Abstract
We describe a two-gene cluster encoding a bacteriocin, xenocin, and the cognate immunity protein in the insect-pathogenic bacterium Xenorhabdus nematophila, which infects and kills larval stages of the common crop pest Helicoverpa armigera. The two genes, xcinA and ximB, are present in the genome as a single transcriptional unit, which is regulated under SOS conditions. The stress-inducible promoter was activated by mitomycin C, glucose, and Fe3+ depletion and at an elevated temperature when it was tested in Escherichia coli cells. Expression of the xenocin protein alone in E. coli inhibited the growth of this organism. The growth inhibition was abolished when the immunity protein was also present. A recombinant xenocin-immunity protein complex inhibited the growth of E. coli indicator cells when it was added exogenously to a growing culture. Xenocin is an endoribonuclease with an enzymatically active C-terminal domain. Six resident bacterial species (i.e., Bacillus, Enterobacter, Enterococcus, Citrobacter, Serratia, and Stenotrophomonas species) from the H. armigera gut exhibited sensitivity to recombinant xenocin when the organisms were grown under iron-depleted conditions and at a high temperature. Xenocin also inhibited the growth of two Xenorhabdus isolates. This study demonstrates that Fe3+ depletion acts as a common cue for synthesis of xenocin by X. nematophila and sensitization of the target strains to the bacteriocin.
Bacteriocins constitute the most abundant and diverse family of microbial defense systems that are produced to control the growth of closely related, competing bacterial species inhabiting a common ecological niche (32). Microbes encounter in their natural habitats a variety of stressful conditions that cause damage to cellular DNA. For survival of the population under such conditions, the microbes respond by activating a DNA repair regulatory network, called the SOS response. This network involves coordinated induction of multiple genetic loci to maintain the integrity of the bacterial genome (18, 28). Activation of genes responsible for synthesis and release of bacteriocins is one of the results of the bacterial SOS response. The genes encoding the bacteriocins are mostly plasmid encoded and are organized as pairs; the first open reading frame (ORF) encodes a killer protein, and the second ORF encodes an immunity protein, which provides the host cell with a defense against the toxic action of the killer protein (21). The antibiotic proteins are named after the bacterial species that produce them; e.g., the “colicins” are derived from Escherichia coli (16), the “pyocins” are derived from Pseudomonas aeruginosa (32), and “klebocin” is derived from Klebsiella pneumoniae (10). Most bacteriocins have a common molecular organization consisting of three functional domains, the receptor binding, membrane translocation, and cytotoxic domains. In gram-negative bacteria, the lethal action of bacteriocins involves the following essential steps. After release from the host cell, the bacteriocin binds to specific surface receptors on the target cells, such as vitamin B12 receptor BtuB or iron siderophore receptor FepA (4). This binding is followed by import into the cells with the help of either Ton proteins (ExbB, ExbD, and TonB) or Tol proteins (TolA, -B, -Q, and -R) (24, 37) for interaction with the target molecule. Bacteriocins use a variety of strategies to induce cell death through their cytotoxic domains. The pore-forming bacteriocins create voltage-gated channels, causing depolarization of the cytoplasmic membrane (23, 47), while nucleases cleave tRNA or rRNA at specific sites to inhibit protein synthesis (27, 42) or degrade nucleic acids nonspecifically in the target cells (17, 19).
Xenorhabdus nematophila is a gram-negative gammaproteobacterium belonging to the family Enterobacteriaceae (7, 15). In nature this organism lives in a specific symbiotic association with entomopathogenic nematodes belonging to the family Steinernematide (2). The bacterium-nematode complex is pathogenic to insects (1, 5, 15) and is used as a biological control agent against different groups of insects, including Lepidoptera, Coleoptera, and Diptera (11, 12, 30, 38). The infective juvenile larvae of the nematode carry the bacterium into the insect and release it in the gut or hemocoel depending upon the route of infection (1, 15). X. nematophila is known to produce potent cytotoxins (15) and orally toxic proteins that kill larval stages of Helicoverpa armigera (20, 44), a polyphagous lepidopteran pest that infests important crop plants and accounts for huge losses in agricultural output. After the death of the larval prey, the bacterium secretes a variety of antibiotic molecules, including antibacterial peptides, antibiotics like xenorhabdins, xenocoumacins, etc., and bacteriocins (34), that eliminate most of the microbial species present in the dead larvae and prevent putrefaction, which is necessary for optimum nematode growth. In this study we describe a novel bacteriocin (xenocin) gene cluster in the genome of the entomopathogenic bacterium X. nematophila.
MATERIALS AND METHODS
Bacterial strain, media, and culture conditions.
X. nematophila strain 19061 was grown at 28°C. E. coli strain DH5α (Bethesda Research Laboratories) was used as the host for cloning. The BL21(DE3)/pLysS and M15 strains were used in expression studies and as indicator cells. The plasmid vector pGEM-T Easy from Promega (Madison, WI) was used for PCR cloning. pBSK(+) was used for preparing a partial genomic library. Luria-Bertani (LB) medium and M9 minimal medium (with no added iron) were used to grow bacterial strains. Ampicillin, kanamycin, and chloramphenicol were used at concentrations of 100, 35, and 25 μg/ml, respectively.
Isolation of the xenocin gene cluster from the Xenorhabdus genome.
In an unrelated study to isolate the phospholipase C gene of X. nematophila, a 500-bp DNA fragment was obtained by PCR amplification. Sequence analysis of this fragment revealed homology with a hemolytic protein of Neisseria meningitidis (accession number AAF42109). The 500-bp DNA fragment hybridized with a ∼4.5-kb band in the partially digested genomic DNA of X. nematophila. A partial genomic DNA library was produced by cloning 4- to 6-kb HindIII-digested genomic DNA fragments in the pBSK+ vector. Genomic DNA clones were screened with a 500-bp radiolabeled DNA probe (3). Positive clones were grown in LB medium, and plasmid pBSK4.3 (Table 1) was isolated and sequenced.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid/strain | Characteristics | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 φ80dlacZΔM15 | Invitrogen |
| M15 | NaIs Strs Rifs Thi− Lac− Ara+ Gal+ MtI− F− RecA+ Uvr+ Lon+ | Qiagen |
| BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm(DE3)/pLysS(Cmr) | Novagen |
| Plasmids | ||
| pBSK(+) | 2.9-kb cloning vector; Ampr | Stratagene |
| pGEM-T Easy | 3-kb vector for cloning PCR fragments; Ampr | Promega |
| pQE30 | 3.4-kb expression vector; Ampr | Qiagen |
| pET28 | 5.3-kb expression vector; Kanr | Novagen |
| pBSK4.3/BSK4.3 | pBSK(+) containing 4.3-kb insert from X. nematophila genome | This study |
| pGEM1/GEM1 | pGEM-T Easy containing xcinA and ximB genes with native promoter | This study |
| pBSK1/BSK1 | pBSK(+) containing xcinA and ximB genes with native promoter | This study |
| pGEM2/GEM2 | pGEM-T Easy containing xcinA alone with native promoter | This study |
| pBSK2/BSK2 | pBSK(+) containing xcinA alone with native promoter | This study |
| pJC1/JC1 | pGEM-T Easy containing xcinA and ximB genes | This study |
| pJC2/JC2 | pQE30 containing xcinA amd ximB genes | This study |
| pJC3/JC3 | pET28 containing 318-bp catalytic domain | This study |
| pJC4/JC4 | pET28 containing catalytic domain and 270-bp partial ximB (N′-terminal 86 amino acids) | This study |
| pLacZ/LacZ | pGEM-T Easy containing lacZ coding sequence | This study |
| pXcinPr/XcinPr | pGEM-T Easy containing 300-bp upstream sequence of xcinA | This study |
| pXPro-lacZ/XPro-lacZ | pGEM-T Easy containing xcinA upstream sequence fused to lacZ coding sequence | This study |
| pXimPr/XimPr | pGEM-T Easy containing 300-bp upstream sequence of ximB | This study |
| pIPro-lacZ/IPro-lacZ | pGEM-T Easy containing ximB upstream sequence fused to lacZ coding sequence | This study |
Bioinformatic analysis.
The 4.3-kb sequence was analyzed bioinformatically using the ORF-finding services of NCBI (http://www.ncbi.nlm.nih.gov). The protein data banks were searched using the BLASTP program (http://www.ncbi.nlm.nih.gov/BLAST). Different annotated colicin sequences were aligned by using ClustalW (Mac vector 7.0). Identities of promoter sequences associated with the ORFs were determined by using the software BPROMO (www.softberry.com) and www.fruitfly.org.
RNA preparation and Northern hybridization.
Overnight cultures of X. nematophila were diluted 100-fold into fresh LB medium, and grown until the optical density at 600 nm was 0.5. Mitomycin C (0.3 μg/ml) was added to the growing cultures to induce bacteriocin genes, and 10-ml samples were removed at different times. Total RNA was prepared using an RNeasy kit (Qiagen) according to the manufacturer's protocol. For Northern hybridization 10 μg of total RNA was denatured at 65°C for 10 min in 5× RNA loading buffer, 10 ml of which consisted of 80 μl of 500 mM EDTA (pH 8), 720 μl of 37% (12.3 M) formaldehyde, 2 ml of 100% glycerol, 3 ml of formamide, 4 ml of 10× gel buffer [200 mM 3-(morpholino)propanesulfonic acid (MOPS) (free acid), 50 mM sodium acetate, 10 mM EDTA (pH 7.0)], and 16 μl of a bromophenol blue solution. RNA samples were resolved in 1.2% agarose gels prepared in MOPS buffer with 1.8% (vol/vol) formaldehyde. DNA that included ORF1 (xcinA) and ORF2 (ximB) were radioactively labeled by nick translation (Invitrogen) and used as probes after purification. RNA was transferred overnight on a nylon membrane (Amersham Life Science, Illinois) in the presence of 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was performed at 65°C for xcinA and at 58°C for ximB overnight. The membrane was washed, and the blots were developed using a phosphorimager (Typhoon 9210; Amersham).
RT-PCR analysis.
X. nematophila cells were subjected to different stress conditions, including mitomycin C (as described above), 25 mM Desferal (Sigma Aldrich), and a high temperature (37°C), and total RNA was isolated as described above. The RNA was treated with DNase (RQ1 RNase-free DNase; Promega) as recommended by the manufacturer and checked by PCR for DNA contamination before reverse transcription (RT)-PCR was performed using a Qiagen one-step RT-PCR kit. A 50-μl reaction mixture containing 100 ng RNA (DNA free), each primer at a concentration of 20 mM, 5× Qiagen one-step RT-PCR buffer, a deoxynucleoside triphosphate mixture (containing each deoxynucleoside triphosphate at a concentration of 10 mM), and 2 μl of Qiagen one-step RT-PCR enzyme mixture was incubated at 50°C for 30 min. For PCR DNA was denatured at 95°C in the first cycle, and this was followed by 30 cycles of amplification.
Promoter identification by primer extension analysis.
To determine the 5′ limits of the RNA expressing the xcinA and ximB genes, a primer extension reaction was performed using the avian myeloblastosis virus reverse transcriptase system (Promega). Twenty-four-base pair primers PriXc and PriIm (Table 2) were synthesized using the 5′ ends of the xcinA and ximB genes, respectively. The primers were end labeled with [γ-32P]ATP and T4 polynucleotide kinase using the manufacturer's protocol. The labeled primers were purified by filtration through a G50 column. Total RNA was isolated from uninduced X. nematophila cells and from cells after induction with mitomycin C, as described above. The primers were annealed with purified, denatured RNA at 45°C for 2 h. The RNA with annealed primers was precipitated with ethanol, and the extension reaction was carried out at 42°C for 1.5 h. The extension product was resolved in an 8% polyacrylamide denaturing gel for size determination.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| XenocinF1 | 5′ GGT ACC TGA GTA TCT TGT ATG GTT AAA CAT 3′ |
| 1 | 5′ GGA TCC ATG TGT CCA ATA TAC GGT GAT 3′ |
| 2 | 5′ AAG CTT AAG GTA TTT TTT AAT ATT GCG 3′ |
| 3 | 5′ GGA TCC ATG GGA ATT CAA TTA AAA 3′ |
| 4 | 5′ AAG CTT CTA TTT CTT CGG AAC AAT 3′ |
| 5 | 5′ AAG CTT GCT GAG CAA GAA CAA GAA CTA 3′ |
| 6 | 5′ GGA TCC TCG ATA ATC AAA TGA TAT TTG 3′ |
| XcinProF | 5′ GGT ACC TGA GTA TCT TGT ATG GTT AAA CAT 3′ |
| XcinProB | 5′ GGA TCC AAT AAA TTC CTT ATT TTT ATA AAT 3′ |
| XimProF | 5′ GGT ACC TGT CCA ACA GAA TGG ACA 3′ |
| XimProB | 5′ GGA TCC GAA TTT TCT CCA TTA AAG 3′ |
| PriXc | 5′ ACC ATC ACC GTA TAT TGG ACA CAT 3′ |
| PriIm | 5′ TGA ATT CCC ATG AAT TTT CTC CAT 3′ |
| LacZF | 5′ GGA TCC ATG ACC ATG ATT ACG GAT TCA |
| LacZB | 5′ AAG CTT TTA TTT TTG ACA CCA GAC CAA 3′ |
Determination of promoter activity by reporter lacZ gene fusion.
For fusion of upstream regions of the xcinA and ximB genes with the lacZ coding sequence, the lacZ coding sequence was PCR amplified from the genome of E. coli K-12 using forward primer LacZF and backward primer LacZB containing BamHI and HindIII sites, respectively, and the resulting fragment was cloned in the pGEM-T Easy vector, producing pLacZ. The upstream regions of xcinA and ximB (300 bp each upstream of the start codon) were also PCR amplified using primers XcinPrF and XcinPrB and primers XimPrF and XimPrB, respectively, and 4.3 kb of genomic DNA as the template. The amplified fragments with a HindIII site at the 5′ end and a BamHI site at the 3′ end were cloned into the pGEM-T Easy vector, and the pXcinPr and pXimPr constructs were obtained. Both plasmids were digested with SphI (site present in the pGEM-T Easy vector) and BamHI (site present in the insert), releasing the upstream regions, which were ligated to the pLacZ plasmid with complementary ends. The fusion constructs were transformed in DH5α cells, and positive clones were selected. Plasmids pXPr-lacZ and pIPr-lacZ from the selected clones were sequenced.
Strains containing the promoter-lacZ fusions were grown in LB medium at 30°C to log phase and subjected to different stress conditions, including a higher temperature (42°C); for SOS conditions, log-phase cells were washed and resuspended in M9 medium in the presence of mitomycin C (0.3 μg/ml). To study the effect of nutrients like glucose, iron, etc., cells were grown to log phase in LB medium and resuspended in M9 medium under specific nutrient conditions. The cultures were incubated for 4 to 6 h, and 1-ml samples were removed at regular intervals to measure β-galactosidase activity, which was expressed in Miller units (13).
Cloning and expression of xenocin, catalytic domain, and immunity proteins.
ORF1 and ORF2 encoding xenocin and immunity protein, respectively, were obtained by PCR amplification for cloning. Primer 1 (the positions of the primers are shown in Fig. 3) with a BamHI site at the 5′ end and a backward primer, primer 4 with a HindIII site at the 3′ end, were used to amplify the two contiguous genes from the 4.3-kb genomic DNA fragment. The amplified product (2.848 kb) was ligated into the pGEM-T Easy vector, producing the pJC1 plasmid. Plasmid DNA was digested with BamHI and HindIII, and the 2.848-kb fragment was ligated to the pQE30 vector, resulting in plasmid pJC2. Plasmid pJC2 was transformed in M15 cells, and the resulting strain, strain JC2, was used for expression and purification of both proteins with a six-His tag under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter. The catalytic domain of xenocin was cloned alone and with the immunity domain (N-terminal 86 residues of the immunity protein). For the catalytic domain primers 5 and 2 and for the catalytic and immunity domains primers 5 and 6 with a BamHI site in the forward primer and a HindIII site in the backward primer were used for PCR amplification. The 318- and 588-bp products were first cloned in the pGEM-T Easy vector, excised by digestion with BamHI and HindIII, and ligated to the pET28(a) vector, and the resulting constructs, pJC3 and pJC4, were transformed in E. coli BL21(DE3)/pLysS cells.
FIG. 3.
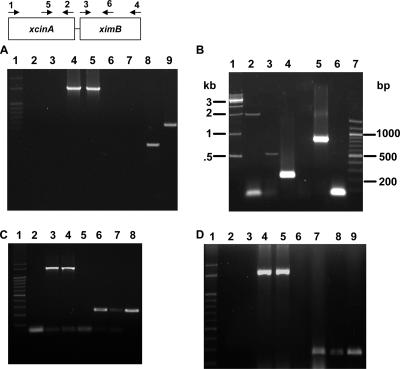
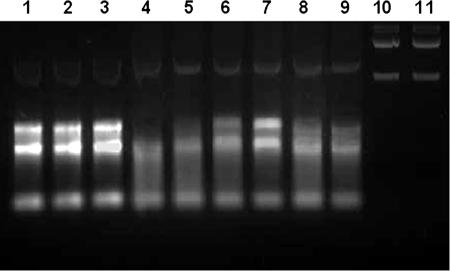
Analysis of xcinA and ximB mRNA by RT-PCR. (A) Comparison of X. nematophila strain 19060 and xenocin-sensitive isolate X(sensitive) after induction with mitomycin C. Lane 1, markers; lanes 2 and 3, RNA from X(sensitive) and strain 19060, respectively, without reverse transcriptase; lanes 4 and 5, control 16S RNA from X(sensitive) and strain 19060, respectively; lanes 6 and 7, RNA from X(sensitive) amplified with primers 5 and 2 (lane 6) and with primers 5 and 6 (lane 7); lanes 8 and 9, RNA from X. nematophila 19060 amplified with primers 5 and 2 (lane 8) and with primers 5 and 6 (lane 9). (B) X. nematophila cells were induced with mitomycin C for 3 h, and total RNA was isolated. Lane 1, 1-kb ladder; lane 2, primers 1 and 2; lane 3, primers 5 and 6; lane 4, primers 5 and 2; lane 5, primers 3 and 4; lane 6, primers 1 and 4; lane 7, 100-bp ladder. (C) Analysis of xcinA mRNA induced by 25 mM Desferal. Lane 1, 100-bp ladder; lane 2, RNA from induced cells without reverse transcriptase, amplified with primers 5 and 2; lanes 3 and 4, 16S RNA from uninduced and induced loading controls, respectively; lanes 5 and 6, RNA from uninduced (lane 5) and induced (lane 6) cells amplified with primers 5 and 2; lanes 7 and 8, RNA from uninduced (lane 7) and induced (lane 8) cells amplified with primers 3 and 6. (D) Analysis of xcinA mRNA by heat induction. Lane 1, 100-bp ladder; lanes 2 and 3, RNA from uninduced and induced cells without reverse transcriptase, respectively; lanes 4 and 5, 16S RNA controls; lanes 6 and 7, RNA from cells grown at 30°C (lane 6) and at 37°C (lane 7) amplified with primers 5 and 2; lanes 8 and 9, RNA from cells grown at 30°C (lane 8) and at 37°C (lane 9) amplified with primers 3 and 6.
Purification of recombinant activity domains, xenocin, and immunity proteins.
Strains JC2 and JC4 were grown at 30°C in LB medium containing appropriate antibiotics. The proteins were induced with 1 mM IPTG when the optical density of the culture reached 0.5, and incubation was continued for 6 h. Cells were harvested and washed with 40 ml of cold 50 mM sodium phosphate buffer (pH 8) containing 300 mM NaCl (buffer A). The cell pellet was suspended in 25 ml of buffer A, and cells were disrupted by sonication at 4°C. The cell lysate was centrifuged at 12,000 × g for 30 min at 4°C in an RC5 plus centrifuge, and the six-His-tagged xenocin-immunity protein complex and the catalytic domain-immunity domain complexes were purified from JC2 and JC4, respectively, as follows. The supernatant from the centrifugation step was loaded on an Ni-nitrilotriacetic acid (NTA) agarose column preequilibrated with buffer A at 4°C. The column was washed extensively with buffer A containing 50 mM imidazole, and the protein complex was eluted with buffer A containing 300 mM imidazole. Fractions containing pure protein complex were concentrated using Centricon (Millipore PM10). The xenocin-immunity protein complex was dialyzed overnight against 100 volumes of 50 mM sodium phosphate buffer (pH 7), and the final preparation was stored at −20°C in the presence of 15% glycerol. To separate xenocin from the immunity protein, the protein complex was denatured with 6 M guanidine-HCl in 50 mM sodium phosphate buffer (pH 8) containing 150 mM NaCl (buffer B) at room temperature for 2 h and loaded on an Ni-NTA spin column preequilibrated with buffer B. The immunity protein was obtained in the flowthrough fraction; the xenocin with a hexahistidine tag was eluted next from the column with 300 mM imidazole in buffer A. Purified proteins were dialyzed overnight against 100 volumes of 20 mM sodium phosphate buffer (pH 7).
To obtain the individual domain proteins, the Ni-NTA-purified catalytic domain-immunity domain complex was dialyzed against 20 mM glycine-HCl buffer (pH 3.0) overnight, followed by purification with a Sepharose-SP column (HiTrap SP; Amersham Biosciences) as described previously (39). First the catalytic domain was eluted with an NaCl gradient (0 to 2 M, pH 3), and then the immunity domain was eluted with 20 mM sodium phosphate buffer (pH 7.0). The domains were dialyzed against sodium phosphate buffer (pH 7.0) for functional assays.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry.
To confirm the identity of the protein, 2 μg of cloned and purified catalytic domain protein was digested with 80 ng of trypsin in 25 mM ammonium bicarbonate buffer (pH 8.5) at room temperature. After 5 h of incubation, the reaction was stopped with 60 ng of trypsin inhibitor, and a 2-μl sample was spotted onto a 96-well matrix-assisted laser desorption ionization plate. Each spot was overlaid with a 1:1 mixture of α-cyano-4-hydroxycinnamic acid (10 mg/ml) in 50% acetonitrile and 0.1% trifluoroacetic acid, and the spectra were recorded.
In vitro RNase activity.
RNase activity was measured by using total bacterial RNA from E. coli strain BL21(DE3)/pLysS as the substrate. The reaction mixture (20 μl) contained 1.2 μg of RNA in 50 mM Tris HCl buffer (pH 7.5), 50 mM NaCl, 5 mM EDTA and the protein sample to be tested. After 1.5 h of incubation at 37°C, 2.5 μl of the loading buffer (40% sucrose, 0.125 M EDTA, 0.5% sodium dodecyl sulfate; pH 8) was added, and the mixture was heated at 95°C for 2 min and resolved on a 1% agarose gel containing ethidium bromide.
Endogenous activities of domains, xenocin, and immunity proteins.
To study the toxic effect of xenocin and its neutralization by the immunity protein, a 3.148-kb DNA fragment containing both the xcinA and ximB genes with native promoters was amplified using primer XenocinF1 (300 bp upstream of the start codon of the xcinA locus) and primer 4, (primer positions are shown in Fig. 3) and cloned in the pBSK(+) vector, producing the pBSK1 construct. The xcinA gene alone with its native promoter was also amplified using primer XenocinF1 and primer 2. The 2.030-kb DNA product was also cloned in the pBSK(+) vector, yielding plasmid pBSK2. Both plasmids were transformed in DH5α cells. For protein expression, strains BSK1, BSK2, JC3, and JC4 were grown at 37°C in LB medium with appropriate antibiotics with shaking (∼200 rpm). Overnight cultures were subcultured in fresh medium and incubated until the optical density was 0.5. The cultures were diluted 1:100 in fresh medium and induced with either 0.3 μg/ml of mitomycin C (BSK1 and BSK2) or 1 mM IPTG (JC3 and JC4). The optical densities of the cultures were monitored to determine growth.
Exogenous growth inhibition assay.
The bacteriostatic activity of BSK1 cells after exposure to mitomycin C was tested. A culture was grown in LB medium to log phase at 30°C, induced with mitomycin C (0.3 μg/ml), and incubated for 5 h. The cells were centrifuged and sonicated to prepare the cytoplasmic fraction. The culture supernatants and cytoplasmic fractions were sterile filtered, and bacteriostatic activity was tested using indicator plates. Similarly, crude soluble extracts of cells containing the pJC2 plasmid, expressing the xenocin-immunity protein complex under control of the T7 promoter, was also tested on indicator plates. LB agar plates with or without antibiotics were overlaid with 3 ml of soft nutrient agar containing indicator E. coli strains containing pLysS or DH5α grown in M9 medium, and the protein complex was applied to the surface or to sterile disks. In some experiments the target cells with pLysS were prepared in M9 medium containing 2 μg/ml FeSO4. The plates were incubated overnight at 37°C, and the sizes of clearance zones were recorded.
Isolation of gut bacteria from H. armigera and sensitivity assay.
H. armigera third- and fourth-instar larvae were collected in the field and starved for 2 h. Each larva was dissected, and the gut was removed and resuspended in 1 ml of 1× phosphate-buffered saline. The cleaned guts were homogenized, serially diluted in 1× phosphate-buffered saline, and plated on tryptic soy broth plates. Morphologically distinct bacterial colonies were isolated and grown in M9 medium for 18 h, and bacterial lawns were prepared as described previously. The bacteriostatic effect of the recombinant xenocin-immunity protein complex was tested using bacterial lawns in a plate assay. The sensitive strains were identified by comparison with the 16S rRNA gene sequences in the database. Genomic DNA was isolated from each bacterial isolate, and the 16S rRNA gene was amplified using degenerate primers for the conserved regions of the gene. The amplified product was ligated in the pGEM-T Easy vector and transformed in DH5α cells. Positive clones were sequenced and identified with the help of bacterial Ribosomal Database Project II (http://rdp.cme.msu.edu/). Two Xenorhabdus isolates whose colony sizes differed were obtained from a Steinernema carpocapsiae-infected insect larva (unpublished results) and identified as described above.
Nucleotide sequence accession number.
The sequence of the 4.3-kb DNA fragment has been deposited in the NCBI data bank under accession number DQ 084383.
RESULTS
Analysis of the 4.3-kb genomic DNA fragment.
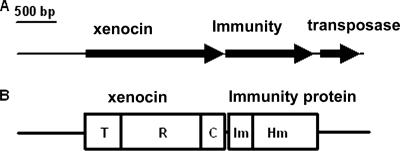
Nucleotide sequence analysis of the ∼4.3-kb genomic DNA fragment obtained from a clone reacting with the 500-bp DNA probe showed that there were three ORFs. The major ORF, 1,730-bp ORF1, corresponded to a 64-kDa product, and the second ORF, 1,106-bp ORF2, encoded a 42-kDa protein and was followed by the third ORF, ORF3, which was 448 bp long and encoded a transposase protein in a different reading frame (Fig. 1). Protein-protein BLAST analysis using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST) of the deduced 64-kDa protein showed that there was 31% similarity to cloacin DF13 and bacteriocin of K. pneumoniae and 30% similarity to colicins E3 and E6 of E. coli. The protein contained three characteristic domains, the N-terminal translocation domain, the C-terminal catalytic domain, and the central receptor binding domain (31). The 64-kDa protein was similar to RNase-type bacteriocins. Alignment of the putative 64-kDa bacteriocin with similar proteins using ClustalW indicated that the C-terminal catalytic domain was more conserved than the N-terminal translocation and receptor binding domains (see Fig. S1 in the supplemental material). The similarity at the level of the catalytic domain was much higher (77 and 75% with E. coli E6 and E3, respectively; 73% with cloacin DF13; and 72% with K. pneumoniae bacteriocin). ORF2 encoded a 42-kDa protein with 86 well-conserved, N-terminal amino acids forming a functionally active immunity domain that was fused to a putative hemolysin domain at the C terminus with homology to a hemolysin-like protein of N. meningitidis (accession number AAF42109). The functional significance of the hemolysin domain is being investigated; no hemolytic activity was observed with rabbit or sheep red blood cells or insect hemocytes. This domain does not appear to play any role in host cell lysis for the release of xenocin. A third ORF in the different frame was very similar to an ORF encoding a transposase of E. coli (3e-43).
FIG. 1.
(A) Genomic organization of X. nematophila xenocin gene cluster (4,337 bp). The arrows indicate transcriptional polarities of the ORFs encoding xenocin, the immunity protein, and the transposase protein. (B) Domain map of the encoded proteins. T, translocation domain; R, receptor binding domain; C, catalytic domain; Im, immunity domain; Hm, hemolysin domain.
Transcriptional analysis of the xcinA and ximB genetic loci.
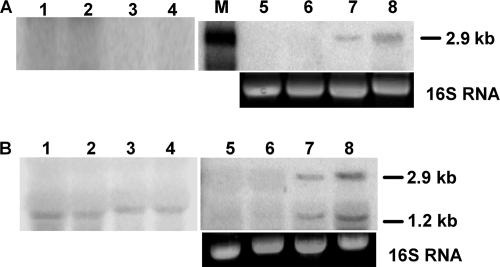
Induction of the xcinA and ximB genes in response to mitomycin C was monitored by performing a Northern blot analysis of RNA with specific DNA probes. When an xcinA-specific probe was used, the intensity of a ∼2.9-kb band reacting with the probe increased with induction time (Fig. 2A, lanes 5 to 8), while a corresponding band was absent in uninduced cells (Fig. 2A, lanes 1 to 4). Maximum induction occurred after 3 h of exposure to mitomycin C (Fig. 2 A, lane 8). The 2.9-kb band suggested that the mRNA produced under SOS conditions is bicistronic, containing both the xcinA and ximB loci. When the total RNA was hybridized with the ximB probe, a smaller band (∼1.2 kb) reacted at all time points during incubation of the uninduced cells, indicating that a basal level of an ximB transcript was produced under normal growth conditions (Fig. 2B, lanes 1, 2, 3, and 4). Upon induction of the cells with mitomycin C, two species, a 2.9-kb mRNA and a 1.2-kb mRNA reacting with the ximB probe, were observed (Fig. 2B, lanes 5, 6, 7, and 8). The intensity of both bands increased with time of incubation, reaching the maximum at 3 h (Fig. 2B, lane 8), which suggested that under SOS conditions two ximB gene transcripts are produced from two different promoters, providing excess immunity protein.
FIG. 2.
Northern blot of xcinA and ximB genes. Total RNA was obtained from uninduced and mitomycin C-induced X. nematophila cells at different times. Ten micrograms of RNA was used in each lane. (A) Blot probed with labeled xcinA gene. Lanes 1 to 4, RNA from uninduced cells at 0, 1, 2, and 3 h, respectively, with ethidium bromide-stained bands corresponding to the 16S RNA of the gel as a loading control; lanes 5 to 8, RNA from mitomycin C-induced cells at 0, 1, 2, and 3 h, respectively; lane M, 3-kb RNA marker. (B) Blot probed with labeled xcimB gene. Lanes 1 to 4, RNA from uninduced cells at 0, 1, 2, and 3 h, respectively; lanes 5 to 8, RNA from mitomycin C-induced cells at 0, 1, 2, and 3 h, respectively, with ethidium bromide-stained bands corresponding to 16S RNA in the gel used as a loading control.
The transcripts were also analyzed by RT-PCR to confirm the organization of the xcinA and ximB genes in the genome of X. nematophila. No amplification occurred with RNA from a Xenorhabdus isolate (large-colony variant) (Table 3) used as a negative control with primers 5 and 2 or primers 5 and 6, 258 bp internal to the ATG start codon of the ximB gene (Fig. 3A, lanes 6 and 7). In contrast, a 313-bp catalytic domain band and a 588-bp catalytic domain-immunity domain band were amplified with RNA from X. nematophila 19061 (Fig. 3A, lanes 8 and 9). A 1.7-kb product was observed (Fig. 3B, lane 2) with xcinA-specific primers (primers 1 and 2), as expected. A strong 313-bp band was obtained when primers 5 and 2 defining the catalytic domain were used for PCR amplification (Fig. 3B, lane 4). The higher intensity of the band compared to the 1.7-kb xcinA gene band could have been due to lower efficiency of the reverse transcriptase for synthesis of longer mRNA transcripts. A 588-bp amplicon was obtained with primers 5 and 6 (Fig. 3B, lane 3). Amplification of the ximB gene occurred as expected, producing a ∼1,000-bp band with primers 3 and 4 from ORF2 (lane 5). No product was observed with primer 1 and backward primer 4 from the 3′ end of the ximB gene (Fig. 3B, lane 6). This could have been due to the larger size (2.7 kb) of the product. The results demonstrate that transcripts of the xcinA and ximB genes are present in a bicistronic mRNA.
TABLE 3.
Toxicity of recombinant xenocin with bacterial isolates
| Target organism | Sensitivity to recombinant xenocin under the following growth conditionsa:
|
||
|---|---|---|---|
| M9 medium at 30°C | M9 medium at 37°C | M9 medium with Fe3+ | |
| Enterobacter sp. | − | + | − |
| Bacillus sp. | − | + | − |
| Serratia sp | − | + | − |
| Enterococcus sp. | − | + | − |
| Citrobacter sp. | − | + | − |
| Stenotrophomonas sp. | − | + | − |
| Xenorhabdus large-colony variantb | − | + | − |
| Xenorhabdus small-colony variantb | − | ± | − |
The recombinant proteins were induced with 1 mM IPTG.
Xenorhabdus isolates from S. carpocapsiae-infected insect larva grown at 30°C.
Iron is an essential nutrient for microbial growth, and reduced levels of iron are known to induce virulence genes in pathogenic bacteria (35). Since the primary aim of bacteriocin synthesis is to eliminate competing strains, the role of iron in regulating xenocin production was examined. RNA isolated from cells under iron-depleted conditions showed induction of the xcinA locus with primers 5 and 2 (Fig. 3C, lane 6) and increased transcription of the ximB locus when it was tested with primers 3 and 6 (Fig. 3C, lanes 7 and 8). Shifting the cells to 37°C also resulted in an increase in the concentration of the amplified product (Fig. 3D, lanes 6, 7, 8, and 9), indicating that there was upregulation of the genes at a higher temperature.
Determination of transcriptional start points of the xcinA and ximB loci.
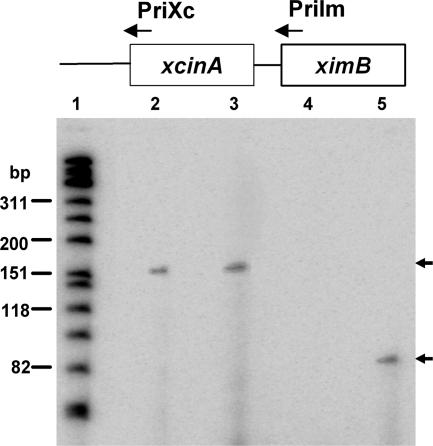
The 5′ ends of the transcripts were determined by a primer extension assay. To assess if the ximB gene is transcribed from an ximB-specific promoter, a 24-bp primer extending from position 1 to position 24 of the ximB gene (PriIm) was used with RNA isolated from X. nematophila. Radiolabeled bands corresponding to ∼151 nucleotides were produced from both uninduced and mitomycin C-induced cells (Fig. 4, lanes 2 and 3), indicating that the ximB gene has its own transcription start site 127 nucleotides upstream of the start codon, producing the shorter (1.2-kb) transcript constitutively. However, when primer PriXc, which was 24 nucleotides internal to the start codon of the xcinA gene, was used, no product was obtained from the uninduced cells (Fig. 4, lane 4), while an ∼82-nucleotide extension product was obtained from the mitomycin C-induced cells (Fig. 4, lane 5). The results corroborate previous findings obtained by Northern blotting, confirming that there are two independent transcriptional units.
FIG. 4.
Promoter identification by primer extension. Total RNA was obtained from uninduced and mitomycin C-induced X. nematophila cells. Each reaction mixture contained 40 μg RNA. Lane 1, radiolabeled φX174 DNA marker; lanes 2 and 3, PriIm primer from the N′ terminus of the ximB gene with RNA from uninduced and induced cells, respectively; lanes 4 and 5, PriXc primer from the N′ terminus of the xcinA gene with RNA from uninduced and induced cells, respectively.
Functional analysis of the upstream promoter region under stress conditions.
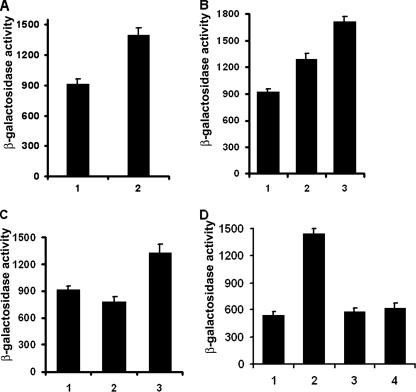
The 300-bp upstream sequences of the xcinA and ximB genes contain the −10 and −35 promoter elements with a Shine-Dalgarno sequence and are designated P1 and P2 (data not shown). Both sequences were predicted to be σ70-specific promoters. The promoter activity under stress conditions was examined by using promoter fusions with the β-galactosidase reporter gene. The results show that mitomycin C caused a 1.5-fold increase in the activity of promoter P1 (Fig. 5A) after 3 h of incubation; similarly, shifting the cultures to a higher temperature (37 or 42°C) also activated the xcinA promoter (Fig. 5B).
FIG. 5.
β-Galactosidase activities of xcinA promoter-lacZ fusions. E. coli strains were grown to log phase in LB medium and resuspended in M9 medium under specific nutrient conditions for induction. (A) Cells grown in M9 medium. Bar 1, no mitomycin C; bar 2, induction with 0.3 μg/ml mitomycin C. (B) Activities at different temperatures. Bar 1, 30°C; bar 2, 37°C; bar 3, 42°C. (C) Cells grown in M9 media. Bar 1, M9 medium control; bar 2, M9 medium with 2 μg/ml FeSO4; bar 3, M9 medium with 2 μg/ml FeSO4 and 25 mM Desferal. (D) Effect of glucose. Bars 1 and 2, no glucose at 0 and 2 h, respectively; bars 3 and 4, cells with 0.2% glucose at 0 and 2 h, respectively. The data are expressed in Miller units.
P1 activity was also regulated by depletion of essential nutrients, like glucose and iron. Addition of 2 μg/ml of FeSO4 to the cells caused a decrease in the promoter activity; when the iron chelator Desferal was added to the culture at a concentration of 25 mM, the promoter activity increased (Fig. 5C). Similarly, addition of glucose to the cells suppressed the promoter activity more than twofold (Fig. 5D). The results indicate that the P1 promoter is activated under SOS and nutrient depletion conditions. Promoter P2 was also functional in E. coli; however, as expected, it was weaker, producing 400 to 500 Miller units of β-galactosidase, while P1 produced 600 to 900 U of the reporter enzyme, in agreement with the Northern blot data. No significant change in the activity of P2 was observed under different stress conditions (data not shown).
Recombinant protein expression and purification.
The xenocin-immunity protein complex was purified by Ni-NTA chromatography. Denaturation of the protein complex with 6 M guanidinium hydrochloride separated the two proteins; xenocin bound with the matrix during a second Ni-NTA chromatography, while the immunity protein alone was obtained in the flowthrough fractions. Xenocin was eluted from the column with 300 mM imidazole. The yields of xenocin and the immunity protein from a 50-ml culture were 2 and 1.2 mg, respectively. Isolation of xenocin-immunity protein as a complex and separation of the individual proteins using guanidinium hydrochloride demonstrated the strong interaction between the two proteins.
In vitro RNase activity of xenocin.
Since ClustalW alignment showed that there was maximum similarity with RNase domains of E3 and E6, the activity of the catalytic domain of xenocin was tested using total RNA isolated from E. coli BL21(DE3)/pLysS cells. The purified catalytic domain protein was able to degrade E. coli RNA (Fig. 6, lane 4), while the purified catalytic domain-immunity domain complex had no effect on the RNA (Fig. 6, lane 2). The enzymatic activity was lost after heat denaturation (Fig. 6, lane 3). The RNase activity was dose dependent, increasing with the protein concentration (Fig. 6, lanes, 4, 5, and 6). Addition of the immunity protein inhibited the enzymatic activity of the catalytic domain. Although the molar ratio of the catalytic domain to the immunity protein in the complex produced in vivo appeared to be 1:1 (data not shown), in the in vitro conditions the ratio required to inhibit the RNase activity completely appeared to be more than 1:3 (Fig. 6, lane 7). This could have been due to less efficient renaturation of the recombinant proteins in in vitro conditions. Addition of bovine serum albumin could not neutralize the RNase activity of xenocin (Fig. 6, lane 9), and the protein had no effect on E. coli DNA, demonstrating its specific action on RNA (Fig. 6, lanes 10 and 11).
FIG. 6.
RNase activity of the purified catalytic domain. Each 20-μl reaction mixture containing 1.2 μg of RNA with the test protein was incubated at 37°C for 1.5 h. Lane 1, RNA without protein; lane 2, RNA with 10 μg of catalytic domain-immunity domain complex; lane 3, 10 μg heat-inactivated catalytic domain; lanes 4, 5, and 6, RNA with 10, 5, and 1 μg catalytic domain, respectively; lanes 7 and 8, 5 μg catalytic domain-immunity domain protein at molar ratios of 1:3 and 1: 1, respectively; lane 9, 5 μg catalytic domain-bovine serum albumin at a molar ratio of 1:2; lane 10, E. coli DNA without protein; lane 11, DNA with 5 μg catalytic domain protein.
Endogenous toxicity of xenocin.
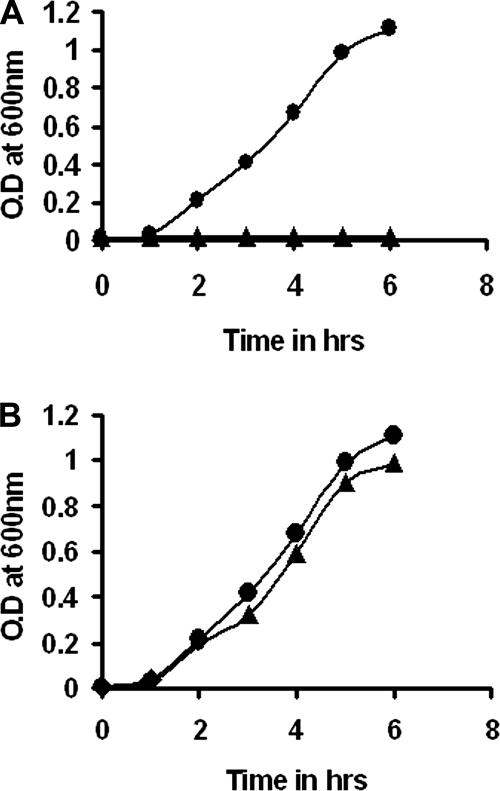
The toxicity of the xenocin and immunity proteins was tested using an E. coli host. The growth curves of the strains are shown in Fig. 7; induction of strain BSK2 containing the xcinA gene by mitomycin C resulted in severe growth retardation of the E. coli host cells, showing the detrimental effect of the protein (Fig. 7A). Strain BSK1, containing both genes, was able to grow normally like the uninduced culture (Fig. 7B). The results demonstrated the neutralizing activity of the protein encoded by the ximB gene. Likewise, the catalytic domain alone was sufficient to inhibit host cell (E. coli) growth, and the immunity domain (the first 86 residues or complete immunity protein) was able to abolish the toxicity in vivo (data not shown), reflecting the minimum activity domains of the two proteins.
FIG. 7.
Growth curves for strains BSK1 and BSK2. Cultures were grown to log phase and subcultured in fresh medium with and without mitomycin C. Bacterial growth was monitored by determining the optical density at 600 nm (O.D at 600 nm). (A) •, uninduced strain BSK2; ▴, strain BSK2 induced with mitomycin C. (B) •, uninduced strain BSK1; ▴, induced BSK1 cells.
Exogenous bacteriostatic activity.
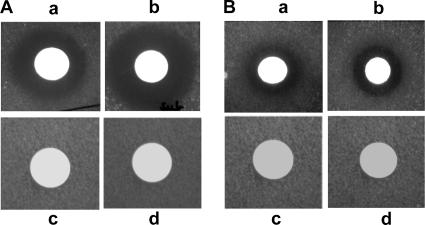
When the BSK1 strain was induced by mitomycin C, it produced xenocin in both the cytoplasm and culture filtrate. Both fractions inhibited growth of E. coli BL21(DE3)/pLysS indicator cells when they were grown in M9 medium (Fig. 8A, panels a and b). No activity was detected when the indicator strain was grown in LB medium or nutrient broth (data not shown). Supplementation of M9 medium with iron abolished growth inhibition of the target cells (Fig. 8A, panels c and d), establishing the necessity of nutrient depletion (specifically iron depletion) for susceptibility of the target cells. When the purified recombinant xenocin-immunity protein complex was added exogenously, it was also able to inhibit the growth of DH5α and BL21(DE3)/pLysS cells (Fig. 8B, panels a and b). A zone of clearance was observed only when the indicator strains were grown in M9 minimal medium. Cytoplasmic fractions from induced cells containing vector alone and sodium phosphate buffer (pH 7), used as control, showed no zone of clearance (Fig. 8B, panels c and d). The results demonstrate the ability of xenocin to recognize and enter the susceptible target cells like other bacteriocins.
FIG. 8.
Exogenous bacteriostatic activity. (A) Zone of clearance of BL21(DE3)/pLysS target cells grown in M9 medium with (a) cytoplasmic and (b) supernatant fractions of BSK1 cells after induction with mitomycin C. BL21(DE3)/pLysS target cells were also grown in M9 minimal medium containing iron and tested with (c) cytoplasmic and (d) supernatant fractions of mitomycin C-induced BSK4.3 cells. (B) Zone of clearance of (a) DH5α and (b) BL21(DE3)/pLysS target cells with supernatant of JC2 cells after induction with IPTG, (c) induced supernatant of M15 cells containing vector alone, and (d) buffer with BL21(DE3)/pLysS target cells.
Recombinant xenocin-immunity protein complex inhibited the growth of six bacterial strains isolated from H. armigera, an insect pest susceptible to X. nematophila. Bactericidal activity was observed only when the strains were grown in M9 medium (Table 3) and not when they were grown in LB medium or nutrient broth. Uninduced recombinant cells had no effect on the isolated strains. Since M9 medium contained glucose (0.2 to 2%) but no iron, glucose depletion does not seem to be necessary for target cell sensitivity, while iron depletion is. This was confirmed by addition of iron to M9 medium, which abolished the sensitivity of isolated bacteria to the recombinant protein. Similarly, growth at a high temperature was also essential for the strains to be sensitive to recombinant xenocin (Table 3). When the two Xenorhabdus isolates were compared, the large-colony variant was more sensitive than the small-colony variant. These results demonstrate that iron depletion and elevated temperatures act as specific signals for inducing sensitivity in the target cells against xenocin.
DISCUSSION
This is the first report describing a bacteriocin protein, xenocin, of X. nematophila. The chromosomal origin of xenocin-encoding genes in X. nematophila is unique, as the bacteriocin genes are generally plasmid borne and transmitted horizontally with a high frequency among related species (18, 22). The presence of a putative transposase sequence downstream of the cluster reflects its acquisition through a chromosomal insertion. A phage-based lytic bacteriocin with phage tail morphology has been described previously for X. nematophila (6); like xenocin, it was also induced at a high temperature and by mitomycin C. Previously, bacteriocins were thought to have evolved from phages, but recently sequence comparisons have unequivocally established their separate origin (9).
Several lines of evidence, including data from Northern blotting, RT-PCR, and primer extension analysis, demonstrated that under normal growth conditions the two genes behave as separate transcriptional entities, and only ximB is expressed constitutively from the P2 promoter. Under SOS conditions, induction of both the xcinA and ximB genes from the stronger P1 promoter provides the toxin-immunity protein complex for killing the competing strains. Since the immunity protein protects the host cell from the “killer” protein produced by “self” or competing species, its expression through differentially controlled promoters ensures its constant presence in the cell in adequate amounts.
Interestingly, sensitivity of the target bacterial strains to xenocin was observed only when the strains were grown in minimal medium (0.2 to 2% glucose and no iron) and not when they were grown in enriched media like Luria or nutrient broth, indicating the critical importance of xenocin in the antibacterial warfare of X. nematophila when the levels of nutrients are low. Thus, during infection of the larval host by X. nematophila, the iron-depleted environment (14, 25) acts as a common cue to trigger both xenocin synthesis and display of the cognate receptor on the target cell surface, ensuring rapid killing of the competitors. Cloacin DF13, another RNase-type bacteriocin, was shown to use iron siderophore-aerobactin receptors of target cells (46). Although it is well known that nutrient depletion is one of the factors forcing bacteria to produce bacteriocins, the specific role of Fe3+ depletion in induction of bacteriocin is reported here for the first time for X. nematophila. Since iron depletion made the target cells sensitive to xenocin, it is assumed that iron-repressed proteins act as the toxin receptors on the sensitive cells; at this stage we do not know the identity of the receptor protein(s) recognized by xenocin. All the E group colicins bind to the vitamin B12 receptor BtuB and are imported into the cell through the Tol system (42). However, our results suggest that although xenocin belongs to the E3-type colicins, it perhaps binds to iron-repressed proteins and consequently might be imported through the Ton system in the target cell (8, 33). This hypothesis is supported by the presence of a putative “Ton box”-like consensus sequence (EAMAI) in the N terminus of the protein (29). More work is needed to confirm these results.
X. nematophila produces antibacterial peptides to control the growth of other bacterial strains associated with the larval prey (1, 26, 43, 45). These antibiotics serve two critical functions: (i) they prevent putrefaction of the larval carcass by eliminating competing bacterial strains, and (ii) they provide the host strain unrestricted access to the available nutrients. The sensitivity of a number of bacterial strains from the gut of H. armigera demonstrated the wide range of activity of xenocin across different genera belonging to both the gram-positive and gram-negative groups. In light of the narrow activity range of bacteriocins (36, 41), our results have important practical implications for the control of pathogenic soil bacteria. Thus, xenocin could be part of the antibacterial arsenal of X. nematophila (40) required to keep a dead larval carcass free of other bacterial species for its own consumption. Finally, who wins the microbicidal war in an insect body probably depends on factors contributed by both the host and the resident bacteria. The results suggest that xenocin production by X. nematophila could be an important tool for successful predation of insect larvae, leading to a robust bacterium-nematode symbiotic system.
Supplementary Material
Acknowledgments
Jitendra Singh thanks the Council of Scientific and Industrial Research, New Delhi, for the award of a senior research fellowship.
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp. bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditae and Steinernematidae. J. Gen. Microbiol. 1283061-3065. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and G. B. Dunphy. 1993. Symbiotically associated entomopathogenic bacteria, nematodes and their insect hosts, p. 1-23. In N. Beckage, S. Thompson, and B. Federici (ed.), Parasites and pathogens of insects, vol. 2. Academic Press, Inc., New York, NY. [Google Scholar]
- 3.Ausbel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 4.Barnard, T. J., M. E. Watson, Jr., and M. A. McIntosh. 2001. Mutations in the Escherichia coli receptor FepA reveal residues involved in ligand binding and transport. Mol. Microbiol. 41527-536. [DOI] [PubMed] [Google Scholar]
- 5.Bedding, R. A., and R. J. Akhrust. 1975. Nematodes and their biological control of insect pests. Nematologia 21109-110. [Google Scholar]
- 6.Boemare, N. E., M.-H. Boyer-Giglio, J.-O. Thaler, R. J. Akhurst, and M. Brehelin. 1992. Lysogeny and bacteriocinogeny in Xenorhabdus spp., bacteria associated with entomopathogenic nematodes. Appl. Environ. Microbiol. 583032-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boemare, N. E., and R. J. Akhrust. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp. (Enteriobacteriaceae). J. Gen. Microbiol. 134751-761. [Google Scholar]
- 8.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16295-307. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., H. Pilsl, and P. Groß. 1994. Colicins: structures, modes of action, transfer through membranes and evolution. Arch. Microbiol. 161199-206. [DOI] [PubMed] [Google Scholar]
- 10.Chhibber, S., A. Goel, N. Kapoor, M. Sexana, and D. V. Vadhera. 1988. Bacteriocin (Klebocin typing) of clinical isolates of Klebsiella pneumoniae. Eur. J. Epidemiol. 4115-118. [DOI] [PubMed] [Google Scholar]
- 11.Converse, V., and P. S. Grewal. 1998. Virulence of entomopathogenic nematodes to the western masked chafer Cyclocephala hirta. (Coleoptera: Scarabaeidae) J. Econ. Entomol. 91428-432. [DOI] [PubMed] [Google Scholar]
- 12.Converse, V., and R. W. Miller. 1999. Development of the one-on-one quality assessment assay for entomopathogenic nematodes. J. Invertebr. Pathol. 74143-148. [DOI] [PubMed] [Google Scholar]
- 13.Cutting, S. M., and P. B. Vander Horn. 1996. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley, Chichester, United Kingdom.
- 14.De Gregorio, E., P. T. Spellman, G. M. Rubin, and B. Lemaitre. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 9812590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 6021-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillor, O., B. C. Kirkup, and M. A. Riley. 2004. Colicins and microcins: the next generation antimicrobials. Adv. Appl. Microbiol. 54129-146. [DOI] [PubMed] [Google Scholar]
- 17.Hsia, K.-C., C.-L. Li, and H. S. Yuan. 2005. Structural and functional insight into sugar-nonspecific nucleases in host defense Curr. Opin. Struct. Biol. 15126-134. [DOI] [PubMed] [Google Scholar]
- 18.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigm and paradoxes. Microbiology 1421569-1580. [DOI] [PubMed] [Google Scholar]
- 19.James, R., C. N. Penfold, G. R. Moore, and C. Kleanthous. 2002. Killing of E. coli cells by E group nuclease colicins. Biochimie 84381-389. [DOI] [PubMed] [Google Scholar]
- 20.Khandelwal, P., and N. B. Bhatnagar. 2003. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 692032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleanthous, C., and D. Walker. 2001. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26624-631. [DOI] [PubMed] [Google Scholar]
- 22.Konisky, J. 1982. Colicins and other bacteriocins with established modes of action. Annu. Rev. Microbiol. 36125-144. [DOI] [PubMed] [Google Scholar]
- 23.Lakey, J. H., and S. L. Slatin. 2001. Pore-forming colcins and their relatives. Curr. Top. Microbiol. Immunol. 257131-161. [DOI] [PubMed] [Google Scholar]
- 24.Lazzaroni, J.-C., J.-F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84391-397. [DOI] [PubMed] [Google Scholar]
- 25.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25697-743. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., G. Chen, and J. M. Webster. 1997. Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophila (Enterobacteriaceae). Can. J. Microbiol. 43770-773. [DOI] [PubMed] [Google Scholar]
- 27.Masaki, H., and T. Ogawa. 2002. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie 84433-438. [DOI] [PubMed] [Google Scholar]
- 28.Michel, B. 2005. After 30 years of study, the bacterial SOS response, still surprises us. PloS Biol. 31174-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mora, L., N. Diaz, R. H. Buckingham, and M. D. Zamaroczy. 2005. Import of the transfer RNase colicin D requires site-specific interaction with the energy-transducing protein TonB. J. Bacteriol. 1872693-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimatsu, T., and J. J. Jackson. 1998. Interaction of insecticides, entomopathogenic nematodes and larvae of the western corn rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 91410-418. [DOI] [PubMed] [Google Scholar]
- 31.Ohno-Iwashita, Y., and K. Imahori. 1980. Assignment of the functional loci in colicin E2 and E3 molecules by the characterization of their proteolytic fragments. Biochemistry 19652-659. [DOI] [PubMed] [Google Scholar]
- 32.Parret, A. H., and R. De Mot. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other γ-proteobacteria. Trends Microbiol. 10107-112. [DOI] [PubMed] [Google Scholar]
- 33.Pugsley, A. P. 1984. The ins and outs of colicins. Part I: production and translocation across membranes. Microbiol. Sci. 1168-175. [PubMed] [Google Scholar]
- 34.Pugsley, A. P. 1993. The complete general secretary pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 36.Riley, M. A. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Microbiol. 32255-278. [DOI] [PubMed] [Google Scholar]
- 37.Riley, M. A., and J. E. Wertz. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84357-364. [DOI] [PubMed] [Google Scholar]
- 38.Schroeder, P. C., C. S. Ferguson, A. M. Shelton, W. T. Wilsey, M. P. Hoffmann, and C. Petzoldt. 1996. Greenhouse and field evaluations of entomopathogenic nematodes (Nematoda: Heterorhabditidae and Steinernematidae) for control of cabbage maggot (Diptera: Anthomyiidae) on cabbage. J. Econ. Entomol. 891109-1115. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Z., K. Chak, and H. S. Yuan. 2005. Identification of an essential cleavage site in ColE7 required for import and killing of cells. J. Biol. Chem. 28024663-24668. [DOI] [PubMed] [Google Scholar]
- 40.Sicard, M., J. Tabart, N. E. Boeare, O. Thaler, and C. Moulia. 2005. Effect of phenotypic variation in Xenorhabdus nematophila on its mutualistic relationship with the entomopathogenic nematode Steinernema carpocapsa. Parasitology 131687-694. [DOI] [PubMed] [Google Scholar]
- 41.Šmarda, J., and D. Šmajs. 1998. Colicins: exocellular lethal proteins of Escherichia coli. Folia Microbiol. 43563-582. [DOI] [PubMed] [Google Scholar]
- 42.Soelaiman, S., K. Jakes, N. Wu, L. Chunmin, and M. Shoham. 2001. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 81053-1062. [DOI] [PubMed] [Google Scholar]
- 43.Sundar, L., and F. N. Chang. 1993. Antimicrobial activity and biosynthesis of indole antibiotic produced by Xenorhabdus nematophilus. J. Gen. Microbiol. 1393139-3148. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana, M., H. Hori, N. Suzuki, T. Uechi, D. Kabayashi, H. Iwahana, and H. K. Kaya. 1996. Larvaecidal activity of the symbiotic bacterium Xenorhabdus japoicum from the entopathogenic nematode Steinernema kushidia against Anomala cuprae, J. Invertebr. Pathol. 68152-159. [DOI] [PubMed] [Google Scholar]
- 45.Thaler, J. O., S. Baghdigian, and N. Boemare. 1995. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain of Xenorhabdus nematophilus. Appl. Environ Microbiol. 612049-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Tiel-Menkveld, G. J., J. M. Mentjox-Vervuurt, B. Oudega, and F. K. De Graf. 1982. Siderophore production by Enterobacter cloacae and a common receptor protein for the uptake of aerobactin and cloacin DF13. J. Bacteriol. 150490-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zakharov, S. D., and W. A. Cramer. 2002. On the pathway and mechanism of colicin import across the outer membrane. Biochim. Biophys. Acta 1565333-346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.