Abstract
Mycobacterial genomes are endowed with many eukaryote-like nucleotide cyclase genes encoding proteins that can synthesize 3′,5′-cyclic AMP (cAMP). However, the roles of cAMP and the need for such redundancy in terms of adenylyl cyclase genes remain unknown. We measured cAMP levels in Mycobacterium smegmatis during growth and under various stress conditions and report the first biochemical and functional characterization of the MSMEG_3780 adenylyl cyclase, whose orthologs in Mycobacterium tuberculosis (Rv1647) and Mycobacterium leprae (ML1399) have been recently characterized in vitro. MSMEG_3780 was important for producing cAMP levels in the logarithmic phase of growth, since the ΔMSMEG_3780 strain showed lower intracellular cAMP levels at this stage of growth. cAMP levels decreased in wild-type M. smegmatis under conditions of acid stress but not in the ΔMSMEG_3780 strain. This was correlated with a reduction in MSMEG_3780 promoter activity, indicating that the effect of the reduction in cAMP levels on acid stress was caused by a decrease in the transcription of MSMEG_3780. Complementation of the ΔMSMEG_3780 strain with the genomic integration of MSMEG_3780 or the Rv1647 gene could restore cAMP levels during logarithmic growth. The Rv1647 promoter was also acid sensitive, emphasizing the biochemical and functional similarities in these two adenylyl cyclases. This study therefore represents the first detailed biochemical and functional analysis of an adenylyl cyclase that is important for maintaining cAMP levels in mycobacteria and underscores the subtle roles that these genes may play in the physiology of the organism.
Cyclic AMP (cAMP) is an important intracellular second messenger that is involved in several signaling pathways in both bacteria and eukaryotes (2, 5). Studies have shown a role for cAMP not only in physiological processes such as catabolite repression and sporulation (16) but also in the regulation of several virulence pathways, e.g., in Pseudomonas, Vibrio, and Candida species (1, 17, 19, 40, 41). The synthesis of cAMP is dependent on the activity of adenylyl cyclases, which can be membrane-bound or soluble proteins (7). The largely differing amino acid sequences of nucleotide cyclases allow their classification into six classes, among which the class III nucleotide cyclases have the widest phyletic distribution (7, 37).
Adenylyl and guanylyl cyclases of the class III family are proteins that form head-to-tail dimers and generate either two (homodimeric enzymes) or one (heterodimeric enzymes) active site at the dimer interface (20). Nucleotide cyclases catalyze the conversion of ATP or GTP to cAMP or GMP, respectively, in a metal-dependent manner, and biochemical and structural studies have identified specific sequence motifs that are involved in substrate (ATP or GTP) or metal (Mg2+) binding (20). In contrast to the limited domain compositions of mammalian nucleotide cyclases, the bacterial counterparts are often fused to a variety of domains, highlighting their ability to be regulated allosterically (37). Their presumable involvement in diverse biological phenomena in bacteria is perhaps due to this remarkable biochemical and structural versatility.
Recent studies have shown the presence of a large number of class III nucleotide cyclases in the genomes of several bacteria such as actinobacteria, alphaproteobacteria, and cyanobacteria (37). The genomes of Mycobacterium tuberculosis strains H37Rv and CDC1551 encode 16 and 17 class III cyclases, respectively (24, 33). The presence of multiple cyclases within a genome is in striking contrast to the well-studied paradigm of the single-copy class I cyclase gene in Escherichia coli. Studies on the role of these mycobacterial cyclases, and indeed cAMP, in the biology of the organism are therefore critical to a comprehensive understanding of cyclic nucleotide monophosphates in more complex signaling networks. For example, the deletion of Rv3676, a homolog of the cAMP receptor protein from M. tuberculosis, leads to the attenuation of M. tuberculosis in macrophage infections and in the mouse model of tuberculosis (29), suggesting that several important genes are under cAMP regulation in M. tuberculosis. Furthermore, inactivating point mutations within the cAMP or DNA binding domains of Rv3676 were found in several strains of Mycobacterium bovis BCG, and this apparent lack of Rv3676 function was suggested to be one of the additional reasons for the attenuation of these BCG strains (42). Microarray-, proteomic-, and SELEX-based approaches have also helped in the identification of several cAMP-regulated genes (3, 11, 29). Those studies suggest that cAMP levels must be finely regulated by the adenylyl cyclases, and the presence of multiple adenylyl cyclases in the genome implies a certain amount of redundancy in the proteins that are involved in cAMP production. However, given the diversity of these enzymes in terms of their domain organization, mycobacteria may have evolved a specific need for different cyclases at different times, and this can be studied only by a systematic analysis of the role of each cyclase gene, one at a time.
The Rv1647 cyclase from M. tuberculosis is conserved in Mycobacterium leprae (ML1399), even though 12 other cyclases found in M. tuberculosis are deleted or are pseudogenes in M. leprae (35). In addition, Rv1647 orthologs can be found in M. bovis, Mycobacterium avium, Mycobacterium smegmatis, as well as Mycobacterium marinum (38). We therefore hypothesized that Rv1647 and its orthologs serve a housekeeping role in mycobacteria and maintain cAMP levels in the organism. In this study, we investigated the role of MSMEG_3780, the Rv1647 ortholog, in the biology of mycobacteria. Our results provide evidence to suggest that MSMEG_3780 maintains basal levels of cAMP in M. smegmatis during the logarithmic phase of growth. Certain stress conditions alter intracellular levels of cAMP in M. smegmatis, and the reduction in intracellular levels of cAMP seen upon the acidification of cultures could also be correlated with a reduced transcription of the MSMEG_3780 gene. This reduction in intracellular cAMP levels was restored upon complementation of the knockout strain with the MSMEG_3780 or the Rv1647 gene, suggesting that M. smegmatis could be used to understand fundamental aspects of cAMP and cAMP signaling in mycobacteria.
MATERIALS AND METHODS
Cloning, expression, and purification of MSMEG_3780.
The MSMEG_3780 gene as annotated by TIGR was cloned using primers MS3780FLfwd (5′-ACAGGCTCGAGTGTTCCTCGACGC-3′) and MS3780rvs (5′-CGCGCGCGGCCGCACCACATCGACTATGCGTCAG-3′). PCR was carried out on the genomic DNA of M. smegmatis mc2155 (35), and the product was digested with XhoI and NotI and cloned into the compatible SalI-NotI sites of pPROEx-HT-C to generate plasmid pPRO-MSMEG_3780FL. The catalytic domain of the protein, from amino acids 149 to 392, was obtained by cloning the EcoRI-NotI fragment from pPRO- MSMEG_3780FL into pPROEx-HT-A to generate plasmid pPRO-MSMEG_3780CD. The protein was expressed and purified from E. coli BL21(DE3) cells as described previously for the Rv1647 protein (35). Adenylyl cyclase assays were performed as described previously, and cAMP was measured by radioimmunoassay (36).
GST pull-down assays.
The EcoRI-NotI fragment from pPRO-MSMEG_3780CD was cloned into similarly digested vector pGEX-6P-1 to generate plasmid pGEX-MSMEG_3780CD. Plasmid pGEX-Rv1647CD was generated by subcloning the XhoI-NotI fragment from pPRO-16471-328 (35) into similarly cut vector pGEX-6P-1. Glutathione S-transferase (GST), GST-MSMEG_3780, and GST-Rv1647 proteins were purified according to a procedure described previously (18). GST or GST fusion proteins (3 μg) on glutathione beads were interacted independently with 5 μg of His-tagged prey protein in 100 mM buffer at pH 9.0 (25) containing 5 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 5 mM 2-mercaptoethanol, 12 mM Mn, and 10% glycerol at 4°C for 1 h, and unbound proteins were removed by washing the beads five times with 1 ml of interaction buffer. Bound proteins were assessed by immunoblotting with a hexahistidine-specific monoclonal antibody (GE Health Sciences).
Measurement of cAMP levels in M. smegmatis.
Mycobacterial strains were grown in 7H9TG (Middlebrook 7H9 broth supplemented with 0.2% glycerol and 0.05% Tween 80) at 37°C, with shaking at 250 rpm, or on Middlebrook 7H10 agar. Whenever necessary, hygromycin (150 μg/ml for E. coli and 50 μg/ml for mycobacteria) was used.
Cells were centrifuged at 13,000 × g; cells washed with a solution containing 10 mM Tris-HCl (pH 7.5), 0.89% NaCl, and 0.05% Tween 80 (TBST); the pellet was resuspended in 0.1 N HCl; and the suspension was boiled for 10 min. Samples were then flash-frozen and lysed by bead beating for 1 min. Aliquots of the lysate were taken for estimation of cAMP by radioimmunoassay (35). Spent medium (supernatant of the 13,000 × g spin) was acidified by twofold dilution in 0.2 N HCl and boiled for 10 min, and suitable aliquots were monitored for cAMP directly by radioimmunoassay.
For measurement of cAMP levels under stress conditions, 2-day-old cultures started from single colonies were diluted 1:100 into fresh medium and grown for 30 h. Cells were then centrifuged, washed in chilled sterile TBST, and resuspended in either fresh control medium or medium containing 50 mM MES [2(N-morpholino)ethanesulfonic acid] (pH 5.5), 5 mM H2O2, and 1 mM sodium nitroprusside, 10 mM sodium phosphate (pH 7.2) containing 0.9% NaCl (phosphate-buffered saline [PBS]), 0.02% glycerol, or 0.05% sodium dodecyl sulfate (SDS) (22). Cultures were grown for 1.5 h or 5 h at 37°C and/or at 42°C for 1.5 h. Cultures were then centrifuged, and the cell pellet was taken for measurement of cAMP levels.
Generation of the ΔMSMEG_3780 strain.
A strain with a deletion of the MSMEG_3780 gene (ΔMSMEG_3780) was generated using the suicide vector approach (27). The 5′ knockout fragment consisting of 1,013 bases upstream of MSMEG_3780 was amplified by PCR using primers MS3780-5′KO-HindIII-fwd (5′-ACAGTTGAAGCTTCTCGGCCTCGGTCAG-3′) and MS3780-5′KO-XbaI-rvs (5′-GGGTCTTCTAGACACGCGCTCGGTCAGCG-3′). A fragment of 1,101 bases downstream of the catalytic domain of MSMEG_3780 was amplified using primers MS3780-3′KO-XbaI-fwd (5′-ACTCCCTCTAGATTGCCAGCCGCCCGG-3′) and MS3780-3′KO-NotI-rvs (5′-TCGGCGGCGGCCGCGTTGACACGTTTGCCG-3′). The 5′ fragment was cloned into vector pBKS-II(+) using the HindIII and XbaI sites to obtain plasmid pBKS-MSMEG_3780-5′KO, and the 3′ fragment was cloned into pPROEx-HT-B using the XbaI and NotI sites to generate plasmid pPRO-MSMEG_3780-3′KO. DNA sequencing confirmed the identity of the cloned sequences compared to that reported in the genome. The HindIII-XbaI and XbaI-NotI fragments from pBKS-MSMEG_3780-5′KO and pPRO-MSMEG_3780-3′KO, respectively, were ligated into HindIII- and NotI-cut vector p2NIL to generate plasmid p2NIL-MSMEG_3780-5′3′KO. The PacI fragment from pGOAL19 containing three markers (β-galactosidase, hygromycin resistance, and sucrose susceptibility) was cloned into p2NIL-MSMEG_3780-5′3′KO to generate plasmid p2NIL-MSMEG_3780-5′3′KO-PacI.
Plasmid p2NIL-MSMEG_3780-5′3′KO-PacI (1 μg) was electroporated into M. smegmatis mc2155, and single crossovers and double crossovers were obtained essentially as described previously (27). Double crossovers were further tested by genomic PCR and Southern blotting as described below to obtain the ΔMSMEG_3780 strain.
To construct the ΔMSMEG_3780 strain complemented with the MSMEG_3780 gene driven by its own promoter, a StuI-XbaI fragment from pPRO-MSMEG_3780FL was cloned into compatible HincII-XbaI sites of vector pBKS-II(+) to generate pBKS-MSMEG_3780FL. The insert was removed from this plasmid as a KpnI-XbaI fragment and cloned into similarly digested integration vector pMH94h (a kind gift of W. R. Bishai, Johns Hopkins Center for Tuberculosis Research) to generate pMH94h-MSMEG_3780, which was electroporated in the ΔMSMEG_3780 strain. Positive integrants (ΔMSMEG_3780C) carrying the required insert were screened by colony PCR and validated by Southern hybridization. PCRs were carried out with primers MS3780FLfwd and MS3780rvs on genomic DNA prepared from the wild-type strain and the ΔMSMEG_3780 and ΔMSMEG_3780C strains to confirm the presence of the deletion in the ΔMSMEG_3780 strain and the complementation of MSMEG_3780. Southern blotting was carried out by electrophoretic separation of 2 μg of genomic DNA of wild-type, knockout, and complement strains digested with SacI and by transferring the DNA to Hybond nylon membranes (GE Healthcare). The probe was prepared with a random hexamer labeling kit (Promega) using an EcoRI-SmaI fragment present upstream of the catalytic domain of MSMEG_3780. The blot was probed at 60°C for 16 h and washed once in 2× SSC (1× SSC is 0.15 mM NaCl plus 0.015 M sodium citrate) for 15 min at 60°C prior to exposure to a phosphorimager.
Preparation of RNA from M. smegmatis.
Cells (5 × 108 CFU) were lysed by bead beating in 200 μl of Tri reagent (Sigma-Aldrich) prewarmed to 65°C. The lysate was mixed by pipetting, followed by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was treated with chloroform (40 μl) followed by centrifugation at 12,000 × g for 15 min at 4°C. The RNA was precipitated with 100 μl of isopropyl alcohol, and the RNA pellet was treated with RNase-free DNase (1 to 5 units). RNA (5 μg) was used for reverse transcription using 200 units of Moloney murine leukemia virus reverse transcriptase enzyme in the presence of 10 units of RNase inhibitor using random primers. PCR was carried out using 1 μl of the reverse transcriptase reaction mixture and primers MS3779RTF (5′-GACCCTTTTCGCCCTGGCCGT-3′) and MS3779RTR (5′-CTGCCGCGCCGTAACCTGAC-3′). The sigA transcript levels were assessed using primers MSsigART428F (5′-ACGAGGAAGAGTCCGAGGCCCTG-3′) and MSsigART536R (5′-GCTCCACCTCTTCTTCGGCGTTGA-3′).
Fractionation of M. smegmatis cells for Western blot analysis.
Cultures of M. smegmatis cells were grown at 37°C and centrifuged at 5,000 × g for 15 min at 4°C, and the cell pellet was washed three times with wash buffer (10 mM Tris-HCl [pH 7.5], 250 mM sucrose, and 0.05% Tween 80), frozen in liquid nitrogen, and stored at −70°C until processing. The frozen pellet was resuspended in a solution containing 3 ml of 20 mM Tris-HCl (pH. 7.5), 5 mM 2-mercaptoethanol, 2 mM sodium dihydrogen phosphate, 2 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine; sonicated; and centrifuged at 5,000 × g to remove unbroken cells and cell wall debris. Lysates were centrifuged at 100,000 × g for 4 h at 4°C (L8-M ultracentrifuge; Beckman). The supernatant was separated as the cytosol, and the pellet was kept as the membrane fraction. The pellet fraction was resuspended in lysis buffer, and both fractions were stored at −70°C until use. Western blot analysis was performed using an antibody raised against the GST-MSMEG_3780 catalytic domain at a dilution of 1:5,000 by using enhanced chemiluminescence.
Cloning and characterization of promoters.
The genomic region of MSMEG_3780 containing the putative promoter was amplified by PCR using primers MS3780prmScaSpe (5′-TACTAGTACTCAGCGCACGGCGGAAGC-3′) and MS3780prmXbaStu (5′-ACTCTAGAGGCCTCATTCGGCATCCCTGTCCA-3′). The product was digested with XbaI and SpeI and cloned into plasmid pBKS-II(+) to generate plasmid pBKS-PMSMEG_3780, and the sequence of the insert was verified. The Vibrio harveyi luciferase gene was obtained as a 2.4-kb BamHI-HindIII insert from plasmid pLUX (30) (a kind gift of Richard Friedman, University of Arizona) and cloned into BamHI-HindIII-digested vector pBKS-II(+) to generate plasmid pBKS-luxAB. The MSMEG_3780 promoter (obtained as an XbaI-BamHI insert from plasmid pBKS-PMSMEG_3780) and the luciferase gene (luxAB, obtained as a 2.4-kb BamHI-HindIII insert from plasmid pBKS-luxAB) were cloned into XbaI-HindIII-digested vector pMV-10-25 (8) to generate plasmid pMV-PMSMEG_3780-luxAB.
Primers MS3780prmHindIII-trunc (5′-GCGGGCAAAGCTTCGACGCGCACGATTG-3′) and MS3780prmXbaStu were used to amplify ∼250 bp of the annotated MSMEG_3780 gene from M. smegmatis genomic DNA. The amplicon was digested with HindIII and XbaI and cloned into similarly digested vector pBKS-II(+) to generate pBKS-PMSMEG_3780truncated, and the sequence was verified. The KpnI-SpeI fragment from pBKS-PMSMEG_3780truncated and the SpeI-HindIII fragment containing luxAB from plasmid pBKS-luxAB were then cloned into KpnI-HindIII-cut vector pMV10-25 to generate pMV-PMSMEG_3780truncated-luxAB.
To generate the shortest promoter fragment, pBKS-PMSMEG_3780truncated was digested with SmaI and HincI to release a 180-bp fragment. The vector was then religated to generate pBKS-PMSMEG_3780short. A KpnI-XbaI fragment from this plasmid was cloned into KpnI-SpeI-digested pMV-PMSMEG_3780-luxAB to generate plasmid pMV-PMSMEG_3780short-luxAB. The region encompassing the putative Rv1647 promoter (∼400 bp) was amplified by PCR using primers Rv1647PKpnIF (5′-CGCAGTTTGGGTACCTTCTTCCCGCCCTAG-3′) and Rv1647PBamHIR (5′-CGCGAGGCCGGATCCCGATCCCGCAGCAG-3′) and cloned into KpnI-BamHI-digested vector pBKS-II(+) to generate plasmid pBKS-PRv1647, and the sequence was confirmed. A KpnI-BamHI fragment was isolated from pBKS-PRv1647 and ligated into similarly cut pMV-PMSMEG_3780-luxAB to generate plasmid pMV-PRv1647-luxAB.
The sigA promoter of approximately 330 bp (12) was amplified by PCR from genomic DNA using primers MSsigA_Kpnfwd (5′-AAGTGGTACCCGATGCTGAAGAACCGG-3′) and MSsigA_Xhorvs (5′-TGCCTCGAGCACCCTTTCGGTCTTACGAATTC-3′). The PCR product was digested with KpnI and XhoI and cloned into the similarly cut vector pBKS-II(+) to generate pBKS-PMSsigA. This plasmid was digested with KpnI-SpeI and cloned into similarly digested pMV-PMSMEG_3780-luxAB to generate pMV-PSigA-luxAB.
The luxAB genes were obtained from plasmid pBKS-luxAB as a KpnI-XbaI fragment and cloned into KpnI-NheI-digested vector pMV10-25 to generate plasmid pMV-Phsp60-luxAB such that the hsp60 promoter (30) drives the expression of luciferase.
All promoter plasmids were electroporated into M. smegmatis cells. Aliquots of the culture (usually 10 μl) were added to 80 μl of TBST, and 10 μl of 1% decanal (Sigma) prepared in 100% ethanol was added. Luciferase activity was measured in a VICTOR3 1420 multilabel counter (Perkin-Elmer) in a 96-well Opti plate (Perkin-Elmer). Sterile TBST without bacteria was used as a blank, and this value was subtracted from the test values.
Generation and characterization of the Rv1647 complement strain.
A region encompassing the catalytic domain of Rv1647 (from amino acids 67 to 316 as annotated currently by TIGR and from amino acids 9 to 328 as per our earlier construct pPRO-16471-328) (35) was amplified by PCR from genomic DNA of M. tuberculosis H37Rv using primers Rv1647CSpeIfwd (5′-GCACTAGTCCTGGCGATCCGGAATTCGGC-3′) and Rv1647NotIrvs (5′-TAGGCGGCCGCGTTCTACTGTGCATCGGGACT-3′). The 800-bp amplicon was digested using SpeI and NotI and cloned into similarly cut vector pPRO-A to generate pPRO-Rv1647Ccat. Plasmid pPRO-Rv1647Ccat (35) was digested with a BamHI-XbaI fragment, and the ∼800-bp fragment was ligated into a KpnI-BamHI fragment from pMV-PMSMEG_3780-luxAB and cloned into KpnI-XbaI-digested pMH94h to generate pMH94hRv1647C. This plasmid allows the expression of Rv1647 under the MSMEG_3780 promoter, with a translational fusion to residue 144 (as annotated by TIGR) of the MSMEG_3780 protein. To confirm the integration of the Rv1647 gene, PCR was carried out with primers Rv1647CSpeIfwd and Rv1647-938R (5′-TCGCTAACAGTCTTGATGCGAT-3′) on genomic DNA prepared from M. smegmatis wild-type, knockout, and ΔMSMEG_3780RvC strains. The 16S rRNA gene was amplified from the genomic DNA of the three strains using primers Myco16s-fwd (5′-CGAGCGTTGTCCGGAATTA-3′) and Myco16s-rvs (5′-TCCACCGCTACACCAGGAAT-3′) to confirm the integrity of the genomic DNA. Southern blot analysis was performed on SacI-digested genomic DNA prepared from the complement strain to confirm the integration of the Rv1647 catalytic domain at the att site. The probe used was a 250-bp fragment released by HindIII-SpeI digestion of pBKS-PMSMEG_3780truncated.
Membrane fractions were prepared from the complement strain and subjected to Western blot analysis using affinity-purified antibody to Rv1647 (35) and a monoclonal antibody to gyrase A characterized previously (23).
RESULTS
Cyclases and cAMP levels in M. smegmatis.
M. smegmatis has orthologs of eight cyclases from M. tuberculosis (38), six of which are predicted to be homodimeric adenylyl cyclases (MSMEG_0228, MSMEG_3243, MSMEG_3780, MSMEG_4279, MSMEG_4924, MSMEG_5018, and MSMEG_6154). In addition to a cyclase that is unique to its genome (MSMEG_3243), M. smegmatis has an ATPase domain-containing cyclase (MSMEG_0228) that has an ortholog only in M. leprae and M. avium but not in M. tuberculosis. This variety of cyclases with diverse domain fusions suggests that M. smegmatis could serve as a model for studying cAMP-based signaling in mycobacteria with respect to several cyclases that it shares with other pathogenic members.
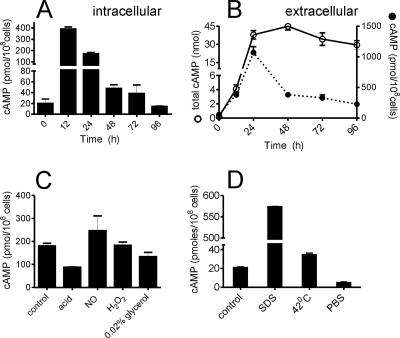
Given the large number of putative adenylyl cyclases, we decided to measure cAMP levels in M. smegmatis under various conditions. Intracellular cAMP levels in wild-type M. smegmatis were high during the early phases of growth (Fig. 1A). cAMP was also efficiently extruded by M. smegmatis into the growth medium, and concentrations of cAMP reaching ∼4 μM accumulated in the culture medium of the cells (Fig. 1B) at 24 h and thereafter. The total amount of cAMP in the medium did not decrease during culture, indicating very little degradation of cAMP in the extracellular medium (Fig. 1B). The amount of cAMP found extracellularly appears to be dependent on the intracellular concentration of cAMP. Thus, extracellular levels of cAMP normalized to cell number (Fig. 1B) showed their peak at 24 h, after the intracellular levels had peaked at 12 h (Fig. 1A). Moreover, cAMP levels per million living cells declined extracellularly in the late log and stationary phases, reflective of the reduced intracellular concentrations of cAMP at these times. One can only speculate as to the function of these high extracellular levels of cAMP; perhaps mycobacteria use this second messenger to signal to each other, as is seen in Dictyostelium (10).
FIG. 1.
cAMP levels in M. smegmatis during growth and stress conditions. (A) Intracellular cAMP levels during growth of wild-type M. smegmatis. (B) Total amount of extracellular cAMP (solid line) present in a culture (total volume, 10 ml). Extracellular cAMP (dotted line) of the same cultures is expressed per 108 cells. C. M. smegmatis cells were cultured for 30 h, resuspended in the medium indicated, and cultured for 5 h, following which the intracellular cAMP level was measured. (D) M. smegmatis cells were cultured for 30 h, and cells were resuspended in fresh medium and cultured for another 1.5 h at 37°C or at 42°C. Alternatively, cells were resuspended in medium containing 0.05% SDS or in PBS and cultured for 1.5 h. The intracellular cAMP level was measured at the end of 1.5 h. All data shown represent the means ± standard errors of the means (SEM) of duplicate determinations, with each assay being performed three times.
Early reports on cAMP in mycobacteria showed no change in cAMP levels during growth of the bacteria when expressed as cAMP concentration per gram (dry weight) (26). We chose to express our data per viable cell in order to remove artifacts that might arise from altered cell wall composition during growth of the organism, which would contribute to measurements of dry weight and therefore mask changes in cAMP levels in the bacterial cell. Assuming a cellular volume of ∼1 μm3, the molar concentrations of intracellular cAMP are ∼3 mM, which is comparable to levels reached in mammalian cells upon forskolin stimulation of the adenylyl cyclases.
We also measured cAMP levels under different stress conditions to investigate if the level of this second messenger is sensitive to environmental conditions that M. smegmatis or M. tuberculosis could experience. No change in viability was seen under the stress conditions utilized in Fig. 1C. In contrast to the response that has been well studied in E. coli, where the cAMP level increases dramatically upon catabolite repression (13), no change in cAMP levels were evident in carbon-starved M. smegmatis cells (Fig. 1C). Neither NO nor H2O2 caused a change in intracellular cAMP levels, but acid stress reduced cAMP levels by ∼50%. Additional stresses were tested, but under these conditions, M. smegmatis could tolerate a shorter duration (1.5 h) without a significant loss in viability. A dramatic increase in intracellular cAMP levels was seen in cells treated with SDS, while a reduction in cAMP levels was seen in cells treated with PBS (Fig. 1D).
Given the high levels of cAMP in M. smegmatis, we wished to assess the contribution of individual cyclases to the total cAMP levels. The MSMEG_3780 gene is an adenylyl cyclase that is present in all mycobacterial genomes, and its orthologs in M. tuberculosis and M. leprae have been recently biochemically characterized (35). However, their role in the physiology of mycobacteria remains unknown. We describe below the importance of MSMEG_3780 in contributing to intra- and extracellular cAMP levels during growth and other environmental insults.
Characterization of the MSMEG_3780 cyclase.
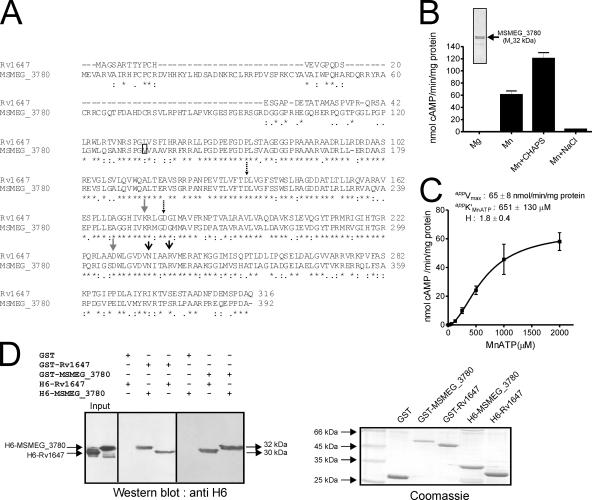
The MSMEG_3780 gene is predicted to encode a protein of 392 amino acids in length (TIGR annotation), with a predicted molecular mass of 42.8 kDa, while annotation by the Sanger Centre suggests that the N-terminal residue of the protein begins at residue 133 by TIGR annotation, which would result in a protein with a molecular mass of ∼30 kDa (Fig. 2A). In MSMEG_3780, Asp-212 and Asp-256 are predicted to be metal-binding residues, Asn-313 and Arg-317 are the transition-state stabilizing residues, and Lys-252 and Asp-306 contribute to the substrate-specifying residues, i.e., ATP binding residues (Fig. 2A).
FIG. 2.
Expression, purification, and characterization of MSEMG_3786 adenylyl cyclase and interaction with Rv1647. (A) Sequence similarity of the Rv1647 and the MSMEG_3780 proteins. The boxed residue represents the annotated start of MSMEG_3780 according to the Sanger Centre annotation. The predicted amino acid sequences were aligned using ClustalW. Dashed arrows represent substrate-specifying residues; gray arrows represent metal-binding residues; black arrows represent transition-state stabilizing residues. (B) Purified MSMEG_3780 (∼500 ng) was assayed in the presence of either 10 mM Mg or 10 mM Mn as a free metal along with 1 mM ATP. CHAPS (0.5%) or NaCl (500 mM) was added in some assays, along with 10 mM Mn and 1 mM MnATP as a substrate, and assays were carried out at pH 9. The inset shows purified MSMEG_3780 used for adenylyl cyclase assays. Values represent the means ± SEM of duplicate determinations, with each assay being performed three times. (C) MSMEG_3780 activity was assayed in the presence of various concentrations of MnATP in the presence of 10 mM free Mn. The level of cAMP produced was measured by radioimmunoassay. All data shown represent the means ± SEM of triplicate determinations, with each assay being performed twice. (D) A GST pull-down assay was performed as described in Materials and Methods. Hexahistidine-tagged proteins that interacted with the GST fusion protein were detected by Western blot analysis using an anti-His6 antibody. An equal aliquot of proteins that were taken for interaction was subjected to SDS-polyacrylamide gel electrophoresis, and the gel was stained to ensure that approximately equal amounts of protein were used for the experiment. The result shown is representative of experiments performed twice.
Similar to observations with Rv1647, full-length MSMEG_3780 (TIGR) overexpressed in E. coli was prone to proteolysis (35). Cloning, expression, and purification of the catalytic domain of MSMEG_3780 from residues 149 to 392 showed that the enzyme had significant adenylyl cyclase activity in vitro (Fig. 2B). Many of the biochemical properties of MSMEG_3780 were similar to those of Rv1647 (35). For example, the pH optimum for adenylyl cyclase activity was pH ∼9, and the activity was also stimulated 1.5- to 2-fold by CHAPS but not by 500 mM NaCl (Fig. 2B), indicating some distinct biochemical properties. The apparent Km for MnATP (∼600 μM) was also comparable to that observed for Rv1647 (Fig. 2C).
To provide further biochemical evidence for the structural and possible functional equivalence of MSMEG_3780 and Rv1647, we performed GST pull-down experiments. We had successfully utilized this heterodimerization assay to understand the oligomeric status of Rv1625c and its mutants (18). As shown in Fig. 2D, GST-MSMEG_3780 could associate not only with itself but also with Rv1647. Similarly, GST-Rv1647 could heterodimerize with MSMEG_3780 and homodimerize with histidine-tagged Rv1647.
Nucleotide cyclases form heterodimers only with structurally similar proteins (15). Therefore, given the shared biochemical properties of Rv1647 and MSMEG_3780, it appears that a study of MSMEG_3780 not only could provide information on cAMP levels in M. smegmatis but also could suggest the function of the Rv1647 ortholog in M. tuberculosis. In order to understand the role of MSMEG_3780 further, we generated and characterized a deletion mutant as described below.
Generation and characterization of the ΔMSMEG_3780 strain.
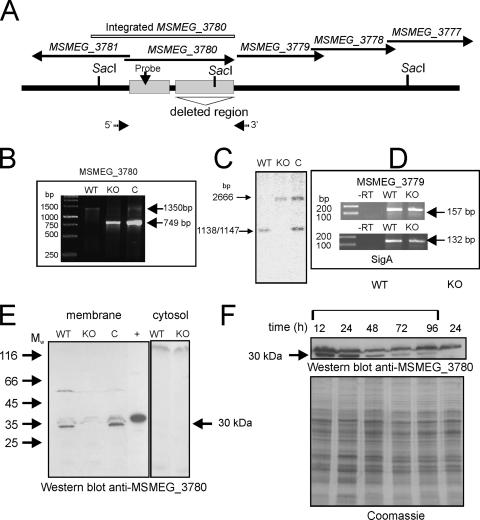
The ΔMSMEG_3780 knockout strain, lacking the entire catalytic domain of MSMEG_3780, was generated with an in-frame deletion in MSMEG_3780 to avoid polar effects on other genes that could be present in an operon with MSMEG_3780. The knockout strain was also complemented with a genomic insertion at the att1 site of a fragment encompassing ∼500 bp of upstream sequences of the annotated MSMEG_3780 gene (to include the putative promoter of MSMEG_3780) and the full-length gene (Fig. 3A, B, and C). In order to ensure that the transcription of the downstream MSMEG_3779 gene was not compromised in the ΔMSMEG_3780 strain, reverse transcription-PCR was performed with RNA prepared from the wild-type and knockout strains using primers specific for MSMEG_3779. As shown in Fig. 3D, the expression levels of MSMEG_3779 RNA in the wild-type and the knockout strains were similar, thus indicating the absence of polar effects of the deletion.
FIG. 3.
Generation and characterization of the ΔΜSMEG_3780 strain and expression of MSMEG_3780 during growth. (A) Diagrammatic representation of the genome region of MSMEG_3780 indicating the presence of SacI sites used for analysis and the region encompassing the catalytic domain of MSMEG_3780 that was deleted in the knockout strain. Arrows indicate the 5′ and 3′ primers used for PCR analysis. (B) PCR was performed using primers flanking the MSMEG_3780 gene with template genomic DNA prepared from wild-type (WT), knockout (KO), or complement (C) strains. The wild-type strain should amplify a fragment of ∼1,300 kb, while the knockout strain should amplify a fragment of ∼800 bp in length. The complement strain would show both fragments as a result of the insertion of the promoter-MSMEG_3780 fragment at the att site. (C) Southern blot analysis was performed on genomic DNA prepared from the three strains following digestion with SacI. The probe was prepared from a region containing the promoter of the MSMEG_3780 gene, which was also present in the fragment that was inserted at the att site in the complement strain. (D) Reverse transcription (RT)-PCR analysis of the MSMEG_3779 gene of RNA prepared from the wild-type and knockout strains. RNA was prepared from the two strains and reverse transcribed, and PCR was performed using primers specific to the MSMEG_3779 gene or sigA for normalization. (E) Western blot analysis was performed with cytosolic and membrane fractions prepared from wild-type, ΔΜSMEG_3780, and complemented strains (50 μg protein) using a specific antibody raised to the GST-MSMEG_3780 protein. The + indicates a lane loaded with 20 ng of purified His-tagged MSMEG_3780. Mw, weight-average molecular weight (in thousands). (F) Membrane protein was prepared from the wild-type and ΔΜSMEG_3780 strains and analyzed by Western blotting using MSMEG_3780 antibody. The Coomassie-stained gel shows the normalization of total protein taken for the Western blot analysis.
The expression of MSMEG_3780 during growth was assessed by Western blot analysis. Western blot analysis of lysates from the wild-type and knockout strains using an antibody against MSMEG_3780 showed a prominent band of ∼30 kDa in the membrane fraction of wild-type cells, which was absent in membranes prepared from the ΔMSMEG_3780 strain (Fig. 3E). The band reappeared in the complement strain. Therefore, the protein in M. smegmatis associates with the particulate fraction in cells despite the absence of any predicted transmembrane domain in its coding sequence. The Rv1647 protein in M. tuberculosis was also found in the particulate fraction (35). The activation of the enzymatic activities of these two proteins by detergents suggested a preference for a hydrophobic environment, and perhaps, this is provided by the membrane environment in the cell. Rv1647 could be identified in a proteomic analysis of membrane proteins from M. tuberculosis (14), in agreement with the localization of Rv1647 that we reported previously (35). This also strengthens our observation that MSMEG_3780 is also located in the membrane (Fig. 3E).
The molecular mass of the protein as expressed in M. smegmatis was ∼30 kDa, much smaller than the ∼42.8 kDa according to TIGR annotation. We previously reported that the mass of Rv1647 in M. tuberculosis lysates using an affinity-purified antibody was ∼30 kDa (35). Significant similarity between the Rv1647 and MSMEG_3780 proteins starts from residues L43 (Rv1647) (TIGR annotation) and L121 (MSMEG_3780) (TIGR annotation) (Fig. 1A). Proteins expressed from these residues would have a predicted molecular mass of ∼29 kDa, close to the molecular mass that was observed in Western blot analyses. As described below, we were indeed able to show that a promoter for MSMEG_3780 lies within the region annotated by TIGR as being within the coding sequence of MSMEG_3780.
The expression of MSMEG_3780 was monitored during growth, and the highest levels of the MSMEG_3780 protein were seen at 12 h and 24 h (Fig. 3F), at times when cAMP levels were also found to be high.
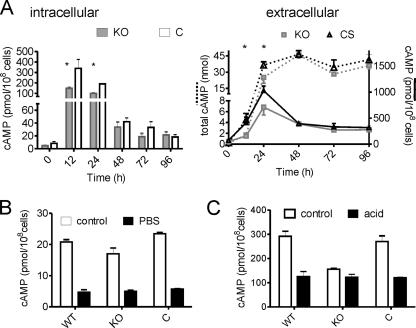
The wild-type and ΔMSMEG_3780 strains showed no difference in their growth rates and cell or colony morphologies in vitro. However, we did observe differences in cAMP levels in the two strains. During the early and mid-log phases of growth, levels of cAMP in the ΔMSMEG_3780 strain were significantly lower than those in the wild-type or complement strains (Fig. 4A), indicating that the activity of MSMEG_3780 could contribute to the generation of cAMP at this time. The cAMP levels in the complemented strain were similar to those of wild-type M. smegmatis. Moreover, levels of extracellular cAMP were lower at 12 h and 24 h in the ΔMSMEG_3780 strain, which is reflective of the reduced intracellular levels of cAMP in this strain (Fig. 4A).
FIG. 4.
cAMP levels in the ΔΜSMEG_3780 strain. (A) Intra- and extracellular cAMP levels during growth of wild-type, ΔΜSMEG_3780 (knockout [KO]), and complemented (C) strains. * represents values that are significantly different (P < 0.05) in the knockout strain compared to those of wild-type (WT) and complemented strains (CS). Data shown are the means ± SEM of duplicate determinations, with the experiment being repeated three times. (B) Intracellular cAMP levels were measured in the indicated strains under conditions similar to those detailed in the legend of Fig. 1C. (C) Intracellular cAMP levels were measured in the indicated strains under conditions similar to those detailed in the legend of Fig. 1D.
We also evaluated the contribution of the MSMEG_3780 gene to cAMP levels during various stresses. A reduction in cAMP levels was seen in both wild-type and knockout strains when cultured in PBS, indicating that cAMP levels under these conditions were regulated by the activities of perhaps a number of adenylyl cyclases and not just MSMEG_3780 (Fig. 4B). Interestingly, however, the reduction in intracellular cAMP levels was not seen in the ΔMSMEG_3780 strain upon acid stress (Fig. 4C). This reduction in intracellular cAMP levels was again restored upon complementation of the knockout strain, indicating that the fall in intracellular cAMP levels in acid-stressed bacteria was indeed regulated by MSMEG_3780 (Fig. 4C). The knockout strain grew at rates comparable to those of the wild-type strain over 48 h in acidified medium but with both strains growing slower in acidified medium than in normal medium (data not shown).
The intracellular pH of M. smegmatis remains stable at ∼6.5 even upon lowering the external pH to 4 (28), suggesting that the decrease in intracellular cAMP production may not be because of a reduction in the catalytic activity of MSMEG_3780, which is poorly active at low pHs. Therefore, we believe that a major factor in reducing intracellular cAMP levels upon acid stress is the decreased transcription of the MSMEG_3780 gene. We therefore decided to identify and study the promoter of MSMEG_3780 and assess the role of transcriptional regulation in controlling total cellular cAMP levels.
Promoter of the MSMEG_3780 gene.
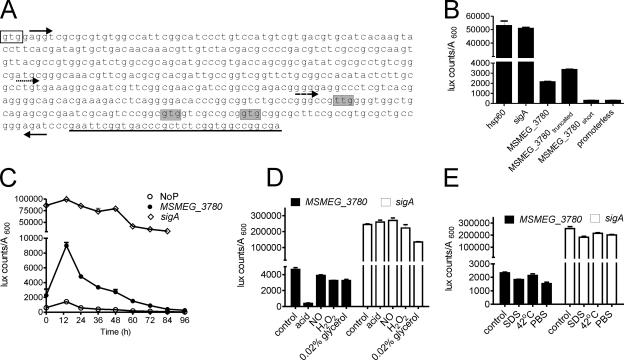
In order to test if nucleotide sequences within the gene based on the annotation at TIGR (i.e., encoding amino acid residues 1 to 133) harbored a promoter, we cloned a fragment from nucleotides (nt) 5 to 448, according to TIGR annotation, upstream of the Vibrio harveyi luciferase (luxAB) gene (30) (Fig. 5A). Significant luciferase activity could be detected with the cloned region of the MSMEG_3780 gene (Fig. 5B), indicating the presence of a promoter in this region. A shorter construct, from nt 251 to 448, showed promoter activity comparable to that of the longer promoter, while a fragment from nt 361 to 448 showed low luciferase activity equivalent to that of a promoterless construct, thereby defining the 3′ end of the promoter (Fig. 5B). We therefore predict that the start codon of MSMEG_3780 could be downstream of nt 251, and indeed, proteins made from any of the residues shown in Fig. 5A would correspond in size to the MSMEG_3780 protein detected by Western blotting. The activity of the MSMEG_3780 promoter was markedly lower than that of sigA or hsp60 (Fig. 5B), suggesting that MSMEG_3780 is a minor protein component in the cell.
FIG. 5.
Identification of the MSMEG_3780 promoter and its activity during growth and stress conditions. (A) Sequence of a part of the MSMEG_3780 gene indicating promoters used for analysis. Forward arrows indicate the 5′ end of the MS_3780, MS_3780truncated (dotted arrow), or MS_3780short (dashed arrow) promoter, and the reverse arrow indicates the 3′ ends of the promoter constructs. The clear box indicates the translational start codon as annotated by TIGR, while shaded nucleotides indicate alternative start codons based on the promoter analysis. Underlined bases indicate sequences that encode the beginning of the catalytic domain of MSMEG_3780. (B) Regions containing the putative promoter of MSMEG_3780 were cloned upstream of the luxAB gene, and luciferase activity was measured. The activities of the hsp60 and sigA promoters and a promoterless construct were also monitored. (C) Activities of the promoterless construct (NoP) and MSMEG_3780 and sigA promoters during growth. M. smegmatis cells were transformed with the indicated plasmids, and luciferase activity was monitored as described in Materials and Methods. (D) M. smegmatis cells were cultured for 30 h, and cells were resuspended in medium as indicated and cultured for another 5 h, following which luciferase activity was monitored. (E) M. smegmatis cells were cultured for 30 h, and cells were resuspended in medium for another 1.5 h at 37°C or at 42°C. Alternatively, cells were resuspended in medium containing 0.05% SDS or in PBS and cultured for 1.5 h, followed by the estimation of luciferase activity. All data shown are the means ± SEM of duplicate determinations of experiments performed three times.
We monitored the activity of the MSMEG_3780 promoter during growth, and in agreement with the Western blot analysis, promoter activity was highest at 12 h and reduced dramatically thereafter (Fig. 5C). The activity of the sigA promoter remained constant at all stages of growth. Therefore, it appears that MSMEG_3780 could be responsible for the maintenance of cAMP levels in the bacterium in the logarithmic phase of growth. Since we observed that the transcriptional regulation of MSMEG_3780 could contribute to cAMP levels, we also assessed MSMEG_3780 promoter activity during various stresses.
A dramatic reduction in MSMEG_3780 promoter activity was seen upon acid stress, suggesting that the reduced transcription of MSMEG_3780 could account for the reduction in cAMP levels in acidified cultures. MSMEG_3780 promoter activity remained constant under other stress conditions after 5 h, and no changes in sigA promoter activity were seen even under conditions of acid stress (Fig. 5D). MSMEG_3780 promoter activity was reduced (∼30%) under both SDS and PBS stresses in 1.5 h, and this reduction was also seen with the sigA promoter (Fig. 5E). Therefore, the increase in intracellular cAMP levels upon SDS treatment could not be correlated with the altered transcription of MSMEG_3780 but could be due to a general modulation of the catalytic activity of adenylyl cyclases in the bacteria since many of these proteins are membrane associated (38) and perhaps activated by mild detergents.
The Rv1647 promoter is acid sensitive, and the gene can complement the ΔMSMEG_3780 strain.
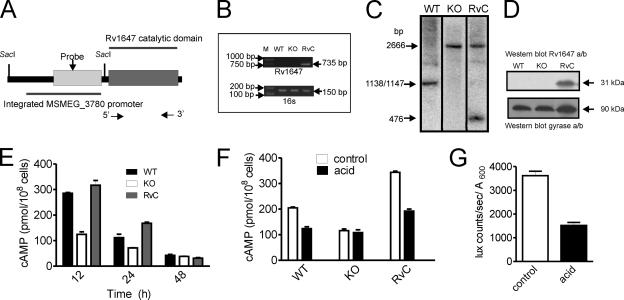
In the studies described thus far, we have observed changes in the expression levels of MSMEG_3780 that could be correlated with alterations in cAMP levels. In order to assess whether Rv1647 in M. tuberculosis shares these attributes, we studied the functional similarity between the Rv1647 and MSEMG_3780 proteins by generating a ΔMSMEG_3780 strain complemented by the expression of Rv1647 (Fig. 6A and B). Southern blotting confirmed the presence of the Rv1647 gene in the ΔMSMEG_3780 strain (Fig. 5C). The expression of Rv1647 in the complemented strain was seen by Western blot analysis using an antibody to Rv1647 (Fig. 6D). Complementation by the Rv1647 protein in the ΔMSMEG_3780 strain could indeed restore cAMP levels during early logarithmic growth in the strain (Fig. 6E), and acid stress also reduced cAMP levels in the complemented strain (Fig. 6F).
FIG. 6.
Characterization of the ΔMSMEG_3780 strain complemented with Rv1647. (A) Diagrammatic representation of the DNA inserted into the ΔMSMEG_3780 strain containing the MSMEG_3780 promoter driving the expression of the catalytic domain of the Rv1647 gene. The SacI sites used for analysis are shown, and arrows indicate the positions of primers used for PCR. (B) PCR analysis of genomic DNA prepared from the wild-type (WT), knockout (KO), and complement (RvC) strains using Rv1647-specific primers. Also shown is a PCR of the genomic DNA with primers specific to the 16S gene. (C) Southern blot analysis of the DNA prepared from wild-type, knockout, and complement strains was performed following SacI digestion. The probe used was a region present in the MSMEG_3780 promoter. The fragments hybridizing in wild-type and knockout genomic DNAs are identical to those shown in Fig. 3, while a convenient SacI site that was introduced into the Rv1647 fragment used for integration allowed the detection of a unique fragment in complement genomic DNA. (D) Western blot analysis using membranes prepared from wild-type, knockout, and complement strains at 30 h of growth. Total protein (50 μg) in the blot was normalized using an affinity-purified antibody to Rv1647 and a monoclonal antibody to the gyrase A subunit. (E) Intracellular cAMP levels were measured in the wild-type, knockout, and complement strains. Data shown are the means ± SEM of duplicate determinations of experiments performed twice. (F) Intracellular cAMP levels in wild-type, knockout, and complement strains following acid stress for 5 h. Values shown are the means ± SEM. (G) The Rv1647 promoter was fused to luxAB, and the promoter activity was measured in control cells and cells cultured under acidic conditions for 5 h. Data shown represent the means ± SEM of duplicate determinations of experiments performed twice.
In order to show that Rv1647 gene transcription could be regulated by acid stress, we cloned the putative Rv1647 promoter (encompassing the 107-bp intergenic region between the Rv1646 and Rv1647 genes to 298 bp into the Rv1647 gene) upstream of luciferase and monitored the regulation of its activity under conditions of acid stress. Indeed, the promoter of Rv1647 was also downregulated upon acid stress (Fig. 6G). In a recent study where a global analysis of genes in M. tuberculosis that were sensitive to pH 5.5 was reported (31), the Rv1647 gene was not found to show a change in mRNA levels. That microarray analysis was performed 2 h after subjecting cells to acid stress, indicating that Rv1647 mRNA could be stable for many hours, as the transcription of the Rv1647 gene is reduced dramatically in as little as 1 h of acid stress (Fig. 6G).
In conclusion, the results presented here have shown that an adenylyl cyclase from M. tuberculosis could functionally complement its ortholog in M. smegmatis, thereby validating the use of M. smegmatis as a model system to study the roles of cAMP and adenylyl cyclases in mycobacteria.
DISCUSSION
A number of questions as to the role of cAMP in mycobacteria remain, and the importance of this second messenger in the physiology and perhaps pathophysiology of these bacteria remains largely speculative (39). Here, we have studied the functional importance of a highly conserved adenylyl cyclase gene in mycobacteria and used M. smegmatis as a model system to address questions related to the synthesis of cAMP by MSMEG_3780 and levels of cAMP in M. smegmatis during bacterial growth and under conditions of stress. Under normal growth conditions, levels of cAMP appear to be high in the log phase, and at least some of this cAMP is generated by MSMEG_3780, since cAMP levels at 12 h and 24 h of growth were lower in the ΔMSMEG_3780 strain. The marked reduction in intracellular cAMP levels in the stationary phase (<10% of the levels seen at 24 h of growth) could not be accounted for by a reduction in MSMEG_3780 expression, since levels of cAMP fell even in the knockout strain. This indicates that either a very efficient degradation mechanism is operative in the cell or there is low adenylyl cyclase activity at later stages of growth. We have identified the product of the Rv0805 gene in M. tuberculosis as being a cAMP phosphodiesterase (32, 34), but no ortholog of Rv0805 has been seen in M. smegmatis. Perhaps other cAMP phosphodiesterases in M. smegmatis need to be identified.
The Rv1625c and Rv1264 adenylyl cyclase knockouts generated in M. tuberculosis did not show dramatic phenotypic differences from wild-type H37Rv (9, 15), nor did they show reduced virulence. In the latter studies, no measurements of cAMP were made, and therefore, the contribution of these genes to regulating cAMP levels in M. tuberculosis is not known.
Certain stress conditions (PBS, acid stress, and SDS) regulated the levels of intracellular cAMP. Therefore, genes (possibly involved in adaptation to these stresses) that are regulated under these conditions could be cyclic AMP receptor protein (CRP) responsive in mycobacteria. Indeed, the importance of cAMP and CRP in regulating the expression of acid shock proteins in E. coli was reported previously (6), suggesting the possibility that acid-inducible or -repressed genes in mycobacteria could also be regulated by cAMP levels in the cell. It is interesting that the increase in cAMP levels normally associated with carbon starvation in E. coli was not seen in M. smegmatis. The major intrinsic protein family of glycerol facilitators was identified in the M. smegmatis genome, while no members of this family could be detected in the genome of M. tuberculosis (43), and it is likely that these transporters facilitate the uptake of glycerol in M. smegmatis, protecting it from severe carbon stress. There are far fewer genes involved in carbohydrate uptake in M. tuberculosis than in M. smegmatis (4 versus 28, respectively) (43), suggesting that cAMP responses to carbon deprivation could vary in these two bacteria.
It is intriguing that a large amount of cAMP is extruded by M. smegmatis into the culture medium. It was suggested previously that cAMP secreted by pathogenic Mycobacterium microti is responsible for the inhibition of phagosome-lysosome fusion (21), thereby implicating cAMP as being a virulence factor or a “toxin” in mycobacteria. This mechanism of elevating cAMP levels in host cells is distinct from the strategy employed by pathogens such as Bordetella pertussis and Bacillus anthracis, which secrete potent class IV adenylyl cyclase directly into the host cell (1). Whether cAMP has a novel, as-yet-unidentified role as an extracellular signaling molecule in mycobacteria is not known. Indeed, cAMP is known to act as the first messenger in the swarming behavior of Dictyostelium (10), making it attractive to suggest that cAMP could have a role to play in quorum sensing and perhaps biofilm formation in mycobacteria.
The secretion of cAMP could also be a mechanism of regulating intracellular levels of cAMP, along with the degradation of cAMP by phosphodiesterases such as Rv0805. The Rv0805 gene is predicted to be regulated by the CRP (4), indicating feedback regulation of Rv0805 transcription and thereby intracellular cAMP concentrations. However, the overexpression of the Rv0805 gene in M. smegmatis reduced intracellular levels of cAMP by ∼30% (34), indicating that the synthetic capacity of cAMP exceeds the ability of Rv0805 to degrade cAMP. Given that the formation of cAMP by the cell requires ATP and is therefore energy demanding, the high amounts of cAMP produced by mycobacteria should have a vital and perhaps as-yet-unidentified role to play.
Rv1647 and it orthologs are present in all mycobacterial genomes sequenced so far (35). Thus, the present studies are the first to describe in detail a strain of M. smegmatis that has a deletion in this highly conserved adenylyl cyclase, the product of the MSMEG_3780 gene. Despite the conservation of this gene across several mycobacteria, it appears to be dispensable for growth in vitro. Interestingly, however, transcription of MSMEG_3780 varies during in vitro growth of cells, and the MSMEG_3780 gene, as does its counterpart in M. tuberculosis, contains an acid-sensitive promoter. Therefore, the generation of a strain of M. tuberculosis that is deficient in the Rv1647 gene would shed light on the role that this gene may have to play not only in the normal physiology of the bacteria but also during infection and in the persistence of the pathogen during tuberculosis.
Acknowledgments
We thank Achuth Padmanabhan for help in several experiments and members of the laboratory for many useful discussions and careful reading of the paper. We also thank Vani R. Iyer for technical assistance. We also acknowledge the use of the Proteomics Facility of the Indian Institute of Science for mass spectrometric analysis.
This study was supported by the Department of Biotechnology, Government of India.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Ahuja, N., P. Kumar, and R. Bhatnagar. 2004. The adenylate cyclase toxins. Crit. Rev. Microbiol. 30187-196. [DOI] [PubMed] [Google Scholar]
- 2.Antoni, F. A. 2000. Molecular diversity of cyclic AMP signalling. Front. Neuroendocrinol. 21103-132. [DOI] [PubMed] [Google Scholar]
- 3.Bai, G., M. A. Gazdik, D. D. Schaak, and K. A. McDonough. 2007. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect. Immun. 755509-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, G., L. A. McCue, and K. A. McDonough. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 1877795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, D. A., and J. M. Kelly. 2004. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol. Microbiol. 521229-1242. [DOI] [PubMed] [Google Scholar]
- 6.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147709-715. [DOI] [PubMed] [Google Scholar]
- 7.Danchin, A. 1993. Phylogeny of adenylyl cyclases. Adv. Second Messenger Phosphoprotein Res. 27109-162. [PubMed] [Google Scholar]
- 8.Delogu, G., A. Bua, C. Pusceddu, M. Parra, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Expression and purification of recombinant methylated HBHA in Mycobacterium smegmatis. FEMS Microbiol. Lett. 23933-39. [DOI] [PubMed] [Google Scholar]
- 9.Dittrich, D., C. Keller, S. Ehlers, J. E. Schultz, and P. Sander. 2006. Characterization of a Mycobacterium tuberculosis mutant deficient in pH-sensing adenylate cyclase Rv1264. Int. J. Med. Microbiol. 296563-566. [DOI] [PubMed] [Google Scholar]
- 10.Dormann, D., and C. J. Weijer. 2006. Chemotactic cell movement during Dictyostelium development and gastrulation. Curr. Opin. Genet. Dev. 16367-373. [DOI] [PubMed] [Google Scholar]
- 11.Gazdik, M. A., and K. A. McDonough. 2005. Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J. Bacteriol. 1872681-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez, M., L. Doukhan, G. Nair, and I. Smith. 1998. sigA is an essential gene in Mycobacterium smegmatis. Mol. Microbiol. 29617-628. [DOI] [PubMed] [Google Scholar]
- 13.Gstrein-Reider, E., and M. Schweiger. 1982. Regulation of adenylate cyclase in E. coli. EMBO J. 1333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu, S., J. Chen, K. M. Dobos, E. M. Bradbury, J. T. Belisle, and X. Chen. 2003. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 21284-1296. [DOI] [PubMed] [Google Scholar]
- 15.Guo, Y. L., U. Kurz, A. Schultz, J. U. Linder, D. Dittrich, C. Keller, S. Ehlers, P. Sander, and J. E. Schultz. 2005. Interaction of Rv1625c, a mycobacterial class IIIa adenylyl cyclase, with a mammalian congener. Mol. Microbiol. 57667-677. [DOI] [PubMed] [Google Scholar]
- 16.Horinouchi, S., Y. Ohnishi, and D. K. Kang. 2001. The A-factor regulatory cascade and cAMP in the regulation of physiological and morphological development in Streptomyces griseus. J. Ind. Microbiol. Biotechnol. 27177-182. [DOI] [PubMed] [Google Scholar]
- 17.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 18.Ketkar, A. D., A. R. Shenoy, U. A. Ramagopal, S. S. Visweswariah, and K. Suguna. 2006. A structural basis for the role of nucleotide specifying residues in regulating the oligomerization of the Rv1625c adenylyl cyclase from M. tuberculosis. J. Mol. Biol. 356904-916. [DOI] [PubMed] [Google Scholar]
- 19.Klengel, T., W. J. Liang, J. Chaloupka, C. Ruoff, K. Schroppel, J. R. Naglik, S. E. Eckert, E. G. Mogensen, K. Haynes, M. F. Tuite, L. R. Levin, J. Buck, and F. A. Muhlschlegel. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 152021-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linder, J. U., and J. E. Schultz. 2003. The class III adenylyl cyclases: multi-purpose signalling modules. Cell. Signal. 151081-1089. [DOI] [PubMed] [Google Scholar]
- 21.Lowrie, D. B., P. S. Jackett, and N. A. Ratcliffe. 1975. Mycobacterium microti may protect itself from intracellular destruction by releasing cyclic AMP into phagosomes. Nature 254600-602. [DOI] [PubMed] [Google Scholar]
- 22.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31715-724. [DOI] [PubMed] [Google Scholar]
- 23.Manjunatha, U. H., S. Mahadevan, S. S. Visweswariah, and V. Nagaraja. 2001. Monoclonal antibodies to mycobacterial DNA gyrase A inhibit DNA supercoiling activity. Eur. J. Biochem. 2682038-2046. [DOI] [PubMed] [Google Scholar]
- 24.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10204-219. [DOI] [PubMed] [Google Scholar]
- 25.Newman, J. 2004. Novel buffer systems for macromolecular crystallization. Acta Crystallogr. D Biol. Crystallogr. 60610-612. [DOI] [PubMed] [Google Scholar]
- 26.Padh, H., and T. A. Venkitasubramanian. 1976. Cyclic adenosine 3′,5′-monophosphate in mycobacteria. Indian J. Biochem. Biophys. 13413-414. [PubMed] [Google Scholar]
- 27.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 1461969-1975. [DOI] [PubMed] [Google Scholar]
- 28.Rao, M., T. L. Streur, F. E. Aldwell, and G. M. Cook. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 1471017-1024. [DOI] [PubMed] [Google Scholar]
- 29.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menendez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 561274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, E. A., A. Clark, and R. L. Friedman. 2005. Bacterial luciferase is naturally destabilized in Mycobacterium tuberculosis and can be used to monitor changes in gene expression. FEMS Microbiol. Lett. 243243-249. [DOI] [PubMed] [Google Scholar]
- 31.Rohde, K. H., R. B. Abramovitch, and D. G. Russell. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2352-364. [DOI] [PubMed] [Google Scholar]
- 32.Shenoy, A. R., M. Capuder, P. Draskovic, D. Lamba, S. S. Visweswariah, and M. Podobnik. 2007. Structural and biochemical analysis of the Rv0805 cyclic nucleotide phosphodiesterase from Mycobacterium tuberculosis. J. Mol. Biol. 365211-225. [DOI] [PubMed] [Google Scholar]
- 33.Shenoy, A. R., K. Sivakumar, A. Krupa, N. Srinivasan, and S. S. Visweswariah. 2004. A survey of nucleotide cyclases in Actinobacteria: unique domain organization and expansion of the class III cyclase family in Mycobacterium tuberculosis. Comp. Funct. Genomics 517-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shenoy, A. R., N. Sreenath, M. Podobnik, M. Kovacevic, and S. S. Visweswariah. 2005. The Rv0805 gene from Mycobacterium tuberculosis encodes a 3′,5′-cyclic nucleotide phosphodiesterase: biochemical and mutational analysis. Biochemistry 4415695-15704. [DOI] [PubMed] [Google Scholar]
- 35.Shenoy, A. R., N. P. Sreenath, M. Mahalingam, and S. S. Visweswariah. 2005. Characterization of phylogenetically distant members of the adenylyl cyclase family from mycobacteria: Rv1647 from M. tuberculosis and its ortholog ML1399 from M. leprae. Biochem. J. 387541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shenoy, A. R., A. Srinivas, M. Mahalingam, and S. S. Visweswariah. 2005. An adenylyl cyclase pseudogene in Mycobacterium tuberculosis has a functional ortholog in Mycobacterium avium. Biochimie 87557-563. [DOI] [PubMed] [Google Scholar]
- 37.Shenoy, A. R., and S. S. Visweswariah. 2004. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 56111-21. [DOI] [PubMed] [Google Scholar]
- 38.Shenoy, A. R., and S. S. Visweswariah. 2006. Mycobacterial adenylyl cyclases: biochemical diversity and structural plasticity. FEBS Lett. 5803344-3352. [DOI] [PubMed] [Google Scholar]
- 39.Shenoy, A. R., and S. S. Visweswariah. 2006. New messages from old messengers: cAMP and mycobacteria. Trends Microbiol. 14543-550. [DOI] [PubMed] [Google Scholar]
- 40.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, R. S., M. C. Wolfgang, and S. Lory. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 721677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spreadbury, C. L., M. J. Pallen, T. Overton, M. A. Behr, S. Mostowy, S. Spiro, S. J. Busby, and J. A. Cole. 2005. Point mutations in the DNA- and cNMP-binding domains of the homologue of the cAMP receptor protein (CRP) in Mycobacterium bovis BCG: implications for the inactivation of a global regulator and strain attenuation. Microbiology 151547-556. [DOI] [PubMed] [Google Scholar]
- 43.Titgemeyer, F., J. Amon, S. Parche, M. Mahfoud, J. Bail, M. Schlicht, N. Rehm, D. Hillmann, J. Stephan, B. Walter, A. Burkovski, and M. Niederweis. 2007. A genomic view of sugar transport in Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Bacteriol. 1895903-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]