. Carmody, and M. Otto. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 1901498-1505. [DOI] [PubMed] [Google Scholar]
- 67.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 1821688-1693. [DOI] [PubMed] [Google Scholar]
- 68.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 27513863-13871. [DOI] [PubMed] [Google Scholar]
- 69.Weinrick, B., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, Y. Fang, and R. P. Novick. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 1868407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36230-243. [DOI] [PubMed] [Google Scholar]
- 71.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]